Abstract
Activating mutations of NRAS are common in acute myeloid leukemia, chronic myelomonocytic leukemia, and myelodysplastic syndrome. Like all RAS proteins, NRAS must undergo a series of post-translational modifications for differential targeting to distinct membrane subdomains. Although farnesylation is the obligatory first step in post-translational modifications of RAS, to date, successes of therapies targeting farnesyl protein transferase are modest. Other RAS modifications, such as palmitoylation, are required for optimal plasma membrane association of RAS proteins. However, the relative importance of these latter modifications of RAS in leukemogenesis is not clear. We have previously shown that expression of oncogenic NRAS using a bone marrow transduction and transplantation model efficiently induces a chronic myelomonocytic leukemia- or acute myeloid leukemia-like disease in mice. Here we examined the role of palmitoylation in NRAS leukemogenesis using this model. We found that palmitoylation is essential for leukemogenesis by oncogenic NRAS. We also found that farnesylation is essential for NRAS leukemogenesis, yet through a different mechanism from that of palmitoylation deficiency. This study demonstrates, for the first time, that palmitoylation is an essential process for NRAS leukemogenesis and suggests that the development of therapies targeting RAS palmitoylation may be effective in treating oncogenic NRAS-associated malignancies.
Introduction
RAS proteins are small GTPases that act as molecular switches, transducing extracellular signals from activated receptors at the cell surface through various signaling pathways to the nucleus, which regulate cell proliferation, survival, and differentiation.1 Three RAS genes encode 4 widely expressed isoforms: HRAS, NRAS, and the splice variants KRAS4A and KRAS4B. Mutations causing constitutive activation of RAS are associated with approximately 30% of all human cancers, including both solid tumors and hematologic malignancies, with specific RAS isoforms preferentially associated with specific cancer types. Among myeloid malignancies, activating mutations of NRAS are most common, found in approximately 20% to 40% of acute myeloid leukemia (AML), myelodysplastic syndrome (MPD), and MPD/myeloproliferative disease/MPD, including chronic myelomonocytic leukemia (CMML) and juvenile myelomonocytic leukemia.2
The RAS family members are highly homologous but diverge at hypervariable regions (HVRs) at their C-terminus, although all terminate in a −CAAX motif (C for cysteine, A for aliphatic amino acid, and X for serine or methionine). The HVR is subjected to post-translational modifications (PTMs) that anchor RAS to cellular membranes and target the specific RAS isoforms to functionally distinct microdomains of the plasma membrane (PM) or endomembranes, allowing for interaction with specific pools of activator and effector proteins to generate discrete signal outputs.3,4 RAS proteins are modified first by addition of an isoprenoid lipid to the cysteine residue of the −CAAX motif by farnesyl protein transferase (FTase) or geranylgeranyl transferase (GGTase) in the cytosol, followed by removal of the −AAX tripeptide by RAS converting enzyme (Rce1), and methylation of the newly exposed terminal farnesylated cysteine residue by isoprenylcysteine carboxyl methyltransferase (Icmt) on the cytosolic face of the endoplasmic reticulum (ER).5 Prenylation is the minimal PTM required for membrane association. After −AAX cleavage and methylation, NRAS, HRAS, and KRAS4A are singly or doubly palmitoylated in the Golgi by palmitoyltransferases at cysteine residue(s) immediately upstream of the CAAX motif. Palmitoylated RAS isoforms travel through the classic secretary pathway to the PM, whereas KRAS4B traffics directly from the ER to the PM, relying on a polybasic lysine tract in its HVR as a second means of membrane association/subdomain localization.6,7 The palmitoylation of H, N, and KRAS4A is reversible; palmitoylated RAS at the plasma membrane can be depalmitoylated by a putative acylprotein thioesterase and recycle back to the Golgi.8 Consequently, a pool of depalmitoylated RAS exists at the Golgi and in transit in the case of both normal and oncogenic forms of the protein.
Past studies have examined the roles of PTMs in RAS localization and neoplastic transformation. Prenylation of RAS by FTase is the obligate first PTM for all RAS isoforms and has been shown to be essential for RAS membrane association and neoplastic transformation.9,10 For this reason, much emphasis has been placed on developing RAS farnesyltransferase inhibitors (FTIs). However, successes of therapies targeting FTase are modest to date.11 The reason may be that, in the presence of FTIs, NRAS and KRAS, the predominant oncogenic RAS isoforms in human cancers, can be alternatively prenylated by GGTase.12 This finding indicates that the limited anticancer effects of FTIs were probably the result of inhibition of other farnesylated proteins, such as RhoGTPases, Rheb, and/or CENP-E or CENP-F.13-16 Inhibitors targeting FTase and GGTase in combination have thus far proved too toxic to be clinically useful.17
Because FTIs have not been as successful as initially hoped, interest has turned to postprenylation PTMs of RAS as possible anticancer targets. It has recently been shown that an Rce1 conditional knockout mouse displayed a significantly more rapid disease progression in a KRAS leukemic mouse model, whereas a conditional knockout of Icmt in a similar model delayed but did not prevent disease progression.18,19
Palmitoylation is required for high-avidity plasma membrane binding and for the targeting of RAS to membrane subdomains.20-24 Because palmitoylation is required for localization of NRAS, HRAS, and KRAS4A to the inner face of the PM, a location previously thought to be exclusive for RAS signaling, inhibition of palmitoylation has been an attractive anticancer target.25 However, important roles for RAS signaling from the Golgi, ER, and mitochondrial membranes have been recently described.26-30 In addition, oncogenic HRAS-containing mutations blocking palmitoylation sites (HRAS61LC181S,C184S) localized to the Golgi and internal membranes and transformed NIH3T3 cells with 75% efficiency of oncogenic HRAS61L.27 For these reasons, the role of palmitoylation in NRAS transformation has remained unclear.
The development of leukemia in vivo is a complex process that involves both the effects of oncogenic mutations within susceptible cells and interactions of affected cells with the rest of the in vivo environment. Molecular processes required for leukemogenesis in vivo may or may not be the same as that for cellular transformation in vitro. We have previously shown that oncogenic NRAS effectively induces a CMML or AML-like disease in a mouse bone marrow transduction and transplantation model.31 Here, we used this model to examine the role of palmitoylation in NRAS leukemogenesis. We found that palmitoylation is essential for leukemogenesis by oncogenic NRAS. This study suggests that the development of therapies targeting RAS palmitoylation may be effective in treating NRAS-associated malignancies.
Methods
Construction of retroviral expression vectors
Construction of the murine stem cell virus (MSCV)–GFP-ires-2xmyc-tag-NRASD12 vector has been described previously31 and was used as template to produce all additional constructs described here. NRASD12C181S and NRASD12C186S were created using a mutational 2-step polymerase chain reaction (PCR) system using nested primers to introduce point mutations individually to sense and antisense complimentary overhangs. Subsequently, these individual products were combined as an annealed template in a second PCR reaction to amplify the completed NRASD12 gene containing the desired point mutation. Mutational primers used in separate individual reactions for NRASD12C181S were: 5′ CTC AGG GTA GTA TGG GAT and 5′ ATC CCA TAC TAC CCT GAG. For NRASD12C186S they were: 5′ CAC CAC ACT TGG CAA TCC and 5′ GAT TGC GTG TGG TGA TG. Each mutational primer was paired in its reaction with either 5′ TGA CTG ACT GAA TCG ATG or 5′ CAG GTG GGG TCT TTC ATT, to that anneal to the complimentary strand to amplify. The second PCR reaction used the products of the first round of reactions as the template and used the primers 5′ ATG GAC GAG CTG TAC AAG and 5′ GTC GGA TGC AAC TGC AAG to amplify both products containing the individual new point mutations. Final PCR products and introduced mutations were confirmed by DNA sequencing before subsequent cloning. Sequenced PCR products were first ligated into pCR2.1 TA cloning vector before being excised with NotI and ClaI and inserted into MSCV at these sites. Finally, GFP-ires was isolated from MSCV-GFP-ires-2xmyc-tag-NRASD12 by NotI and inserted into each MSCV-2xmyc-tag-NRASD12C181S or NRASD12C186S to create the final bicistronic expression vector. Plasmids expressing NRASD12 and NRASD12 PTM mutants as N-terminal GFP-fusion proteins were created by taking advantage of a digestion by NcoI of GFP at amino acid 234 found in an earlier sequenced miniprep, which resulted in a NcoI flanked green fluorescent protein (GFP) lacking the most C-terminal 5 amino acids. This was inserted into an NcoI-digested MSCV-GFP-ires-2xmyc-tag-NRASD12 construct. All cDNAs were reconfirmed by DNA sequencing before expression.
Retrovirus production and titering
Retroviruses were produced in BOSC23 cells, cultured, and titered as previously described.32 The viral titer was calculated in transducing units (TUs) by multiplying the percentage of NIH3T3 cells expressing GFP and the total number of cells on the dish at the time of infection. Retroviral titers were matched before bone marrow infection at 1.3 × 105 ± 0.3 × 105 TUs.
Cell culture
NIH3T3 and 32D cl-3 cell lines stably expressing NRASD12, NRASD12C181S, NRASD12C186S, or the control GFP were created by retroviral transduction as described.32,33 All cell lines were sorted by GFP expression to more than 95% homogeneity by fluorescence-activated cell sorting (FACS) using a FACSAria (BD Biosciences). Cell lines were cultured as previously described.32
Cell photographs and serial growth curves
Equal numbers (5 × 105) of NIH3T3 cells expressing vector alone, NRASD12, NRASD12C181S, or NRASD12C186S were plated onto plastic 100-mm tissue culture dishes. Cells were cultured for 6 days (media was changed every 2 days) and photographed using an Olympus E-Volt E500 digital camera attached to an Olympus IX70 inverted microscope (Olympus; original magnification, ×100). For serial growth curves, 2 × 105 cells were plated in 60-mm culture dishes at day 0. Media was changed every 2 days. Cells, in triplicate, were stained with trypan blue to exclude nonviable cells and counted manually under a light microscope each day. NRASD12-expressing cells formed tight spheres after 6 days in culture, making it difficult to determine the cell numbers. For this reason, the growth curve was ended at day 6. The Student t test was used for statistical analysis.
Soft-agar colony-forming assay
A total of 1 mL of 0.6% bottom agar (mixing 1.2% agar with concentrated media [2 times Dulbecco modified Eagle medium + 20% donor bovine serum + 2 times Pen-Strep] with 1:1 ratio) was prepared and introduced to each well of a 6-well tissue culture plate. FACS-purified NIH3T3 cell lines expressing GFP, NRASD12, NRASD12C181S, or NRASD12C186S were diluted to 105 cells/mL, 104 cells/mL, or 103 cells/mL in 1 times Dulbecco modified Eagle medium + 10% donor bovine serum 1 times Pen-Strep. Triplicate 3-mL cells suspended in soft agar were added to the bottom agar for each cell line and incubated in a 37°C CO2 incubator. Colonies were counted under a light microscope at day 14 after plating.
Bone marrow transduction/transplantation
Mouse bone marrow transduction and transplantation experiments were performed as previously described.34 Briefly, bone marrow (BM) cells from 5-fluorouracil (250 mg/kg)–treated 6- to 8-week-old male donor BALB/c mice (Taconic Farms) were infected with retroviruses each day for 2 days before 4 × 105 cells were injected into the tail vein of each lethally irradiated (2 × 4.5 Gy, 4 hours between each dose) female recipient BALB/c mouse as described.34 Retroviral titers were matched before BM infection. Recipient mice were monitored weekly for signs of disease beginning on day 14 after transplantation.
Mice used in this project are housed in the Association for Assessment and Accreditation of Laboratory Animal Care International accredited Foster Animal Research Facility at Brandeis University. All experiments involving mice are approved by the Institutional Animal Care and Use Committee of Brandeis University.
Hematopathologic analysis
Blood was collected from mice by tail bleed, and 3 μL was diluted in 3 mL of Isoton II (Fisher Scientific). White blood cell (WBC) counts were measured using the Coulter Counter model Z1 (Coulter), after lysing the red blood cells with ZAP-O-Globin (Beckman Coulter). Hematocrit was measured by capillary centrifugation on a micro-hematocrit centrifuge (StatSpin). Smears, cytospin, and touch preparations of blood and other murine tissues were stained with Hema 3 stain set (Fisher Scientific) for routine identification of cell morphology. Flow cytometric analysis of GFP-positive WBCs and immunophenotyping of leukemic cells were performed as previously described.31
Subcellular localization analysis of RAS proteins
NIH3T3 cell lines expressing NRASD12 with GFP fused to its N-terminus (GFP-NRASD12) or expressing GFP-fusion versions of NRASD12 PTM mutant proteins (GFP-NRASD12C181S, GFP-NRASD12C186S) were grown overnight on fibronectin-coated (50 μg/mL for 30 minutes at room temperature) glass coverslips. Fresh media was supplanted with 100 μg/mL cycloheximide 3 hours before fixation with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 minutes at room temperature. Cells were permeablized in 0.1% Triton X-100 for 15 minutes at room temperature and blocked with 2% bovine serum albumin in PBS for 30 minutes at room temperature before primary antibody (anti-BIP; Cell Signaling Technology; anti-GOLGA7; ABNOVA) was added at 1:250 dilution in 2% bovine serum albumin/PBS and incubated overnight at 4°C. Alexa Fluor 635-conjugated secondary antibody (Invitrogen) was added to 5 μg/mL final concentration for 1 hour at room temperature. Cells were washed 3 × 10 minutes in PBS after each step. Coverslips were mounted onto slides using Vectashield mounting medium with 4,6-diamidino-2-phenylindole (Vector Laboratories) and fluorescence visualized on a Leica TCS SP2 Spectral Confocal Microscope.
Western blot analysis
Cell lysates were prepared from 90% confluent NIH3T3 cell lines serum starved for 22 hours. Similar lysates were prepared from 32D cl-3 cell-lines starved of serum and WEHI-3B conditional media (as a source of interleukin-3) for 13 hours. Cells were counted and lysed in 1 times sodium dodecyl sulfate-polyacrylamide gel electrophoresis running buffer, sonicated briefly to break up DNA, heated at 100°C for 10 minutes, and centrifuged to remove debris. Lysates were resolved on 6% to 18% gradient polyacrylamide gels, transferred to nitrocellulose membranes, and blotted with the following primary antibodies overnight at 4°C: anti-RAS (RAS10; Upstate Biotechnology), antiactin (AC40; Sigma-Aldrich), anti-myc tag 9E10 monoclonal antibody (from conditional media of a 9E10 hybridoma cell line), and pAkt, Akt, pMek1/2, Mek1/2, pErk42/44, Erk42/44, pS6rp, and S6rp (all 1:1000; Cell Signaling Technology). Horseradish peroxidase-labeled goat anti–mouse IgG or goat anti–rabbit IgG (Pierce Biotechnology) was used as a secondary antibody. Densitometry ratios of expressed NRAS to endogenous RAS were performed using Adobe Photoshop 7.0 (Adobe Systems).
RAS-GTP and Ral-GTP detection assays
Activated RAS and Ral were detected using affinity purification kits (Upstate Biotechnology) according to the manufacturer's instructions. Human RAS-guanosine triphosphate (GTP) or murine Ral-GTP was immobilized on glutathione-agarose beads bound with GST-Raf-1-RAS-binding domain (RBD) or GST-Ral Binding Protein 1 (RalBP1), respectively, then run on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, transferred to nitrocellulose, and detected using RAS or Ral specific antibodies. Total RAS was similarly probed as a loading control in these assays.
Results
Stable expression of palmitoylation-deficient NRASD12 confers morphologic changes and abrogates cell density-dependent inhibition of growth but does not confer anchorage-independent growth to NIH3T3 cells
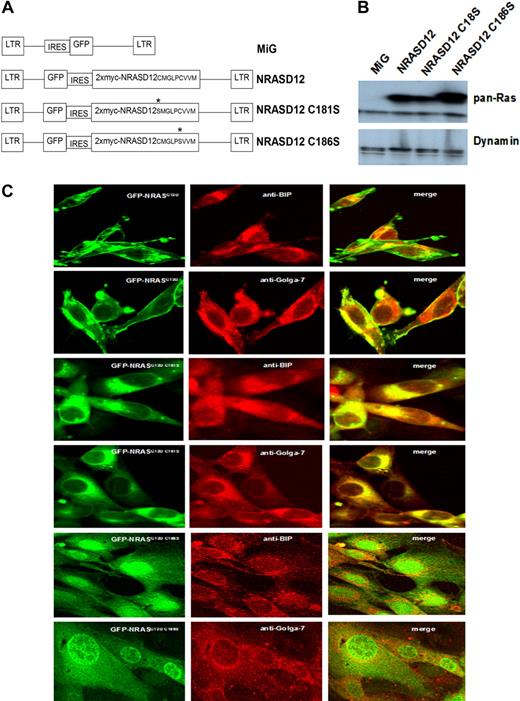
To determine whether palmitoylation is required for NRAS leukemogenesis and similarly confirm the requirement for prenylation, we constructed retroviral vectors expressing myc-tagged NRASD12C181S or NRASD12C186S (well-characterized palmitoylation-defective and prenylation-defective mutants of NRAS, respectively6 ; Figure 1A). We first characterized the PTM mutants of oncogenic NRAS in vitro. NIH3T3 cells were infected by retroviruses containing GFP alone (MiG), NRASD12, NRASD12C181S, or NRASD12C186S. Infected cells (GFP+) were isolated by FACS. The expression of NRAS mutants in NIH3T3 cells was confirmed by Western blotting with an anti-RAS antibody. Each NRAS mutant was expressed at similar levels (Figure 1B).
Expression and localization of oncogenic NRAS and its PTM mutants. (A) Schematic diagram of retroviral expression vectors used to transduce NRASD12, NRASD12C181S, or NRASD12C186S. (B) Immunoblot of lysates of NIH3T3 cells stably expressing the vector control, NRASD12, NRASD12C181S or NRASD12C186S with a pan anti-RAS antibody (top band represents double Myc-tagged-NRAS; and bottom band, endogenous Ras). (C) NIH3T3 cells expressing GFP-fused NRASD12, NRASD12C181S, or NRASD12C186S, costained with fluorescence-conjugated antibodies against Golgi (Golga-7) or ER (BIP) resident proteins, were visualized on a Leica TCS SP2 Spectral Confocal Microscope (original magnification, ×630).
Expression and localization of oncogenic NRAS and its PTM mutants. (A) Schematic diagram of retroviral expression vectors used to transduce NRASD12, NRASD12C181S, or NRASD12C186S. (B) Immunoblot of lysates of NIH3T3 cells stably expressing the vector control, NRASD12, NRASD12C181S or NRASD12C186S with a pan anti-RAS antibody (top band represents double Myc-tagged-NRAS; and bottom band, endogenous Ras). (C) NIH3T3 cells expressing GFP-fused NRASD12, NRASD12C181S, or NRASD12C186S, costained with fluorescence-conjugated antibodies against Golgi (Golga-7) or ER (BIP) resident proteins, were visualized on a Leica TCS SP2 Spectral Confocal Microscope (original magnification, ×630).
To confirm loss of plasma membrane association of the PTM deficient NRAS mutants, we constructed N-terminal GFP-fusion versions of NRASD12, NRASD12C181S, and NRASD12C186S retroviral vectors and expressed them in both 32D cl-3 cells (data not shown) and NIH3T3 cells. As expected, GFP-NRASD12 localized primarily to the plasma membrane and internal membranes that colocalized with immunostained Golgi-resident (Golga-7) and ER-resident (BIP) proteins (Figure 1C). GFP-NRASD12C181S localized entirely to internal membranes, whereas GFP-NRASD12C186S diffused in the cytoplasm and, interestingly, in the nucleus (Figure 1C). These results confirm that for NRAS palmitoylation is required for the association with the plasma membrane and that prenylation is required for any membrane association.
It has been observed that oncogenic KRAS and NRAS have greatly reduced transformation activity compared with oncogenic HRAS in NIH3T3 focus-forming assays.35 Consistent with this finding, we observed that minority populations of NRASD12-expressing cells grown in close proximity to normal NIH3T3 fibroblasts hardly formed foci (data not shown). In addition, NRASD12 did not permit 32D cl-3 myeloid progenitor cells to grow in a factor-independent manner (data not shown).
However, we observed a changed morphology for sorted NRASD12-expressing NIH3T3 cells (Figure 2A). Approximately one week after FACS sorting, NRASD12-expressing cells in culture appeared smaller, more spindle-shaped, and began to grow in a lattice that produced spheres protruding from the flat plane of adherent cells at the lattice nodules. These spheres grew to various sizes before detaching from the dish. Cells from spheres were viable and could merge with other spheres in suspension to form amorphous cell clumps. Interestingly, nonpalmitoylated NRASD12C181S-expressing cells also appeared spindle-shaped and grew in a lattice similar to NRASD12 cells, although these did not produce spheres. In contrast, the prenylation-deficient NRASD12C186S-expressing cells appeared larger and less spindle-shaped, morphologically similar to the MiG control cells.
NIH3T3 cell lines expressing NRASD12 or nonpalmitoylated NRASD12 result in phenotypic changes in morphology and density-dependent inhibition of growth. (A) Morphology of cultured NIH3T3 cells stably expressing the proteins indicated. Equal numbers (5 × 105) of NIH3T3 cells expressing vector alone, NRASD12, NRASD12C181S, or NRASD12C186S were plated onto 100-mm plates and were cultured for 6 days before the pictures were taken (original magnification, ×100). (B) Total numbers of viable cells per plate (means with error bars) for NIH3T3 cells expressing vector alone, NRASD12, NRASD12C181S, or NRASD12C186S were plotted versus time (days). NRASD12C186S cells grew significantly slower than the vector control cells at day 4 (P = .01). NRASD12 and NRASD12C181S cells grew significantly faster than the vector control cells at day 6 (P = .005 and P = .02, respectively).
NIH3T3 cell lines expressing NRASD12 or nonpalmitoylated NRASD12 result in phenotypic changes in morphology and density-dependent inhibition of growth. (A) Morphology of cultured NIH3T3 cells stably expressing the proteins indicated. Equal numbers (5 × 105) of NIH3T3 cells expressing vector alone, NRASD12, NRASD12C181S, or NRASD12C186S were plated onto 100-mm plates and were cultured for 6 days before the pictures were taken (original magnification, ×100). (B) Total numbers of viable cells per plate (means with error bars) for NIH3T3 cells expressing vector alone, NRASD12, NRASD12C181S, or NRASD12C186S were plotted versus time (days). NRASD12C186S cells grew significantly slower than the vector control cells at day 4 (P = .01). NRASD12 and NRASD12C181S cells grew significantly faster than the vector control cells at day 6 (P = .005 and P = .02, respectively).
To characterize any changes in proliferation or cell density-mediated inhibition of growth conferred by blocking PTMs of oncogenic NRAS, we compared growth of NIH3T3 cells stably expressing vector alone, NRASD12, NRASD12 C181S, or NRASD12 C186S in liquid culture (Figure 2B). Growth of MiG-expressing cells stopped after 4 days in culture because of cell density-dependent inhibition, whereas cells expressing NRASD12 continued to proliferate. Interestingly, cells expressing NRASD12C181S, similar to cells expressing NRASD12, were able to overcome normal density-dependent growth controls and continue to proliferate. Cells expressing NRASD12C186S, on the other hand, displayed a significantly reduced proliferation compared with control cells, suggesting that expression of prenylation-defective NRASD12 is toxic to NIH3T3 cells.
To further assess the transforming potential of NRASD12 and its PTM mutants, we performed a soft-agar colony assay. Sorted GFP+ cells were seeded in soft-agar and colonies were counted at day 14 (Table 1). NRASD12-expressing cells formed numerous colonies in soft agar, indicating that oncogenic NRAS can abrogate the anchorage-dependent growth of NIH3T3 cells. In contrast, cells expressing nonpalmitoylated NRASD12C181S formed only a few clusters equivalent to cells expressing NRASD12C186S or GFP alone. These observations demonstrate that expression of palmitoylation-defective NRASD12 causes NIH3T3 cells to display some characteristics of transformation, including morphologic changes and loss of normal density-dependent growth inhibition but cannot confer anchorage-independent growth to these cells.
Anchorage-independent growth of NIH3T3 cells expressing oncogenic NRAS or post-translational modification mutants
| Cell line . | No. of colonies* (mean ± SD) . |
|---|---|
| MiG | 0.7 ± 0.6 |
| NRASD12 | 482.3 ± 48.0 |
| NRASD12C181S | 2.0 ± 1.0 |
| NRASD12C186S | 5.0 ± 1.0 |
| Cell line . | No. of colonies* (mean ± SD) . |
|---|---|
| MiG | 0.7 ± 0.6 |
| NRASD12 | 482.3 ± 48.0 |
| NRASD12C181S | 2.0 ± 1.0 |
| NRASD12C186S | 5.0 ± 1.0 |
Colonies were counted on day 14 after plating.
Palmitoylation and prenylation are each required for NRAS leukemogenesis
We next examined the role of palmitoylation in NRAS leukemogenesis using a mouse bone marrow transduction and transplantation model. We also tested the leukemogenic potential of the prenylation-defective mutant of NRAS because, although prenylation of RAS is the obligate initial PTM and has been shown to be essential for RAS transformation in vitro, the role of prenylation in NRAS leukemogenesis in vivo has not yet been directly tested.
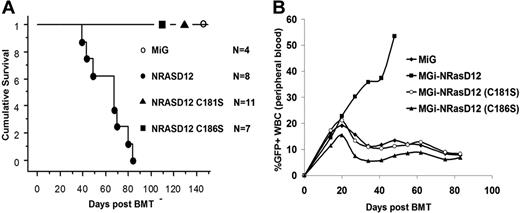
We infected BM cells isolated from 5-fluorouracil–treated mice with titer-matched retroviruses containing NRASD12, NRASD12C181S, NRASD12C186S, or vector control and then transplanted these cells into lethally irradiated syngeneic recipient mice, as previously described.31 As shown previously, all mice receiving NRASD12-transduced BM cells developed a fatal AML (∼ 35%)– or CMML (∼ 65%)–like disease and died in 30 to 90 days after bone marrow transplantation (Figure 3A; and data not shown). However, mice receiving NRASD12C181S or NRASD12C186S-transduced BM cells did not develop any disease and remained healthy for more than 2 years, similar to MiG control mice (Figure 3A; and data not shown). Peripheral blood (PB) collected weekly from these animals displayed no abnormal expansion of any cell type when cell morphologies were examined after differential staining with Hema-3 stain (Fisher; data not shown). Livers and spleens of animals receiving NRASD12C181S- or NRASD12C186S-transduced BM cells were of normal size and weight, similar to MiG mice (data not shown). Cells isolated from livers, spleens, PB, and BM of these animals were assessed by FACS analysis with a panel of myeloid and lymphoid markers (Gr-1, B220, CD19, Thy-1.2, CD86, CD31, CD115 [M-CSFR], Ter-119, Mac-1, CD34, CD38, CD16/32, and c-Kit) and displayed no abnormal hematopoietic expansion, as did MiG mice. Identical results were seen in 2 independent experiments.
Palmitoylation and prenylation are each essential for NRAS leukemogenesis in vivo. (A) Kaplan-Meier plot of cumulative survival of recipient mice transplanted with BM cells infected by NRASD12, NRASD12C181S, or NRASD12C186S containing retroviruses, or the MiG vector. (B) FACS analysis of GFP+ cells in peripheral blood of NRASD12, NRASD12C181S, NRASD12C186S, or GFP vector bone marrow transduction and transplantation mice.
Palmitoylation and prenylation are each essential for NRAS leukemogenesis in vivo. (A) Kaplan-Meier plot of cumulative survival of recipient mice transplanted with BM cells infected by NRASD12, NRASD12C181S, or NRASD12C186S containing retroviruses, or the MiG vector. (B) FACS analysis of GFP+ cells in peripheral blood of NRASD12, NRASD12C181S, NRASD12C186S, or GFP vector bone marrow transduction and transplantation mice.
Weekly analysis of PB of animals receiving NRASD12C181S- or NRASD12C186S-transduced BM revealed a limited increase (analogous to MiG control) in the percentage of GFP+ WBCs over the first 3 weeks after bone marrow transplantation, peaking at 15% to 20% GFP+ WBCs. This result indicates that NRASD12C181S- or NRASD12C186S-transduced BM cells are not deficient for homing to hematopoietic niches (Figure 3B). After 3 weeks, the percentage of GFP+ PB WBCs isolated from MiG, NRASD12C181S, and NRASD12C186S mice began to decline, with similar rates of decline in NRASD12C181S and NRASD12C186S mice as MiG mice, suggesting that expression of palmitoylation- or prenylation-defective NRASD12 is not significantly toxic to hematopoietic cells. These experiments show that palmitoylation and prenylation are essential for NRAS leukemogenesis.
Palmitoylation is required for activation of multiple downstream signaling pathways by oncogenic NRAS
RAS activates multiple downstream signaling pathways, including the phosphoinositide-3 kinase (PI3K), mitogen-activated protein kinase (MAPK)/Erk and Ral pathways. To determine whether and how these signaling pathways are affected by lack of palmitoylation of NRASD12, we examined known activating phosphorylation sites on well-established signaling proteins in the PI3K and MAPK pathways in NIH3T3 cells expressing MiG, NRASD12, NRASD12C181S, or NRASD12C186S by Western blotting with phospho-specific antibodies (Figure 4A-B).
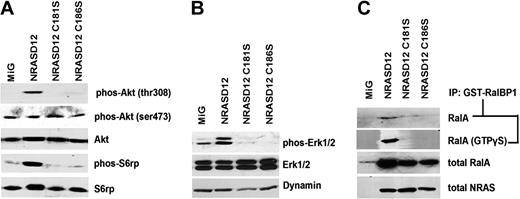
Oncogenic signaling downstream of NRASD12 is disrupted by blocking palmitoylation. Serum-starved lysates of NIH3T3 cells expressing vector, NRASD12, NRASD12C181S, or NRASD12C186S were analyzed by Western blotting. (A) Effect of palmitoylation and prenylation of NRASD12 on phosphorylation of Akt and S6 ribosomal protein. (B) Effect of palmitoylation and prenylation of NRASD12 on phosphorylation of Erk. (C) Effect of palmitoylation and prenylation of NRASD12 on Ral activation. Ral-GTP precipitated from serum-starved NIH3T3 cells expressing vector control, NRASD12, NRASD12C181S, or NRASD12C186S with GST-RalBP1 glutathione agarose beads with or without addition of GTPγS were analyzed by Western blotting with an anti-RalA antibody. Input RalA was also probed. RAS expression was detected as a loading control.
Oncogenic signaling downstream of NRASD12 is disrupted by blocking palmitoylation. Serum-starved lysates of NIH3T3 cells expressing vector, NRASD12, NRASD12C181S, or NRASD12C186S were analyzed by Western blotting. (A) Effect of palmitoylation and prenylation of NRASD12 on phosphorylation of Akt and S6 ribosomal protein. (B) Effect of palmitoylation and prenylation of NRASD12 on phosphorylation of Erk. (C) Effect of palmitoylation and prenylation of NRASD12 on Ral activation. Ral-GTP precipitated from serum-starved NIH3T3 cells expressing vector control, NRASD12, NRASD12C181S, or NRASD12C186S with GST-RalBP1 glutathione agarose beads with or without addition of GTPγS were analyzed by Western blotting with an anti-RalA antibody. Input RalA was also probed. RAS expression was detected as a loading control.
Cells were serum-starved for 22 hours before lysis to minimize RAS signaling resulting from receptor tyrosine kinase activation by serum growth factors. In the PI3K pathway, Akt is a central regulator of cell survival, proliferation, and metabolism.36,37 We detected little or no basal phosphorylation at Akt Thr308, an activating phosphorylation site of Akt and target of PDK1 downstream of PI3K in MiG vector lysates, whereas NRASD12 expression triggered marked phosphorylation at this site (Figure 4A). In contrast, cell lysates isolated from both NRASD12C186S- and NRASD12C181S-expressing cells did not induce phosphorylation at Akt Thr308. Akt phosphorylation at Ser473 contributes to full activation of Akt and is the target of the mTORC2 complex.38,39 We observed that Akt Ser473 was constitutively phosphorylated in MiG cell lysates, and this remained unchanged in lysates of cells expressing NRASD12 or the PTM mutants of NRASD12 (Figure 4A).
Activating phosphorylation of S6rp, a ribosomal protein, is often increased in cells transformed by oncogenic RAS.40 We observed little or no basal activation of S6rp in MiG cell lysates but marked activation of S6rp in lysates of cells expressing NRASD12 (Figure 4A). NRASD12-mediated hyperactivation of S6rp was not seen in lysates of cells expressing NRASD12C181S or NRASD12C186S. Similarly, marked phosphorylation of Erk1/2 was observed in lysates from NRASD12 cells, whereas in NRASD12C186S and NRASD12C181S cell lysates we did not observe activating phosphorylation of Erk1/2 (Figure 4B).
Ral GTPase activation has also been implicated as an important pathway downstream of RAS.41 To assess whether NRAS palmitoylation is necessary for Ral activation, we ran Ral-GTP detection assays using lysates of the cell lines the vector control, NRASD12, NRASD12C181S or NRASD12C186S (Figure 4C). In this assay, active RalA (GTP bound) is precipitated with glutathione-agarose beads bound with GST-tagged RalBP1. The precipitates were then analyzed by Western blotting using an anti-RalA antibody. Once again, we observed marked activation of Ral by NRASD12, and this activity was abolished by either the prenylation or palmitoylation mutation. Lysates treated with a nonhydrolyzable form of GTP (GTPγS) showed similar results, suggesting that nonpalmitoylated NRASD12 is incapable of activating Ral guanine nucleotide exchange factors (RalGEFs). These results demonstrate that palmitoylation, like prenylation, is essential for NRASD12 to activate downstream signaling pathways that are important for cell transformation.
Prenylation, but not palmitoylation, affects GTP loading of oncogenic NRAS
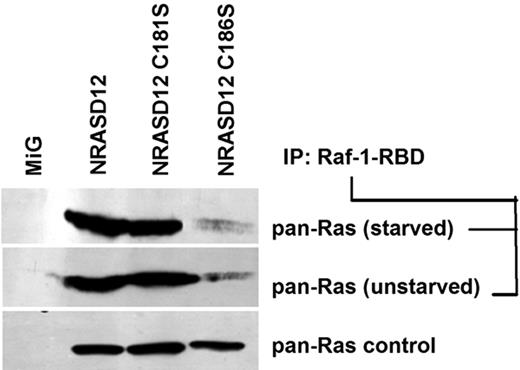
Having observed that multiple oncogenic signaling pathways were not activated by palmitoylation-defective NRASD12, we wondered whether NRASD12C181S retained the ability to bind GTP. GTP-bound RAS proteins from lysates of serum-starved and unstarved NIH3T3 cells expressing NRASD12, NRASD12C181S, NRASD12C186S, or GFP control were analyzed by a RAS-GTP detection assay. We observed that NRASD12 binds GTP in both serum-starved and unstarved lysates, indicating that NRASD12 is constitutively activated (Figure 5; and data not shown). We also found that levels of RAS-GTP from cells expressing NRASD12C181S were similar to those from cells expressing NRASD12 (Figure 5). Interestingly, prenylation-defective NRASD12 showed markedly reduced levels of RAS-GTP, perhaps because the lack of membrane association prevents it from accessing to RasGEFs. These results indicate that, although prenylation is important for GTP loading of NRASD12, palmitoylation in not required for optimal GTP loading of NRASD12 or for its binding the RBD of Raf-1.
Palmitoylation-defective NRASD12C181S retains the ability to bind Raf-1-RBD. RAS-GTP precipitated with glutathione-agarose beads bound with GST-fused RAS-binding domain (residues 1-149) of Raf-1 from serum-starved or unstarved NIH3T3 cells expressing GFP alone, NRASD12, nonpalmitoylable NRASD12C181S, or nonprenylable NRASD12C186S were analyzed by Western blotting with an anti–pan-RAS (RAS10) antibody. RAS from the same lysates was probed as a loading control.
Palmitoylation-defective NRASD12C181S retains the ability to bind Raf-1-RBD. RAS-GTP precipitated with glutathione-agarose beads bound with GST-fused RAS-binding domain (residues 1-149) of Raf-1 from serum-starved or unstarved NIH3T3 cells expressing GFP alone, NRASD12, nonpalmitoylable NRASD12C181S, or nonprenylable NRASD12C186S were analyzed by Western blotting with an anti–pan-RAS (RAS10) antibody. RAS from the same lysates was probed as a loading control.
Discussion
In this report, we demonstrate, for the first time, that palmitoylation is essential for leukemogenesis by oncogenic NRAS, raising the possibility that therapeutics targeting NRAS palmitoylation may be effective in treating NRAS-associated hematologic malignancies as well as other NRAS-related cancers. Palmitoylation-defective NRASD12 localizes entirely to internal membranes in cells and can be activated by GTP binding. Although defective in activating the PI3K, Erk, and Ral pathways, the NRASD12 palmitoylation mutant retains the ability to alter morphology and to abrogate normal density-dependent growth controls when stably expressed in NIH3T3 fibroblast cells. However, this residual activity had no apparent effects in hematopoietic cells.
In addition, we show that prenylation is essential for leukemogenesis by oncogenic NRAS, confirming the importance of this process in RAS oncogenesis in vivo. Prenylation-defective NRASD12 completely loses the ability to associate with cellular membranes, is inefficiently activated by GTP loading, cannot activate downstream oncogenic signaling pathways, and does not transform cells in vitro or in vivo. In addition, NIH3T3 cells express prenylation-defective NRASD12 proliferate at a slower rate than control cells expressing GFP only, suggesting that prenylation-defective NRASD12 is toxic to these cells. Interestingly, a GFP-tagged version of this mutant localized predominately to the nucleus, although this fusion protein is too large to enter the nucleus by passive diffusion. This observation suggests that unmodified RAS might be actively targeted to the nucleus and may serve a yet unknown nuclear function.
It was previously shown that G418-selected NIH3T3 cells stably expressing NRASV12 were not capable of growing in an anchorage-independent manner in soft agar.35 We show here that FACS-purified NRASD12-expressing NIH3T3 cells form spheres in liquid culture, and form colonies in soft agar. This discrepancy may be the result of different NIH3T3 sub-lines used. The same oncogene may have different transforming abilities in different NIH3T3 sub-lines. For example, it has been found that there are permissive and nonpermissive sub-lines of NIH3T3 cells for transformation by abl oncogenes.32,42,43 The NIH3T3 cell line we used can be transformed by various abl oncogenes.32
The observation that active RAS signaling occurs not only on the plasma membrane but also on internal membranes, including the ER and Golgi, has significantly altered the way we view RAS signaling26,27,29,30 ; it has brought to light the idea of compartmentalized signaling, with different resident pools of activators and effectors becoming accessible to RAS at different subcellular locales. Whereas pools of constitutively active H- and NRAS are found on internal membranes and engage Raf-1, only HRAS was reported to retain much of its transforming capacity regardless of palmitoylation status and to become activated in response to growth factors from internal membranes in cultured cell lines.27 Although it is not known whether palmitoylation is also required for leukemogenesis by oncogenic HRAS, we found here that palmitoylation-deficient NRASD12 does not activate the PI3K, Erk, and Ral pathways and loses much of its transforming activity in NIH3T3 cells. The different roles of palmitoylation in H- and NRAS transformation may rely on additional differences between the 2 RAS oncoproteins.
Interestingly, nonpalmitoylated NRASD12 remains constitutively GTP-bound and thus retains its capacity to engage the RAS-binding domain of Raf kinase. However, we found that nonpalmitoylated NRASD12 is incapable of activating Erk. Activation of Raf requires more than simple engagement of its RBD by RAS-GTP. We have noted that, in myeloid progenitor 32Dcl-3 cells stably expressing NRASD12, Raf-1 is phosphorylated on Ser259 (data not shown), a site that has been shown to be phosphorylated by Akt to produce an inhibitory 14-3–3 binding site and block downstream MAPK signaling.44 It is thought that this inhibitory binding site must be dephosphorylated by growth factor-induced phosphatases, such as PP1 and PP2A, which may require association of NRAS with the plasma membrane.45 Similarly, activation of the PI3K and Ral pathways by oncogenic NRAS may also require association with the plasma membrane.
Although targeting prenylation has proven difficult (targeting one of the 2 enzymes responsible for this modification is insufficient, yet targeting both is too toxic), there is reason to think that targeting RAS palmitoylation may prove more successful. Thus far, 23 putative palmitoyl-acyltransferases, each exhibiting a high degree of enzyme-substrate specificity, have been identified.46-48 As such, inhibiting RAS-specific palmitoyl-acyltransferases could prove an effective therapy for leukemias and other cancers involving NRAS.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kevin Mark for technical assistance.
This work was supported by the National Institutes of Health (grant R01HL083515; R.R.).
National Institutes of Health
Authorship
Contribution: B.C. participated in the design of the study, performed research, analyzed data, and drafted the paper; and R.R. designed research, analyzed data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruibao Ren, Rosenstiel Basic Medical Sciences Research Center and Department of Biology, Brandeis University, 415 South St, Waltham, MA 02454; e-mail: ren@brandeis.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal