In this issue of Blood, Tulpule and colleagues1 describe the use of lentiviral vectors to knock down FANCA and FANCD2 in hESC lines, which results in early hematopoietic defects reminiscent of the human disease.
Fanconi anemia (FA) is an autosomal recessive disorder characterized by bone marrow failure, birth defects, and a predisposition to malignancy. A plethora of new scientific information has clearly established that the 13 known FA gene products are involved in the regulation of DNA repair (reviewed in de Winter and Joenje2 ). It is currently believed that 8 of the FA proteins (FANCA, B, C, E, F, G, L, and M) form a nuclear complex that functions “upstream” in the pathway to enzymatically monoubiquitinate a heterodimer pairing between FANCI and FANCD2 (referred to as the ID heterodimer). Following this biochemical activation, the ID heterodimer is targeted to nuclear foci that contain BRCA1, RAD51, and BRCA2/FANCD1, which ultimately participate in homology-directed DNA repair. FANCD2, FANCI, and BRCA2/FANCD1 thus act “downstream” in the pathway from the core complex, and may have additional functions in maintaining genomic integrity as well. This view of the FA pathway certainly explains the hypersensitivity of patients' cells to DNA cross-linking agents, which still forms the basis for diagnosing FA.
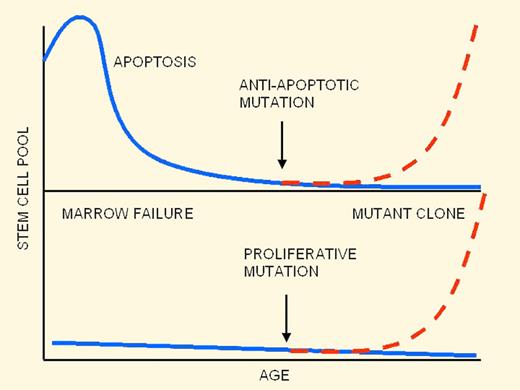
Ontogeny of hematopoietic failure in FA: old theory (top panel) and new theory (bottom panel).
Ontogeny of hematopoietic failure in FA: old theory (top panel) and new theory (bottom panel).
Knowledge gained of the molecular function of the FA gene products has not yet translated, however, into an understanding of the pathophysiology or ontogeny of the bone marrow failure that is the clinical hallmark of the disease. A clear picture has been elusive thus far, largely because mouse knockout models do not exhibit spontaneous marrow failure. Furthermore, because nearly all of the insights obtained have relied on experiments on postnatal hematopoietic cells from the peripheral blood or bone marrow, not much is known about the ontogeny of hematopoiesis in utero.
To summarize many years of research on FA mouse models, individual knockouts of Fanca, Fancc, Fancg, and Fancd2 genes have been generated, but none spontaneously develops bone marrow failure, aplastic anemia, leukemia, or skeletal abnormalities (reviewed in Parmar et al3 ). Although the mice exhibit subtle hematopoietic cell abnormalities including decreased long-term repopulating ability4 and hypersensitivity to oxidative stress5 and inhibitory cytokines such as tumor necrosis factor-alpha,6 progressive pancytopenia is not seen but can be induced after DNA damage with mitomycin C treatment.7
A popular theory8 that accounts for many aspects of the knockout mouse data is summarized in the figure (top panel). In this scenario, FA hematopoietic stem cells are initially normal in number at birth but suffer a steady decline as a result of a high rate of apoptosis, which leads to bone marrow failure. Due to strong selective pressure for stem cells resistant to apoptosis, secondary mutations in apoptosis-regulating genes can lead to clonal escape and expansion of preleukemic cells.
The work from Tulpule et al contradicts several key tenets in this theory and leads to a different picture (bottom panel of the figure). First, Tulpule et al demonstrated that knockdown of FANCA and FANCD2 in hESCs led to a reduction in hematopoietic fates and progenitor numbers that could be rescued by expression of the wild-type FA gene product. In these assays, human embryonic stem cells (hESCs) were induced to differentiate to embryoid bodies (EBs) and then to hematopoietic lineages. Hematopoiesis was impaired from the earliest stages of blood cell formation, which contrasts with the view of progressive stem cell loss during early childhood. A second unexpected finding was an apparent absence of significantly increased apoptosis rates in FA hematopoietic progenitors, suggesting that the observed hematopoietic defect might not be a result of direct cell loss at all.
Other unexpected findings included possible intrinsic developmental differences between “upstream” and “downstream” FA mutants. Knockdown of FANCD2 resulted in a much greater impairment of hematopoietic development from hESCs than knockdown of FANCA, suggesting greater deficiencies in embryonic hematopoiesis and accelerated progression to marrow failure in FA-D2 patients. It is not yet clear whether these differences reflect clinically meaningful differences between patients with mutations in upstream genes versus downstream genes (FANCD2, FANCI, and BRCA2/FANCD1).
For the FA field, the work of Tulpule et al raises a host of new questions. One important issue that suggests itself is how cancer evolves in the FA patient. Because there appear to be markedly different patterns of cancer development in patients with core complex versus downstream FA proteins, this might be a particularly fruitful area to apply the hESC model. Preliminary data from Tulpule and colleagues have suggested that apoptosis may not account for the hematopoietic growth defect in utero. Confirmation of this result should be sought and other mechanisms of growth inhibition tested. The authors have further shown that EB-derived populations exhibited impaired hematopoietic development coupled with primitive type gene expression and reductions in homeotic gene regulators such as CDX1, known to be involved in embryonic hematopoiesis and patterning of the ventral mesoderm.9
A final implication of this work comes from the new induced pluripotent stem cell (iPSC) technology field. The Izpisúa-Belmonte group reported that FA patient fibroblasts could not be reprogrammed to iPSCs in the absence of genetic complementation.10 Only after correction with wild-type FA gene product could iPSCs be generated, which then gave rise to normal numbers of disease-free hematopoietic progenitors. This study has reiterated the suspected crucial role of the FA pathway in the self-renewal of pluripotent stem cells. Coupled with the early impairment of embryonic hematopoiesis in FA, it again points to the powerful selective advantage corrected cells should possess over mutant. Harnessing this advantage to develop effective cell and gene therapy may be the next challenge.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal