Abstract
The number of antigen-specific naive CD8+ T cells is believed to be important in the shaping of adaptive immune responses, and is predictive for the magnitude of priming responses in mouse models. Because of extremely low precursor frequencies, knowledge about these cells comes from indirect techniques and estimations. Here, we present a strategy based on the combination of tetramer staining, magnetic-bead enrichment, and multiparametric cytometry, which permitted direct detection and analysis of CD8+ T cells reactive for 6 different naive epitopes (MART-126-35, HIV-1 Gag p1777-85, hepatitis C virus [HCV] NS31406-1415, HCV Core132-140, NY-ESO-1157-165, and cytomegalovirus [CMV] pp65495-503). Interestingly, we detected higher than 100-fold differences in precursor frequency across these epitopes (from 0.6 × 10−6 to 1.3 × 10−4), but conserved frequencies among humans. Development of a procedure for direct assessment of T-cell precursor frequency in humans has important implications, with particular relevance to vaccine development and monitoring of tumor and self-reactive T cells.
Introduction
Mature circulating CD8+ T cells that have not yet encountered their specific antigen(s) are referred to as naive T cells. Naive T cells continuously circulate throughout the bloodstream and lymphoid organs, making contact with antigen-presenting cells, a process called “immune surveillance.” Upon activation via their T-cell receptors (TCRs) binding to peptide–major histocompatibility class I complex (pMHCIs), CD8+ T cells undergo robust clonal expansion and differentiate into effector and memory cells.1-3 During these different steps, T cells undergo characteristic changes in their gene expression, which is reflected by cell-surface markers. For example, naive T cells may be characterized by basal expression of CD45RA, homing receptors such as CD62L (or L-selectin), chemokine receptors (eg, CCR7), and the costimulatory molecules CD27 and CD28. Although no single molecule can specify a naive T cell, when used in combination, precise identification is possible.4
To adequately function, the immune system must mount an exquisitely specific response to a vast repertoire of potential antigens. To establish the basis for such a diverse response, the genes coding for the α and β chains of the TCR undergo somatic recombination and nucleotide insertion, allowing for the potential generation of 1012 to 1015 different receptors.5 Because of central tolerance and, perhaps as important, the limited niche and resources available for T-cell development, the actual diversity of αβT cells in a given person is in the range of 106 to 107.6 Given the number of CD8+ T cells present in an adult human (∼ 4 × 1010),7 the estimated number of cells per clone is thus in the range of 4000 to 40 000 cells. Many questions remain about the approximately 106-fold gap between the theoretic and observed numbers of different TCRs in the peripheral blood of humans. One thing is clear, even in identical twins possessing the same genetic information, the T-cell repertoires are distinct and in some cases may account for discordance in the development of diseases such as type 1 diabetes or multiple sclerosis.8-10
Standard quantitative assays for measuring specific CD8+ T-cell responses, such as pMHCI-labeled tetramer, intracellular cytokine staining, and enzyme-linked immunospot (ELISPOT), have a limit of detection of 5 × 10−5. Moreover, the latter 2 assays monitor functional activity and thus are not adequate for studying naive antigen-specific CD8+ T cells.11 As a result, current knowledge about precursor frequencies is based on indirect assays such as limiting dilution and microtiter assays, with a major limitation being the requirement for ex vivo stimulation, introducing significant bias and interassay variability.6 There is a real need for direct assessment of the polyclonal naive T-cell repertoire.2,12,13 To this end, 2 recent mouse studies reported an elegant method combining pMHC tetramer staining and magnetic bead enrichment.14,15 These technologies have already been coupled to purify human memory cytomegalovirus (CMV)–specific CD8+ T cells,16 but to date it has not been applied to the detection of naive CD8+ T cells. Here, we report the development of an assay for enumerating epitope-specific T cells from human peripheral blood with a limit of detection of 10−7. We analyzed previously undetectable naive populations specific for HIV-1 Gag p1777-85, hepatitis C virus (HCV) NS31406-1415, HCV Core132-140, NY-ESO-1157-165, and CMV pp65495-503 in the peripheral blood of healthy seronegative donors. We detected higher than 100-fold differences in T-cell precursor frequency across the different naive antigen-specific CD8+ T cells, but surprisingly, there was a tight range of frequencies among humans for a given epitope. The method and data reported may help define new possibilities for identifying appropriate epitopes for tumor immunotherapy and prophylactic vaccination.
Methods
Blood sample processing and HLA-A2 typing
Fresh buffy coat preparations from healthy volunteers were obtained from Etablissement Françcais du Sang (EFS Rungis). Human leukocyte antigen (HLA)–A2 determination was performed by immunophenotyping on whole blood with anti–HLA-A2–fluorescein isothiocyanate antibody (BD PharMingen). Peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll-Paque Plus gradient separation (GE Healthcare). Cells were counted using the Countess Automated Cell Counter (Invitrogen). In one experiment, half of the PBMCs obtained from an HLA-A2–positive donor were frozen for future analysis using standard procedures. Samples were cryopreserved in liquid nitrogen.
Peptide MHC class I tetramers reagents
Three peptide–HLA-A2.1 tetramers, all conjugated with phycoerythrin (PE), were purchased from Beckman Coulter: human CMV pp65495-503 (NLVPMVATV); influenza A Matrix-158-66 (GILGFVFTL); and MART126-35(Leu27) (ELAGIGILTV). Seven peptide–HLA-A2.1 tetramers were prepared in our laboratory: human CMV pp65495-503; influenza A Matrix-158-66; MART126-35(Leu27); HIV-1 Gag p1777-85 (SLYNTVATL); HCV NS31406-1415 (KLVALGINAV); HCV Core132-140 (DLMGYIPLV); and NY-ESO-1157-165 (SLLMWITQV). Briefly, inclusion bodies of heavy chain of HLA-A2.1 and β2-microglobulin protein were prepared in Escherichia coli, purified, and stored at −80°C. At day 0, we achieved the HLA-A2.1–peptide refolding by diluting materials in buffer containing the antigenic peptide, and we maintained the mixture for 4 days at 4°C. All peptides were obtained from PolyPeptide Laboratories with 95% purity. On day 4, we concentrated the protein complexes and performed biotinylation using recombinant BirA enzyme (Avidity), incubating it overnight at 30°C in the presence of protease inhibitors (pepstatin, phenylmethylsulfonyl fluoride, and leupeptin). At day 5, we purified the trimolecular complexes MHC/β2-microglobulin/peptide using a size exclusion chromatography column. After pooling and concentrating the fractions containing the biotinylated MHC peptide complexes (pMHCs), samples were stored at −80°C. Before use, we added PE-, allophycocyanin (APC)–ultravidin (Leinco Technologies), Qdot 655–streptavidin (Invitrogen), APC–cyanin 7 (Cy7)–streptavidin, or PE-Cy7-streptavidin (BD Biosciences) to the biotinylated MHC-peptide monomers at a 4:1 molar ratio for 1 hour at room temperature (RT) to generate fluorescent pMHC tetrameric complexes. Engineered tetramers were stored in the dark at 4°C.
Tetramer-associated magnetic enrichment of antigen-specific CD8+ T cells
Different amounts of PBMCs (20-400 × 106 depending on the experiment) were incubated for 15 minutes with 10 μL of FcR blocking reagent (Miltenyi Biotec), then stained with pMHCI tetramers at 20nM final concentration for 30 minutes in 100 μL of pulldown buffer: phosphate-buffered saline (PBS) 1× (Gibco) plus 1% human serum albumin (PAA Laboratories) plus 5% citrate dextrose anticoagulant (Sigma-Aldrich). After washing, cells were resuspended in 400 μL of buffer. Of the sample, 10 μL was collected for staining (“pre-enriched” fraction). The remaining sample was incubated with 100 μL of anti-PE microbeads (Miltenyi Biotec) for 20 minutes at 4°C, then washed twice and resuspended in 1 mL of buffer. A 5-μL aliquot was collected for counting of the pre-enriched fraction. Cells were filtered using BD Falcon Cell Strainer 70 μm (BD Biosciences) and passed twice over an MS magnetic-activated cell sorting separation column (Miltenyi Biotec). Unbound cells (“depleted” fraction) were collected. After removing the column from the magnet, bound cells (“enriched” fraction) were eluted. To determine the size of the epitope-specific populations within each sample, we used a precise calculation similar to Moon et al.17 The number of total CD8+ T cells within any sample is determined using the following equation: absolute number of CD8+ T cells = (number of CD8+ T cells acquired in the pre-enriched sample) × [(total number of PBMCs in the pre-enriched sample)/(total number of cells acquired in the single cell gate in the pre-enriched sample)]. The absolute number of tetramer-positive T cells is the number of tetramer-positive cells within the single, live, nondump CD3+CD8+ T-cell gate present in the enriched fraction. The frequency of circulating tetramer-positive cells is reported and defined as the absolute number of tetramer-positive T cells/absolute number of CD8+ T cells. An example of the calculation schema is illustrated in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Antibody staining and flow cytometry
Cells were stained in PBS with anti-CD16, anti-CD14 (BD Biosciences), anti-CD19, and anti-CD56 (Biolegend), all conjugated to Pacific blue (dump/lineage channel). In addition, populations were labeled with a combination of anti–CD8-AmCyan, anti–CD45RA-APC, anti–CCR7-PE-Cy7 (BD Biosciences), anti–CD11a–fluorescein isothiocyanate, anti–CD28-Alexa700, anti–CD27-APC-eFluor780 (eBioscience), and anti–CD3–peridinin-chlorophyll-protein complex (PerCP; Biolegend) antibodies for 20 minutes at 4°C (final dilution = 1/100). Of note, the 1/100 dilution, although supersaturated for labeling our populations, did not result in measurable nonspecific staining (data not shown). The choice to use the working dilution suggested by the manufacturer reflects our interest in establishing a standardized protocol with potential clinical application. 4,6 Diamidino-2-phenylindole (DAPI) Nucleic Acid Stain (Invitrogen) was added to each sample just before analysis to exclude dead cells (final concentration = 1/50 000). In some experiments, depending on the color combination, we also used CD45RA-PE-Cy7 (BD Biosciences) and CD45RA–Alexa 700 (Biolegend). Events (2 × 106) were collected from pre-enriched and depleted fractions. The entire sample was collected for analysis of the enriched fraction. All samples were acquired using a LSRII cytometer (BD Biosciences). Data were analyzed using FACSDiva (BD Biosciences) and FlowJo (TreeStar, Inc) software.
IFNγ ELISPOT
The 96-well ELISPOT plates (Millipore) were coated with 100 μL of an anti–human interferon-γ (IFNγ) antibody diluted at 1/1000 (clone 1-D1K; Mabtech) and stored overnight at 4°C. Plates were blocked with culture medium (RPMI 1640 [Gibco] + 10% pooled human AB serum [Labquip Ltd] + 1% l-glutamine [final concentration = 2mM; Sigma-Aldrich] + 2% antibiotics penicillin-streptomycin [final concentration = 100 UI/mL; Gibco]) for 1 hour at 37°C. Cells from pre-enriched and depleted fractions were plated at a final concentration of 2 × 105 per well. Cells were stimulated in duplicate wells using relevant or irrelevant peptides (final concentration = 10 μg/mL) for 20 hours at 37°C. Anti-CD3 antibody was used as positive control (final dilution = 1/1000; Mabtech). For ELISPOT development, plates were washed with 200 μL of PBS–0.05% Tween-20, then incubated for 2 hours at 37°C with 100 μL of an anti-IFNγ biotinylated antibody (Mabtech) diluted at 1/1000 in PBS–0.5% bovine serum albumin (Sigma-Aldrich). Plates were then washed and 100 μL of streptavidin-peroxidase (Vector Labs Standard Vectastain Kit ABC) diluted in PBS–0.1% Tween was added to each well for 1 hour at RT. After washing, 100 μL of 3-amino-9-ethylcarbazole (AEC) substrate (Sigma-Aldrich) diluted in dimethylformamide and phosphate buffer (Vector Labs Standard Vectastain Kit ABC) was added to each well for 4 minutes at RT. The ELISPOT plate evaluation was performed by an independent evaluation service (Zellnet Consulting, Inc).
CTL clone
The cytotoxic T lymphocyte (CTL) clone EM40 was a generous gift from N. Casartelli and O. Schwartz, Institut Pasteur. It was derived from an HIV-1–infected person by repeated stimulations of PBMCs with irradiated autologous Epstein-Barr virus–transformed (B-EBV) B cells loaded with the p17 Gag peptide SLYNTVATL.
Dendritic cell generation and maturation
Dendritic cells were prepared from CD14+-enriched PBMCs by culturing cells for 4 days in the presence of granulocyte and macrophage colony-stimulating factor (GM-CSF; Berlex) and interleukin-4 (R&D Systems), followed by 2 days in medium containing tumor necrosis factor α (Alexis Biochemicals) and prostaglandin E2 (Sigma). Peptide loading was performed on day 7 by incubating mature dendritic cells with purified MART126-35 peptide at a final concentration of 20nM at RT for 1 hour.
Cell culture
Fresh PBMCs were incubated with anti-CD8 microbeads (Miltenyi Biotec) for 15 minutes at 4°C, then washed and passed over an MS column (Miltenyi Biotec). CD8+ cells were enriched by positive separation, and cultured in 24-well plates at 1 × 106 cells per well. Dendritic cells pulsed with peptide were added to each well on day 0 at a 30:1 ratio (T cells/dendritic cells [DCs]). Interleukin-2 cytokine (10 000 IU; R&D Systems) was added to each well on day 2. Cells were harvested on day 7, incubated with 10 μL of FcR blocking reagent for 15 minutes at 4°C in 100 μL of PBS and then stained with MART126-35 and HIV-1 Gag p1777-85 tetramers at 2nM final concentration for 30 minutes at 4°C.
CMV serology
CMV serology was determined on serum samples of healthy donors by enzyme-linked immunosorbent assay for CMV-specific immunoglobulin G (IgG) antibodies (Liaison; Diasorin). CMV-seropositive donors were defined as IgG positive (> 0.2 IU/mL). CMV-seronegative donors were defined as IgG negative (< 0.2 IU/mL).
Results
Development of TAME
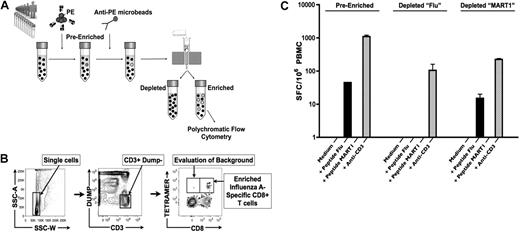
To study naive antigen-specific CD8+ T cells, we developed an enrichment protocol based on pMHC class I tetramer technology, which we refer to as tetramer-associated magnetic enrichment (TAME; Figure 1A). Except where indicated, experiments were performed on fresh PBMCs isolated from HLA-A2+ healthy persons. We first incubated cells with PE-pMHCI tetramers, followed by labeling with anti-PE antibodies linked to magnetic particles, thus permitting enrichment on a magnetic column. Pre-enriched, enriched, and depleted (or flow-through) cell fractions were analyzed by multiparametric cytometry on a flow cytometer. To optimize detection and enumeration of tetramer-positive events, our enrichment protocol includes multiple levels of inclusion and exclusion gating strategies. This strategy permitted the identification of enriched, tetramer-labeled CD8+ T cells with minimal background staining in the CD3+ CD8− T-cell fraction (Figure 1B).
Development of tetramer-associated magnetic enrichment (TAME). (A) PBMCs from healthy donors were stained with HLA-A2.1 tetramers labeled in PE (pre-enriched fraction). Samples were further incubated with anti-PE microbeads, and passed twice over an MS magnetic column. The elution fraction corresponds to the depleted fraction, and retained fraction to the enriched sample. Pre-enriched, enriched, and depleted fractions were then labeled with a flow cytometric panel, and acquisition was performed with a polychromatic flow cytometer. (B) To optimize detection and enumeration of tetramer-positive events, our analysis protocol includes multiple levels of inclusion and exclusion gating strategies to minimize background. As shown, side scatter area versus side scatter width plot is used to exclude cell aggregates. The remaining single-cell events are analyzed on a plot of non–T-cell lineages (all labeled by Pacific blue), and dead cells were stained by 4,6 diamidino-2-phenylindole (dump channel) vs CD3, allowing for T-cell events to be identified. CD8+ T cells specific for influenza A–Matrix158-66 tetramer within the enriched fraction are indicated, with minimal background staining in the CD8− CD3+ T-cell population. (C) PBMCs (2 × 105) from pre-enriched and depleted fractions from influenza A–Matrix158-66 (flu) or MART126-35 TAME were plated for IFNγ ELISPOT analysis. Cells were stimulated by indicated peptides. Data from triplicate wells were averaged, and errors bars indicate SEM. Data shown are representative of 2 independent experiments.
Development of tetramer-associated magnetic enrichment (TAME). (A) PBMCs from healthy donors were stained with HLA-A2.1 tetramers labeled in PE (pre-enriched fraction). Samples were further incubated with anti-PE microbeads, and passed twice over an MS magnetic column. The elution fraction corresponds to the depleted fraction, and retained fraction to the enriched sample. Pre-enriched, enriched, and depleted fractions were then labeled with a flow cytometric panel, and acquisition was performed with a polychromatic flow cytometer. (B) To optimize detection and enumeration of tetramer-positive events, our analysis protocol includes multiple levels of inclusion and exclusion gating strategies to minimize background. As shown, side scatter area versus side scatter width plot is used to exclude cell aggregates. The remaining single-cell events are analyzed on a plot of non–T-cell lineages (all labeled by Pacific blue), and dead cells were stained by 4,6 diamidino-2-phenylindole (dump channel) vs CD3, allowing for T-cell events to be identified. CD8+ T cells specific for influenza A–Matrix158-66 tetramer within the enriched fraction are indicated, with minimal background staining in the CD8− CD3+ T-cell population. (C) PBMCs (2 × 105) from pre-enriched and depleted fractions from influenza A–Matrix158-66 (flu) or MART126-35 TAME were plated for IFNγ ELISPOT analysis. Cells were stimulated by indicated peptides. Data from triplicate wells were averaged, and errors bars indicate SEM. Data shown are representative of 2 independent experiments.
To ensure that we were capturing the majority of tetramer-positive cells during the enrichment, we assayed for the presence of influenza antigen–specific T cells remaining in the depleted fraction. IFNγ ELISPOT was performed on the pre-enriched and depleted fractions. We chose to monitor influenza HLA-A2.1/Matrix-158-66 (flu)–specific CD8+ T cells as most adult persons have been exposed to this pathogen in their life and thus harbor expanded populations of memory cells. Accordingly, we were able to detect influenza-specific IFNγ-producing CD8+ T cells in the pre-enriched PBMCs (Figure 1C). Demonstrating the efficiency of our enrichment protocol, we did not detect IFNγ spot-forming cells in the influenza tetramer–depleted fraction. As a control, MART-1 tetramer–depleted cells were assayed, demonstrating that the enrichment procedure does not influence the ability to activate antigen-specific T cells.
TAME increases detection of antigen-specific T cells by more than 100-fold
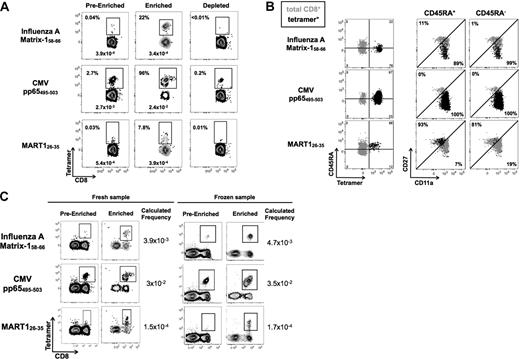
To assess the accuracy and efficiency of the technique, we first evaluated 3 T-cell specificities that may be detected by conventional pMHCI tetramer staining. Influenza HLA-A2.1/Matrix-1 (M158-66) and CMV pp65495-503 from seropositive persons were chosen for the monitoring of memory-specific populations. Melanoma-associated antigen recognized by T cells, MART126-35, also known as MelanA, was chosen for the exceptionally high frequency of naive specific cells in healthy persons.18,19 As expected, we were able to detect tetramer-positive cells before enrichment for all 3 specificities (Figure 2A). After enrichment, antigen-specific populations were clearly defined with few cells lost in the depleted fraction. Based on the CMVpp65495-503 results, we estimate our enrichment protocol to capture approximately 90% of the tetramer-specific cells. In other words, capture on the column is quite efficient and given that the majority of populations of interest are less abundant than CMV, we extrapolate that there is less than 10% loss in the depleted fraction.
TAME increases detection of antigen-specific T cells by more than 100-fold. (A) PBMCs from healthy persons were stained with influenza A–Matrix158-66, CMVpp65495-503, and MART126-35 HLA-A2.1 tetramers labeled with PE. Enrichment was performed and the pre-enriched, enriched, and depleted fractions were analyzed. CD3+ CD8+ lineage-negative cells were gated and the percentage and corresponding frequencies of cells stained by the respective tetramers are reported. (B) Tetramer-enriched populations were further analyzed using anti-CD45RA, anti-CD11a, and anti-CD27 antibodies, permitting accurate discrimination of naive and memory T cells. CD45RA+ or CD45RA− cells were gated and assessed for their expression of CD11a and CD27. Dot plots shown in black represent the phenotype of tetramer-positive populations, and in gray bulk CD8+ T cells are shown as a reference population. All numbers and percentages refer to tetramer-positive population. (C) As the use of frozen cells would be of interest for monitoring banked samples, we confirmed that TAME is not influenced by standard freeze/thawing of PBMCs. Fresh and frozen PBMC from the same donor were stained with influenza A–Matrix158-66, CMVpp65495-503, and MART126-35 HLA-A2.1 tetramers labeled with PE. Enrichment was performed and tetramer frequency is reported. Plots are gated on CD3+ cells. Background staining of non-CD8+ T cells was unchanged by the freezing procedure.
TAME increases detection of antigen-specific T cells by more than 100-fold. (A) PBMCs from healthy persons were stained with influenza A–Matrix158-66, CMVpp65495-503, and MART126-35 HLA-A2.1 tetramers labeled with PE. Enrichment was performed and the pre-enriched, enriched, and depleted fractions were analyzed. CD3+ CD8+ lineage-negative cells were gated and the percentage and corresponding frequencies of cells stained by the respective tetramers are reported. (B) Tetramer-enriched populations were further analyzed using anti-CD45RA, anti-CD11a, and anti-CD27 antibodies, permitting accurate discrimination of naive and memory T cells. CD45RA+ or CD45RA− cells were gated and assessed for their expression of CD11a and CD27. Dot plots shown in black represent the phenotype of tetramer-positive populations, and in gray bulk CD8+ T cells are shown as a reference population. All numbers and percentages refer to tetramer-positive population. (C) As the use of frozen cells would be of interest for monitoring banked samples, we confirmed that TAME is not influenced by standard freeze/thawing of PBMCs. Fresh and frozen PBMC from the same donor were stained with influenza A–Matrix158-66, CMVpp65495-503, and MART126-35 HLA-A2.1 tetramers labeled with PE. Enrichment was performed and tetramer frequency is reported. Plots are gated on CD3+ cells. Background staining of non-CD8+ T cells was unchanged by the freezing procedure.
As these populations could also be evaluated based on pre-enrichment precursor frequencies, it was also possible to determine cell recovery or the yield achieved by TAME. Comparing the calculated frequencies based on analysis of the pre-enriched versus enriched fractions, we conclude that our yield is 70% to 90% (Figure 2A values reported). In addition, we show that the enrichment for the 3 populations is 550×, 35×, and 260× for influenza, CMV, and MART1, respectively. In multiple experiments, we determined the typical yield to be approximately 80% and the enrichment efficiency to be more than 100-fold.
Phenotypic analysis of the tetramer-enriched cells indicates that, as expected, the influenza- and CMV-specific T cells are CD27− and CD11a+—of note, these populations were a mix of CD45RA+ and CD45RA−. In contrast, the MART-1 T cells are CD45RA+, CD27+, and CD11alo, reflective of the high frequency of naive cells in healthy donors for this specificity. When used in combination, these surface markers can be used to accurately define naive T-cell populations (Figure 2B).4 Tetramer-positive cells are shown in black and bulk CD8+ T cells are represented in gray as a reference population. In addition, we demonstrated the ability to perform TAME on frozen cells, which yielded a similar precursor frequency for the populations measured (Figure 2C).
Direct detection of naive CD8+ T cells specific for HIV Gag p1777-85
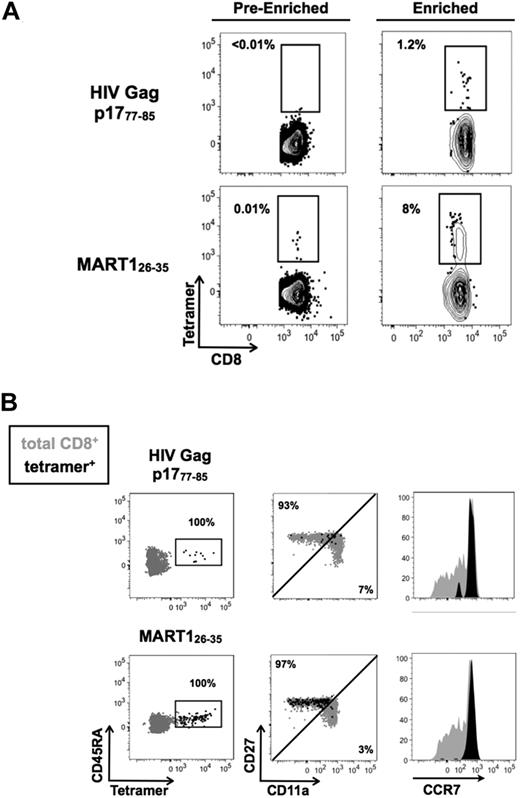
We next applied our quantitative human TAME to the analysis of a rare naive population. We focused on HIV Gag p1777-85 as healthy persons have not previously encountered this epitope, as confirmed by serologic testing. Using TAME, we were able to detect naive CD8+ T cells specific for HIV Gag p1777-85 with a measured precursor frequency of 0.3 × 10−6 (Figure 3A). These cells were not detectable before the enrichment step, but could be clearly identified and analyzed after enrichment. Nearly all of the cells identified shared a naive phenotype (CD45RA+, CD11alo, CD27+, CCR7+; Figure 3B). As an internal control from the same donor, we isolated MART126-35–specific T cells.
Naive CD8+ T cells specific for HIV Gag p1777-85 can be detected in healthy donors. (A) TAME was performed using HIV Gag p1777-85 and MART126-35 HLA-A2.1 tetramers labeled with PE. Percentage of tetramer-specific T cells are indicated before and after enrichment. (B) Enriched tetramer-positive cells were further analyzed using anti-CD45RA, anti-CCR7, anti-CD11a, and anti-CD27 antibodies. Cells were gated on CD45RA and assessed for their expression of other naive T-cell markers. Dot plots shown in black represent the phenotype of tetramer-positive populations; overlaid in gray, bulk CD8+ T cells are shown as a reference population. All percentages refer to tetramer-positive population.
Naive CD8+ T cells specific for HIV Gag p1777-85 can be detected in healthy donors. (A) TAME was performed using HIV Gag p1777-85 and MART126-35 HLA-A2.1 tetramers labeled with PE. Percentage of tetramer-specific T cells are indicated before and after enrichment. (B) Enriched tetramer-positive cells were further analyzed using anti-CD45RA, anti-CCR7, anti-CD11a, and anti-CD27 antibodies. Cells were gated on CD45RA and assessed for their expression of other naive T-cell markers. Dot plots shown in black represent the phenotype of tetramer-positive populations; overlaid in gray, bulk CD8+ T cells are shown as a reference population. All percentages refer to tetramer-positive population.
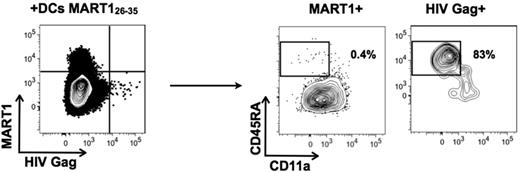
To confirm the specificity of tetramer-reactive cells, we enriched CD8+ T cells from PBMCs, and stimulated them for 7 days with mature dendritic cells (DCs) pulsed with MART126-35 peptide. As described, the MART126-35–enriched population was naive before stimulation (data not shown). After the 7-day coculture with MART126-35–pulsed DCs, the entire population specific for MART1 down-regulated CD45RA and expressed CD11a, whereas the HIV Gag p1777-85–specific cells in the same culture well remained naive (Figure 4). These data suggest that the naive T cells measured in our assays are indeed capable of responding to antigen-specific stimulation.
MART126-35–specific cells specifically mature under stimulation by pulsed DCs. PBMCs were incubated with anti-CD8 microbeads, and enriched cells were cocultured for 7 days with DCs loaded with MART126-35 peptide. T cells were then harvested and simultaneously stained with MART126-35 and HIV Gag p1777-85 tetramers, and cell phenotyping was performed on each tetramer-positive population. Data are representative of 2 independent experiments.
MART126-35–specific cells specifically mature under stimulation by pulsed DCs. PBMCs were incubated with anti-CD8 microbeads, and enriched cells were cocultured for 7 days with DCs loaded with MART126-35 peptide. T cells were then harvested and simultaneously stained with MART126-35 and HIV Gag p1777-85 tetramers, and cell phenotyping was performed on each tetramer-positive population. Data are representative of 2 independent experiments.
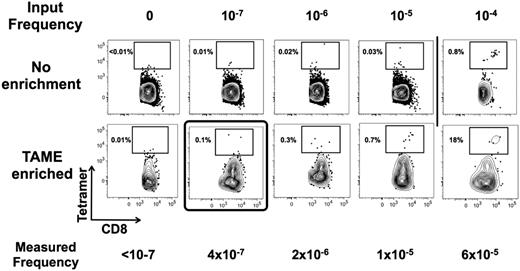
To define the limit of detection of TAME and to develop a strategy for validating the assay for use in immune monitoring protocols, we spiked titrated amounts of an HLA-A2–restricted CTL clone derived from an HIV patient and specific for Gag p1777-85. First, we confirmed the tetramer labeling of the clone and demonstrated that 100% of the cells have an expected antigen-experienced phenotype (data not shown). Next, the T-cell clone was mixed with known amounts of PBMCs isolated from a healthy HLA-A2.1+ donor to achieve a precursor frequency of the T-cell clone of 0, 10−7, 10−6, 10−5, and 10−4. For example, to achieve a precursor frequency of 10−5, we spiked 200 clonal CTLs within 100 million PBMCs (of which 20 million were CD8+ T cells). TAME was performed and pre-enriched versus enriched tetramer-positive cells were compared. In the absence of tetramer enrichment, the limit of detection was between 10−4 and 10−5, consistent with prior evaluation of the tetramer technology. Using TAME, we were able to detect as few as 2 cells spiked into 100 million PBMCs, corresponding to a limit of detection of 10−7 (Figure 5). Given the ability to control for nonspecific staining with this protocol, the true limit is the number of PBMCs used in the protocol. As such, the theoretic limit may be less than 10−7, but for routine clinical monitoring we calculate a working limit of detection to be approximately 10−6, corresponding to observation of 10 to 20 cells in approximately 10 × 106 of CD8+ T cells contained in 50 mL of blood.
TAME permits detection of 1 cell in 107 CD8+ T cells. Titrated amounts of a CTL clone specific for HIV Gag p1777-85 were dispensed into known amounts of PBMCs of a healthy donor. Corresponding frequencies of spiked cells are indicated (input frequency). Samples were then stained with HIV Gag p1777-85 HLA-A2.1 tetramer labeled with PE, and the ability to detect the CTL clone was determined in the pre-enriched and enriched fractions. The limit of detection was determined to be ∼ 10−5 before and ∼ 10−7 after TAME. Measured frequency after enrichment is indicated for comparison. Results are representative of 2 independent experiments.
TAME permits detection of 1 cell in 107 CD8+ T cells. Titrated amounts of a CTL clone specific for HIV Gag p1777-85 were dispensed into known amounts of PBMCs of a healthy donor. Corresponding frequencies of spiked cells are indicated (input frequency). Samples were then stained with HIV Gag p1777-85 HLA-A2.1 tetramer labeled with PE, and the ability to detect the CTL clone was determined in the pre-enriched and enriched fractions. The limit of detection was determined to be ∼ 10−5 before and ∼ 10−7 after TAME. Measured frequency after enrichment is indicated for comparison. Results are representative of 2 independent experiments.
Simultaneous detection of multiple antigen-specific populations
As the volume of blood collected will be the limiting factor in most clinical situations, we evaluated the ability to use TAME for simultaneous detection of several antigen-specific T-cell populations. MART126-35, HIV Gag p1777-85,CMVpp65495-503, and influenza-M158-66-HLA-A2.1 tetramers were prepared using 2 distinct fluorochromes. All were labeled with PE, and the second was labeled with streptavidin linked to APC-Cy7, APC, PE-Cy7, or Qdot 655 (Figure 6A). As such, simultaneous enrichment could be performed using anti-PE microbeads and the different T-cell specificities could be distinguished using the second tetramer label. As shown, within the enriched tetramer-positive population, we could successfully distinguish between the different specificities (Figure 6B). Importantly, the populations all displayed the expected cellular phenotype (Figure 6C), and the calculated precursor frequency was similar to the single tetramer enrichment procedure (supplemental Figure 1 and data not shown).
Simultaneous detection of several antigen-specific populations. (A) A panel including tetramers specific for MART126-35, HIV Gag p1777-85, CMVpp65495-503, and influenza A–M158-66 HLA-A2.1 tetramers was developed. All tetramers were labeled with PE, and a second tetramer labeled by a unique fluorochrome was used, as indicated. (B) Simultaneous enrichment was performed using anti-PE microbeads. Within the global PE-enriched tetramer-positive population, each specificity was identified using the second tetramer label. (C) The cell phenotype of the tetramer-positive cells was then analyzed using anti-CD45RA and anti-CD11a antibodies.
Simultaneous detection of several antigen-specific populations. (A) A panel including tetramers specific for MART126-35, HIV Gag p1777-85, CMVpp65495-503, and influenza A–M158-66 HLA-A2.1 tetramers was developed. All tetramers were labeled with PE, and a second tetramer labeled by a unique fluorochrome was used, as indicated. (B) Simultaneous enrichment was performed using anti-PE microbeads. Within the global PE-enriched tetramer-positive population, each specificity was identified using the second tetramer label. (C) The cell phenotype of the tetramer-positive cells was then analyzed using anti-CD45RA and anti-CD11a antibodies.
Conserved T-cell precursor frequency among healthy donors
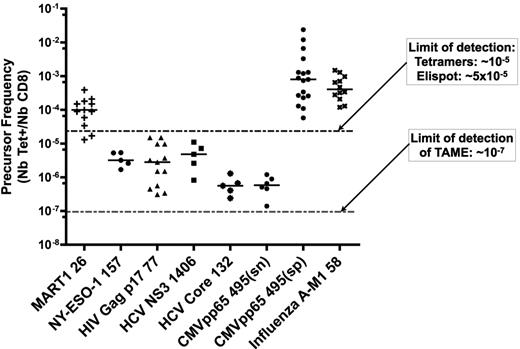
To assess the CD8+ T-cell precursor frequency for commonly studied epitopes, we performed TAME on 5 to 17 healthy donors. T cells could be detected for all specificities (Figure 7). Naive phenotype in the respective populations was confirmed using phenotypic analysis (data not shown). Importantly, the calculated precursor frequency for MART126-35 was consistent with prior reports, with a median of 1.3 × 10−4 (n = 13).18 Undetectable by conventional assays, it was also possible to establish a range for the frequency of NY-ESO-1–, HIV-, HCV-, and CMV-specific T cells in cancer-free and seronegative persons. Median frequencies were 3.6 × 10−6 for the NY-ESO1157-165 self-antigen (n = 5); 1.9 × 10−6 for HIV Gag p1777-85 (n = 13); 0.6 × 10−6 for HCV Core132-140 antigen (n = 5); 5.3 × 10−6 for HCV NS31406-1415 antigen (n = 5); and 0.6 × 10−6 specific T cells for CMVpp65 in seronegative patients (CMVpp65(sn); n = 6). Although we determined high variance when comparing one epitope with another, quite surprisingly, the precursor frequency for a single epitope across different persons was conserved.
Enumeration of epitope-specific CD8+ T cells reveals conserved precursor frequency among healthy donors. After performing TAME with MART126-35, NY-ESO1157-165, HIV Gag p1777-85, HCV NS31406-1415, HCV Core132-140, or CMVpp65495-503 HLA-A2.1 tetramers, CD8+ T cells specific for each of these epitopes were enumerated. CMVpp65495-503 in seropositive persons and influenza A–M158-66 tetramer staining were used as positive controls. Each point represents a single healthy person. CMVpp65(sn) and CMVpp65(sp) indicate seronegative and seropositive donors, respectively, as confirmed by serologic testing.
Enumeration of epitope-specific CD8+ T cells reveals conserved precursor frequency among healthy donors. After performing TAME with MART126-35, NY-ESO1157-165, HIV Gag p1777-85, HCV NS31406-1415, HCV Core132-140, or CMVpp65495-503 HLA-A2.1 tetramers, CD8+ T cells specific for each of these epitopes were enumerated. CMVpp65495-503 in seropositive persons and influenza A–M158-66 tetramer staining were used as positive controls. Each point represents a single healthy person. CMVpp65(sn) and CMVpp65(sp) indicate seronegative and seropositive donors, respectively, as confirmed by serologic testing.
Discussion
TAME permits enumeration and characterization of rare antigen-specific T cells from human peripheral blood
Two recent studies in mice reported a method for the direct detection of naive antigen-specific CD4+ or CD8+ T cells.14,15,17 We have successfully modified this protocol and our study represents the first direct detection of naive antigen-specific CD8+ T cells from human peripheral blood. Of the 6 specificities tested in mice, observed frequencies of specific CD8+ T cells ranged from 6 × 10−5 to 3 × 10−4, corresponding to 80 to 200 cells per animal.15 Here, excluding MART126-35, which is known to be an exception,18 we determined CD8+ T-cell precursor frequencies that ranged from 6.0 × 10−7 to 5.3 × 10−6. With an estimated total number of CD8+ cells of approximately 4 × 1010,7,20 measured frequencies correspond to 24 000 to 200 000 total cells per person, a value that is in fact quite close to the estimations made by indirect methods.7 Although an important point to consider is that, in comparison with mouse studies, we did not examine spleen or lymphoid organs, we expect our assay to reflect the global precursor frequency, as naive T cells continuously circulate.21,22
The development of a method for direct detection of T-cell precursor frequencies in the range of 10−7 will permit analysis of populations that were previously undetectable by standard assays. Compared with limiting dilution assay (also referred to as microcultures), which is currently the principal means of estimating populations with such low frequencies, our technique offers 2 important advantages: it permits direct detection of T cells by mean of their TCR and not by their functional capacity; and perhaps more importantly, TAME does not require in vitro culture, thus removing the intrinsic bias and variation inherent in limiting dilution assay. Nonetheless, we recognize the value of functional assays, as TAME does not establish the effector potential of the detected cells.
Although TAME is a promising tool for fundamental and clinical studies, it is important to recognize the limitations of this approach. Of particular note, it is possible that there may not be an exact correlation between the capacity of a T cell to bind a pMHC I tetramer, and its capacity to become activated when confronted with the respective pMHC I on an APC. Specifically, we must be open to the possibility that T cells detected by tetramer staining may not be capable of responding to the specific stimulus because of unproductive cross-reactivity. Indeed, antagonist peptides exist for a given TCR,23 so it may not be surprising that this technique will ultimately define the theoretic possibility that an “antagonist TCR” to a given peptide exists. Such T cells would not per se be anergic, but are likely responsive to an alternative (possibly related) peptide. Another limitation associated with tetramer-based assays is the restriction to a given HLA; however, based on recent studies and new technologies related to epitope identification, expanding this protocol for other HLA alleles is more feasible than it was in the past.24-27
Epitopes share conserved precursor frequencies among persons
We applied TAME for detection of 6 different naive CD8+ T-cell populations. For all of them, we were able to detect naive antigen-specific T cells from peripheral blood of healthy donors. Perhaps the most surprising result from these studies is that corresponding precursor frequencies were conserved across the healthy donors tested. Although similar observations were made in mice, these studies used inbred strains, which share all major histocompatibility molecules as well as minor H antigens. These results suggest that naive T-cell precursor frequency for a shared epitope is highly regulated, with some extrinsic factor determining the number of cells per pMHC specificity. The availability of cross-reactive low-affinity self-peptide MHC complexes may thus be involved not only in determination of the diversity, but also in active regulation of the pool size for the naive repertoire.18,28,29 Whether this phenomenon occurs in the thymus or in periphery and precise mechanisms remain to be elucidated.
A second interesting finding concerns the naive precursor frequency of CMV-specific T cells in seronegative donors. Of the 6 epitopes tested, this was one of the smallest populations. Considering mouse studies that have reported a correlation between precursor frequency and the magnitude of the response, it is notable that in CMV-seropositive donors, pp65-specific T cells represent one of the largest and most robust effector memory populations.30 We interpret these data to be a result of CMV being a chronic infection with continuous restimulation of CMV-specific CD8+ T cells. If the link between the naive precursor frequency and the robustness of an acute response holds up for natural infection in humans, it would indicate that a very different phenomenon is accounting for the dominance of T-cell populations during chronic infection.
Clinical development
We suggest that the TAME technique has the capacity to be used in the clinical setting. Certainly, it is relevant for monitoring T cells in human subject studies; and perhaps for the first time, it is possible to imagine the development of a validated diagnostic tool. As such, TAME will provide a unique opportunity to directly monitor rare human antigen–specific populations under physiologic and pathologic conditions. To this end, we illustrate the ability to multiplex the assay for monitoring several antigen specificities (Figure 6) and demonstrate the feasibility of starting from frozen PBMCs (Figure 2C). Importantly, starting from 50 mL of peripheral blood, TAME is sensitive enough to monitor CD8+ T cells with a precursor frequency higher than 10−6.
Although our study focused on naive T cells, this approach may be generally applicable for following rare antigen-experienced self-, tumor-, or pathogen-specific T cells. Specifically, we imagine that with proper development of this assay and the advances in tetramer production and analysis,24,26 TAME can be used as a screening tool for detection of cancer or autoantigen-specific T cells. For example, combination of tetramer labeling with phenotypic markers (eg, CD27, CD11a, CD25, PD-1) would permit evaluation if rare expanded populations of T cells have assumed a memory, effector, or tolerized state of differentiation.
Regarding vaccine strategies, we see great potential. In truth, we know nearly nothing about the naive T-cell repertoire. Medium-throughput tetramer production combined with multi-TAME has the potential to provide information about all potential epitope-specific T cells that may react to a given antigen. Phenotypic evaluation may help assess which, if any, are part of the memory pool (possibly due to cross-reactivity with self- or environmental antigens). Comparison with the T-cell specificities that are expanded by vaccination (and/or infection) will then permit assessment (in humans) of what are the true peptide determinants useful in component immunization strategies.
In summary, this work has allowed for the first time direct detection and enumeration of rare naive antigen-specific CD8+ T cells from human peripheral blood. We found significant differences in terms of precursor frequencies for different epitopes, but conserved numbers among humans. This tool will be of importance in both fundamental studies and immune monitoring in clinical trials. Evaluation of the impact of this new biomarker in diverse situations will have important implications in understanding the dynamics of immune responses, especially in relation to vaccine development and tumor-mediated immune evasion.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank P. Bousso, E. Tartour, N. Casartelli, and O. Schwartz for sharing critical reagents and for their helpful advice. We also thank M. L. Chaix and A. Alanio for performing the CMV serology testing, and T. Schumacher and M. Roederer for useful suggestions on data analysis.
This work was supported by grants from Institut National du Cancer (C.A.) and funding from La Ligue contre le Cancer.
Authorship
Contribution: C.A. designed research, performed experiments, analyzed data, and wrote the paper; F.L. provided practical assistance and contributed material for tetramers; H.K.W.L. and M.H. provided practical assistance; and M.L.A. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew L. Albert, Centre d'Immunologie Humaine, Institut Pasteur, 25 rue du Dr Roux, 75724 Paris, France; e-mail: albertm@pasteur.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal