Abstract
CD85j (ILT2/LILRB1/LIR-1) is an inhibitory receptor that recognizes major histocompatibility complex (MHC) class Ia and Ib alleles that are widely expressed on all cell types. On ligand recognition, CD85j diminishes kinase activity by recruiting phosphatases to motifs within its cytoplasmic domain. Within the hematopoietic system, CD85j is expressed with cell-specific patterns and cell surface densities that reflect the different roles of cell contact-mediated inhibition in these lineages. While monocytes ubiquitously have high cell surface expression, B lymphocytes start to express CD85j at intermediate levels during early B-cell maturation and natural killer (NK) cells and T cells exhibit a low level of expression on only a subset of cells. The cell-specific expression pattern is accomplished by 2 complementing but not independent mechanisms. Lymphocytes and monocytes use distinct promoters to drive CD85j expression. The lymphocyte promoter maps 13 kilobases (kb) upstream of the monocyte promoter; its use results in the inclusion of a distant exon into the 5′-untranslated region. A short sequence stretch within this exon has the unique function of repressing CD85j protein translation and is responsible for the subdued expression in lymphocytes. These cell-specific mechanisms allow tailoring of CD85j levels to the distinct roles it plays in different hematopoietic lineages.
Introduction
CD85j (ILT2/LILRB1/LIR-1) is a type I transmembrane protein of the immunoglobulin superfamily of receptors that is broadly expressed by cells of hematopoietic origin. The 110-kDa protein consists of 4 extracellular immunoglobulin-like domains and a 167–amino acid cytoplasmic tail containing 4 immunoreceptor tyrosine-based inhibitory motifs.1,2 CD85j functions to suppress intracellular kinase activity by recruiting the phosphatase Src homology domain-containing phosphatase 1 to phosphorylated tyrosines within its immunoreceptor tyrosine-based inhibitory motifs.1-3 Indeed, CD85j was originally identified as a novel natural killer (NK) cell–inhibitory receptor analogous to the inhibitory killer immunoglobulin-like receptors whose engagement by class I molecules on target cells prevents lysis of normal cells.1 In contrast to killer immunoglobulin-like receptors, which each recognize a limited subset of class I alleles,4 most, if not all, of the classical and nonclassical human major histocompatibility complex (MHC) class I alleles serve as natural ligands for CD85j.5 In addition, the human cytomegalovirus class I homolog UL18 binds CD85j with very high affinity.6
Among the many cell types expressing CD85j are monocytes, B cells, T cells, and NK cells.1,2,7,8 In every cell type tested, in vitro engagement of CD85j leads to dampening of cell activation when cross-linked with the activating stimulus. For example, Fc receptor–mediated7 and B-cell receptor–mediated1,9 signals in monocytes and B cells, respectively, are inhibited by CD85j cross-linking. NK cell–mediated lysis of MHC class I–transfected 721.221 cells is restored by adding CD85j-blocking antibodies.1,10 In T cells, coligation of CD85j and CD3 results in decreased proliferation, cytotoxicity, cytokine production, and actin cytoskeleton rearrangement.3,11,12
In vitro cross-linking of CD85j with the activating stimuli is not a physiologic representation of how CD85j functions in different cellular contexts. In CD8 T cells, MHC class I molecules represent the ligand for both the stimulatory T-cell receptor (TCR) and CD8 coreceptor and the inhibitory CD85j. TCR and CD8 engage MHC class I molecules within a tightly organized and spatially focused synapse that serves to strengthen and stabilize T cell–activating signals and more precisely to direct effector molecules toward the target cell.13 If CD85j is recruited to the synapse, it can be expected to deliver a strong inhibitory signal even at low cell surface concentrations. In NK cells, engagement of inhibitory receptors and their aggregation at the point of contact with target cells is considered a primary event forming an inhibitory synapse and thereby preventing lysis of healthy cells.14
In other cell types, such as B cells and monocytes, whose classic functions do not require MHC class I interactions, in vivo engagement of CD85j may be more dispersed and not directly linked to the activating signal, in particular, if the activating stimulus is a soluble molecule. For these cells, either CD85j will engage in cis-binding to MHC class I molecules expressed on the same cell, as has been shown to occur in monocytes,15 or in trans-binding to MHC class I molecules on neighboring cells during cell-to-cell contacts that do not directly involve or require MHC class I. Because MHC class I is expressed by all nucleated cells, trans interactions could involve a wide range of cell types, including stromal cells, endothelial cells, T cells, and other B cells or monocytes. Obviously, CD85j on NK and CD8 T cells can participate in these cis and trans interactions in addition to the more precise scenarios mentioned earlier.
Given that CD85j function in different cellular contexts probably requires different levels of cell-surface expression, it is intriguing to hypothesize that regulation of CD85j is cell specific. Indeed, and in support of this hypothesis, distinct CD85j expression profiles exist among hematopoietic cells. B cells, monocytes, and dendritic (DC) cells constitutively express CD85j, whereas only a subset of NK cells and T cells express it.1,2,7,8 CD8 T cells are much more likely than CD4 T cells to express CD85j, which, for both, is almost exclusively found on memory cells.8,16 In addition, CD85j expression on CD8 T cells exhibits a strong age dependence, resulting in its expression by a substantial majority of CD8 T cells in the elderly.16 In this study, we examined the hypothesis that mechanisms controlling CD85j expression, which is encoded by the gene LILRB1, are distinct in hematopoietic lineages accounting for different expression levels and accomplishing cell-specific functions. We provide evidence that lymphocytes express CD85j from an up-to-now undescribed promoter distinct from that used by monocytes. In addition, translational efficacy is modulated by a short sequence stretch within exon 1 of LILRB1. LILRB1 promoter choice strongly influences CD85j protein levels in distinct cell types.
Methods
Isolation of human mononuclear cells
Healthy donors were recruited, with informed consent according to the Declaration of Helsinki and according to protocols of Emory University Institutional Review Board, to donate up to 50 mL of whole blood. In most cases, peripheral blood mononuclear cells (PBMCs) were isolated by gradient centrifugation with the use of Lymphocyte Separation Medium (Lonza). Purified NK cells were isolated from whole blood with the use of the Human NK Cell Enrichment Cocktail (StemCell Technologies). Other purified cell subsets were obtained from PBMCs by magnetic bead–assisted sorting. Briefly, PBMCs were incubated with CD8, CD19, or CD14 microbeads as recommended by the manufacturer (Miltenyi Biotec). Desired cell populations were recovered by positive selection with AutoMACS (Miltenyi Biotec).
Flow cytometry
Surface phenotyping of ex vivo–isolated and transfected PBMCs was performed on an LSRII flow cytometer (BD Biosciences). Briefly, cells were incubated with fluorophore-conjugated monoclonal antibodies (mAbs) at 4°C for 15 minutes. Anti–human antibodies used were phycoerythrin-cyanine 7 (PE-Cy7)–conjugated anti-CCR7; fluorescein isothiocyanate (FITC)–, peridinin chlorophyll (PerCP)–-, allophycocyanin (APC)–, and APC-Cy7–conjugated anti-CD3; PerCP-conjugated CD4; PerCP-, APC-, and PE-Cy7–conjugated anti-CD8; APC-conjugated anti-CD14; APC-Cy7–conjugated anti-CD16; PerCP-conjugated anti-CD19; PerCP- and APC-Cy7–conjugated anti-CD20; PE-conjugated anti-CD24; APC-conjugated anti-CD27; PE-Cy7–conjugated anti-CD38; FITC-conjugated anti-CD45RA; FITC-conjugated anti–immunoglobulin D (IgD; all from BD Biosciences); PE-conjugated anti-CD85j (clone HP-F1; Beckman Coulter) and APC-conjugated anti-CD85j (clone HP-F1; eBioscience). After washing, cells were resuspended in 2% paraformaldehyde and analyzed by flow cytometry within 1 to 3 days of staining. Analyses were performed with FlowJo software (TreeStar).
RNA isolation and cDNA synthesis
Total RNA was isolated by Trizol extraction (Invitrogen) from 2 to 4 million cells. RNA pellets were washed with 75% ethanol and dried before cDNA synthesis for real-time polymerase chain reaction (PCR) or 5′-rapid amplification of cDNA ends (5′-RACE) analysis. cDNA for real-time PCR was synthesized with the AMV RT enzyme and random hexamer primers (Roche).
Quantitative real-time PCR
LILRB1 cDNA levels were quantified with Sybr-Green fluorescence (Invitrogen) analyzed on the MXP3000P real-time PCR machine (Stratagene). LILRB1 levels are represented as copy numbers relative to 2 × 105 copies of β-actin, both determined with standard curves. Before quantification of LILRB1 transcript levels from transfected cells, cDNA was treated with DpnI for 1 hour at 37°C to digest plasmid DNA.
Western blotting
Whole-cell extracts were obtained from washed and pelleted cells. Sodium dodecyl sulfate–denatured protein was separated by polyacrylamide gel electrophoresis with the use of Ready-Gels (Bio-Rad) and transferred to Hybond-P polyvinylidene diflouride membrane (Amersham). After blocking with 5% blocking solution (Bio-Rad), blots were probed by overnight incubation at 4°C with 1:200 dilution of anti-CD85j mouse mAb (clone VMP55; Santa Cruz Biotechnology) or 1-hour incubation at room temperature with a 1:5000 dilution of antiactin mouse mAb (Santa Cruz Biotechnology). Primary antibody staining was followed by washing and 1-hour incubation at room temperature with a 1:5000 dilution of horseradish peroxidase–conjugated goat anti–mouse immunoglobulin secondary antibody (Santa Cruz Biotechnology) followed by washing and horseradish peroxidase detection with Immobilon (Millipore). CD85j-probed blots were detected and stripped before β-actin probing and detection.
Plasmids
Transfection studies were performed with plasmids containing the pcDNA3 vector backbone (Invitrogen) and a variety of cDNA sequences amplified from the LILRB1 cDNA clone BC01573117 with the use of platinumTaq polymerase (Invitrogen) and cloned with KpnI and NotI sites included in the sense and antisense primers, respectively. The green fluorescent protein (GFP) control transfections were performed with a plasmid consisting of the XhoI-XbaI fragment from mCD8-GFP18 (gift from D. Schmucker, Dana-Farber Cancer Institute) cloned into pcDNA3. LILRB1 5′-untranslated region (UTR)–GFP fusion constructs were made by cloning the 5′-UTRs from BC015731 and AF283985 upstream of GFP in pmaxGFP (Lonza) with the use of the KpnI and NheI sites. Mutation of sequences within LILRB1 exon 1 was performed on pcDNA3-LILRB1 full-length plasmid with the use of the QuikChange II XL site-directed mutagenesis kit, as described by the manufacturer (Stratagene). Luciferase reporter constructs were generated by cloning LILRB1 promoter sequences into NheI and XhoI sites of pGL4.10 (Promega). All plasmids were confirmed by sequencing (Agencourt) and doubly purified from bacterial culture by HiSpeed Plasmid Maxi Kit (QIAGEN) followed by QIAquick PCR Purification Kit (QIAGEN) before transfection.
Transfection of human PBMCs
Freshly isolated PBMCs or AutoMACS-purified monocytes were transfected with the use of the Nucleofector II (Lonza) as described by the manufacturer. The transfection program used for T cells was V-24 and for monocytes it was Y-01. Cells were stained for analysis by flow cytometry, or processed for luciferase reporter assays, 24 hours after transfection. Plasmid DNA (2 μg) was used for all transfections except for GFP cotransfections for which 2 μg of LILRB1 cDNA plasmid was combined with 1 μg of GFP plasmid, and luciferase reporter assays for which 2.5 μg of DNA was used.
5′-RACE
5′-rapid amplification of CDNA ends (RACE) analysis was performed on total RNA from 4 million cells with the use of the Invitrogen system as described by the manufacturer. Briefly, cDNA was synthesized from total RNA with the use of an LILRB1-specific antisense primer (GSP1). A poly-cytidine tag was added to the 3′-end of cDNA with the enzyme TdT. PCR was performed on tagged and nontagged cDNA with the use of a tag-specific primer and a second nested LILRB1 primer (GSP2). Amplified products were reamplified in a second nested PCR with the use of a third LILRB1 primer (GSP3). Products from the second nested PCR were cloned into pCRII-TOPO (Invitrogen) and sequenced (Agencourt). GSP3 and a fourth LILRB1 primer binding within exon 3 (GSP4) was used for control PCR for total LILRB1 cDNA.
Luciferase reporter assay
Primary cells were transfected, as describe in “Transfection of human PBMCs,” with a DNA mixture containing 0.5 μg of pRL-SV40 vector and 2.0 μg of either the basic pGL4.10 vector or an LILRB1-pGL4.10 construct. Twenty-four hours after transfection, cells were processed and analyzed with the Dual-Reporter Assay System (Promega) read on a TD-20/20 luminometer (Turner Designs).
Statistics
For all comparisons, an analysis of variance with post hoc Tukey test was performed with the use of SigmaStat 3.0 software (Systat Software).
Primers
For a list of the primers used, please see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
CD85j expression differs among peripheral B cells, T cells, NK cells, and monocytes
CD85j is widely expressed within the hematopoietic system, but its expression pattern differs considerably among cell types (Figure 1A-B).1,2,7,8 Expression on monocytes is essentially ubiquitous. By contrast, CD85j is expressed on a subset of NK cells and T cells.1,2 As we16 and others8 have shown, CD85j expression within the T-cell compartment is far more likely for CD8 T cells than for CD4 T cells and is restricted to memory cells and, in particular, CD45RA+ effector cells (Figure 1C). CD85j expression on peripheral B cells is widespread; however, a detailed analysis suggests it too depends on maturation. We examined circulating human transitional B-cell populations that are in the process of completing their maturation after exiting the bone marrow.19,20 Early transitional B cells (T1) express CD85j in lesser frequency than late transitional B cells (T2; Figure 1D). These results are consistent with an earlier report that developing B cells within the bone marrow acquire CD85j expression during maturation.21 Among the other peripheral B-cell populations we examined (naive, nonswitched memory, and switch memory B cells as reviewed by Sanz et al22 ), CD85j was ubiquitously expressed at similar levels (Figure 1E).
Differential CD85j expression on human PBMC subsets. (A) Representative histograms showing CD85j expression on NK cells, CD4 T cells, CD8 T cells, B cells, and monocytes. (B) Frequencies of CD85j+ cells within subpopulations are shown as mean + SD of 12 donors. (C) CD85j expression on CD8 T-cell subsets. Histograms (bottom) show CD85j expression on the CD8 T-cell subpopulations based on the expression of CCR7 and CD45RA as shown in the dot plot at top. Numbers in the histograms (bottom) indicate the percentage of cells expressing CD85j. (D) CD85j expression on peripheral transitional B cells. Histograms (right) show CD85j expression on the populations defined in density plot to the left (gated on CD19+CD20+ cells). T1 indicates early transitional B cells; and T2, late transitional B cells. (E) CD85j expression on peripheral naive and memory B cells. Histograms (right) show CD85j expression on the populations defined by the quadrants in dot plot to the left (gated on CD19+ cells). (F) CD85j cell surface densities (MFI indicates mean fluorescence intensity) by flow cytometry on the CD85j+ cells are shown as mean MFI + SD of 12 donors. (G) CD85j expression on monocytes and B cells are compared by Western blot. Blot is representative of comparisons of 4 donors.
Differential CD85j expression on human PBMC subsets. (A) Representative histograms showing CD85j expression on NK cells, CD4 T cells, CD8 T cells, B cells, and monocytes. (B) Frequencies of CD85j+ cells within subpopulations are shown as mean + SD of 12 donors. (C) CD85j expression on CD8 T-cell subsets. Histograms (bottom) show CD85j expression on the CD8 T-cell subpopulations based on the expression of CCR7 and CD45RA as shown in the dot plot at top. Numbers in the histograms (bottom) indicate the percentage of cells expressing CD85j. (D) CD85j expression on peripheral transitional B cells. Histograms (right) show CD85j expression on the populations defined in density plot to the left (gated on CD19+CD20+ cells). T1 indicates early transitional B cells; and T2, late transitional B cells. (E) CD85j expression on peripheral naive and memory B cells. Histograms (right) show CD85j expression on the populations defined by the quadrants in dot plot to the left (gated on CD19+ cells). (F) CD85j cell surface densities (MFI indicates mean fluorescence intensity) by flow cytometry on the CD85j+ cells are shown as mean MFI + SD of 12 donors. (G) CD85j expression on monocytes and B cells are compared by Western blot. Blot is representative of comparisons of 4 donors.
In addition to distinct CD85j expression patterns, PBMC subpopulations express characteristic levels of CD85j protein. Monocytes express nearly 4 times as much CD85j protein as B cells (Figure 1F-G). NK cells and T cells express CD85j to a lesser degree than B cells, but levels are similar among NK cells and CD4 and CD8 T cells that are CD85j+ (Figure 1F).
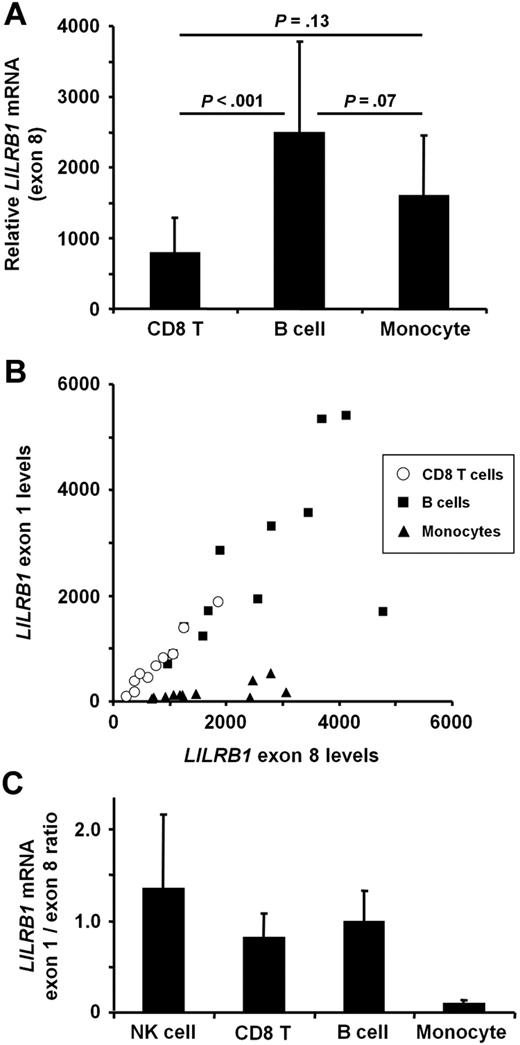
We hypothesized that the distinct CD85j expression profiles within PBMC subsets result from differences in transcription of the LILRB1 gene. As expected, quantitative real-time (qRT)–PCR of exon 8 within the coding region showed LILRB1 transcripts to be lower in CD8 T cells than in B cells and monocytes (Figure 2A). Surprisingly, B cells and monocytes express similar levels of LILRB1 transcripts (Figure 2A) despite a considerable difference at the protein level (Figure 1F-G).
LILRB1 mRNA levels does not account for different subset-specific CD85j protein expression. LILRB1 transcripts were quantified by qRT-PCR in RNA from magnetic bead–separated CD8 T cells, CD19 B cells, and CD14 monocytes, and NK cells were purified by negative selection. (A) Results for a primer set within exon 8 are shown as mean transcript numbers + SDs of 10 to 12 donors per group relative to 2 × 105 β-actin copies. Compared with protein expression, transcript numbers in monocytes were disproportionately low. (B) LILRB1 transcripts were compared for exon 1 to 3 and exon 8 sequences. Results are shown as a scatter plot for CD8 T cells, B cells, and monocytes. (C) Transcript comparisons are quantified as the ratio of LILRB1 exon 1 to 3 to exon 8 copies. Results are shown as mean + SD of 10 to 12 donors per group. NK-cell data are from 3 donors.
LILRB1 mRNA levels does not account for different subset-specific CD85j protein expression. LILRB1 transcripts were quantified by qRT-PCR in RNA from magnetic bead–separated CD8 T cells, CD19 B cells, and CD14 monocytes, and NK cells were purified by negative selection. (A) Results for a primer set within exon 8 are shown as mean transcript numbers + SDs of 10 to 12 donors per group relative to 2 × 105 β-actin copies. Compared with protein expression, transcript numbers in monocytes were disproportionately low. (B) LILRB1 transcripts were compared for exon 1 to 3 and exon 8 sequences. Results are shown as a scatter plot for CD8 T cells, B cells, and monocytes. (C) Transcript comparisons are quantified as the ratio of LILRB1 exon 1 to 3 to exon 8 copies. Results are shown as mean + SD of 10 to 12 donors per group. NK-cell data are from 3 donors.
LILRB1 transcripts in B cells, CD8 T cells, and NK cells contain 5′-UTR sequences that are absent in monocytes
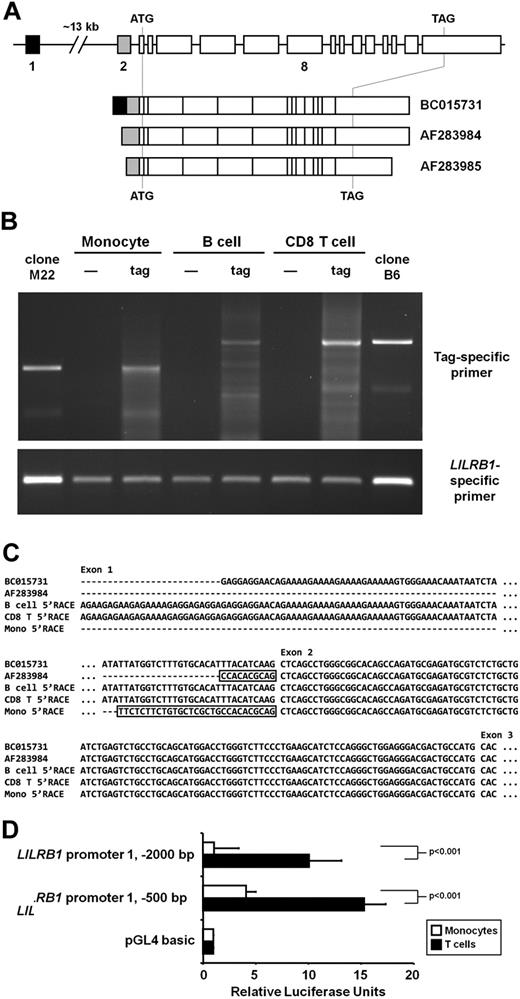
An examination of LILRB1 cDNA sequences submitted to the National Center for Biotechnology Information (NCBI) showed several transcript variants, some of which differ within the 5′-UTRs (see Figure 3A for a schematic). We hypothesized that B cells and monocytes express LILRB1 transcripts with distinct 5′-UTRs that affect CD85j translation. Indeed, submitted sequences differ in the number of potential start codons positioned upstream of the accepted start codon within exon 3 (eg, 7 ATG sequences in BC01573117 compared with 4 ATG sequences in AF28398423 ). Because the primer pair we used to quantify LILRB1 transcripts is specific for a sequence within the coding region, we designed an additional primer pair (subsequently referred to as “exon 1 primers”) targeting the 5′-most exon that is present in some submitted sequences (such as BC015731) but not in others (such as AF283984). The antisense primer of this pair binds to a sequence within exon 3 that is present in all LILRB1 transcripts. To assess whether B cells and monocytes express similar 5′-UTRs, we performed qRT-PCR with the use of both exon 1 primers and exon 8 primers simultaneously on the same cDNA sample. B cells and monocytes yielded strikingly different results. Comparison of LILRB1 exon 1 and exon 8 transcript numbers suggested that most LILRB1 transcripts in B cells included both exons (Figure 2B-C). Monocyte values, however, fall on a much shallower slope, implying that most LILRB1 transcripts from monocytes do not include exon 1 (Figure 2B-C). The same qRT-PCR test performed on cDNA from CD8 T cells and NK cells showed a ratio of LILRB1 exon 1 to exon 8 levels similar to that seen in B cells (Figure 2B-C) and significantly larger than the ratio found in monocytes (P < .001).
LILRB1 transcription initiation sites in CD8 T cells, B cells, and monocytes. (A) Schematics of the LILRB1 locus on human chromosome 19 and selected mRNA sequences currently posted to NCBI. Lines are introns, and boxes are exons (roughly to scale). Exons 1 and 2 are separated by approximately 13 kb as indicated by a gap. The LILRB1 coding region is flanked by a start (ATG) and stop (TAG) codon. (B) PCR of 5′-RACE products generated from monocyte, B-cell, and CD8 T-cell RNA isolated from magnetic bead–separated cells. The enzyme TdT was excluded (—) or included (tag) when tagging the 5′ end of LILRB1 cDNA. PCR products were amplified with a tag-specific sense primer and an LILRB1-specific antisense primer (top) or LILRB1-specific sense and antisense primers (bottom). Templates for lanes indicated by clone M22 and clone B6 were the 5′-RACE clone from monocytes and B cells, respectively, used to obtain the sequences shown in panel C. (C) Sequencing of 5′-RACE products. Total PCR products were TOPO-cloned and sequenced. The sequence corresponding to the major tag-specific band for each cell type in panel B is compared with LILRB1 sequences BC015731 and AF283984. Boxed sequences indicate contiguous sequences upstream of exon 2 in the genome. (D) Luciferase reporter assay of sequences upstream of LILRB1 exon 1. The 500-bp and 2000-bp sequences found immediately upstream of LILRB1 exon 1 on chromosome 19 were amplified and placed upstream of the firefly luciferase ORF of pGL4.10. Freshly isolated primary T cells and monocytes were cotransfected with the reporter constructs and a control Renilla luciferase expression vector. Data represent firefly luciferase activity, normalized to Renilla luciferase activity, relative to that seen with the promoter-less basic pGL4.10 vector. Data from 3 donors are represented as mean + SD.
LILRB1 transcription initiation sites in CD8 T cells, B cells, and monocytes. (A) Schematics of the LILRB1 locus on human chromosome 19 and selected mRNA sequences currently posted to NCBI. Lines are introns, and boxes are exons (roughly to scale). Exons 1 and 2 are separated by approximately 13 kb as indicated by a gap. The LILRB1 coding region is flanked by a start (ATG) and stop (TAG) codon. (B) PCR of 5′-RACE products generated from monocyte, B-cell, and CD8 T-cell RNA isolated from magnetic bead–separated cells. The enzyme TdT was excluded (—) or included (tag) when tagging the 5′ end of LILRB1 cDNA. PCR products were amplified with a tag-specific sense primer and an LILRB1-specific antisense primer (top) or LILRB1-specific sense and antisense primers (bottom). Templates for lanes indicated by clone M22 and clone B6 were the 5′-RACE clone from monocytes and B cells, respectively, used to obtain the sequences shown in panel C. (C) Sequencing of 5′-RACE products. Total PCR products were TOPO-cloned and sequenced. The sequence corresponding to the major tag-specific band for each cell type in panel B is compared with LILRB1 sequences BC015731 and AF283984. Boxed sequences indicate contiguous sequences upstream of exon 2 in the genome. (D) Luciferase reporter assay of sequences upstream of LILRB1 exon 1. The 500-bp and 2000-bp sequences found immediately upstream of LILRB1 exon 1 on chromosome 19 were amplified and placed upstream of the firefly luciferase ORF of pGL4.10. Freshly isolated primary T cells and monocytes were cotransfected with the reporter constructs and a control Renilla luciferase expression vector. Data represent firefly luciferase activity, normalized to Renilla luciferase activity, relative to that seen with the promoter-less basic pGL4.10 vector. Data from 3 donors are represented as mean + SD.
Lymphocytes initiate LILRB1 transcription from a site 13 kilobases upstream of the main site used by monocytes
The results in “LILRB1 transcripts in B cells, CD8 T cells, and NK cells contain 5′-UTR sequences that are absent in monocytes,” when considered together with the genomic structure of the LILRB1 gene, imply that lymphocytes and monocytes do not use the same promoter to transcribe LILRB1. Figure 3A depicts the genomic organization of the LILRB1 gene and indicates the corresponding exons included in LILRB1 transcript sequences submitted to NCBI. As is shown, exon 1 is separated from the other LILRB1 exons by a 13-kilobase (kb) intron. The schematic also shows that only some of the published LILRB1 transcripts include exon 1, whereas others begin with exon 2. Little is known about the mechanisms regulating LILRB1 transcription. The only report of an LILRB1 promoter analysis examined a roughly 1-kb region just upstream of what is called exon 2 in Figure 3A.24 Therefore, we reasoned that monocytes, whose cDNA yielded very low exon 1 signals by qRT-PCR, may initiate LILRB1 transcription from the described promoter upstream of exon 2, whereas lymphocytes initiate LILRB1 transcription from an undescribed promoter upstream of exon 1. To further address this question and identify LILRB1 transcription initiation sites, we performed 5′-RACE analysis of cDNA from monocytes, B cells, and CD8 T cells. As shown in Figure 3B, B cells and CD8 T cells share a major 5′-RACE product that is larger than the major product from monocytes. Sequencing of products from all 3 cell types confirmed that B cells and CD8 T cells initiate LILRB1 transcription with exon 1, whereas monocyte transcripts begin with exon 2 (Figure 3C). The major B-cell and CD8 T-cell product corresponds to a transcription initiation site 26 nt upstream of the first nucleotide of submitted LILRB1 cDNAs that begin with exon 1 (BC015731). Conversely, the major product in monocytes identifies a transcription initiation site 19 nt upstream of submitted LILRB1 sequences beginning with exon 2 (AF283984), and 18 nt upstream of the transcription initiation site identified by Nakajima et al24 The additional upstream nucleotides we identified all correspond to contiguous genomic sequences.
To further support our assertion that lymphocytes and monocytes use distinct promoters to drive CD85j expression, we generated luciferase reporter constructs with genomic sequences upstream of LILRB1 exon 1 and compared activity of these constructs in T cells and monocytes. Indeed, these sequences exhibited strong activity in transfected T cells but only weak activity in monocytes (Figure 3D). In T cells, constructs including the 2000-base pair (bp) or 500-bp sequences directly upstream of LILRB1 exon 1 showed 10- and 15-times stronger activity, respectively, than a luciferase construct lacking a promoter. In monocytes, the 500-bp construct showed only modest activity compared with the basic vector, whereas the activity seen with the 2000-bp construct was negligible. These results strongly suggest that a second, yet undescribed, promoter rests 13 kb upstream of the main LILRB1 exon cluster (exons 2-16) and directs LILRB1 transcription in lymphocytes.
Exon 1 sequences inhibit translation of CD85j
Our initial observation that, despite similar transcript levels, peripheral blood monocytes express far more CD85j protein than B cells suggests that CD85j is not translated as efficiently in B cells. We hypothesized that the unique LILRB1 5′-UTR that results from usage of the upstream promoter by B cells contributes to this diminished protein production. To test this hypothesis, we generated expression vectors containing either LILRB1 coding region cDNA alone or including an exon 1–containing 5′-UTR (from BC015731). CD85j protein production by cells transfected with these vectors was assessed by flow cytometry. Because CD85j is expressed by a subset of CD8 T cells and by nearly all B cells and monocytes, these experiments were carried out in CD4 T cells. Indeed, we found that transfection with the 5′-UTR–containing vector resulted in diminished CD85j expression compared with cells transfected with the LILRB1 coding region vector (Figure 4A). Interestingly, transfection with the full-length LILRB1 cDNA (BC015731) resulted in the same diminished expression, suggesting that the 5′-UTR dominates any effect the 3′-UTR may have on CD85j protein levels.
Exon 1 sequences repress CD85j expression. Human PBMCs were transfected with expression constructs containing variousLILRB1 cDNAs and/or GFP. CD85j and GFP expression in CD4 T cells was analyzed 24 hours after transfection by flow cytometry. (A) Histograms and bar graph comparing CD85j expression by full-length (BC015731) cDNA, 5′-UTR plus coding region, and coding region alone. Results are representative of 6 experiments. MFI indicates mean fluorescence intensity. (B) Histograms and bar graph comparing various portions of the LILRB1 5′-UTR plus coding region and coding region alone. Δex1(AF84) and Δex1(AF85) contain the LILRB1 5′-UTR from AF283984 and AF283985 sequences, respectively. Δex1Δex2 begins with exon 3 and continues through the LILRB1 coding region. (C-F) A plasmid expressing GFP was cotransfected along with a LILRB1 5′-UTR plus coding region or coding region alone plasmid. Results are representative of 6 experiments. (C) Representative histograms showing CD85j and GFP expression in cotransfected CD4 T cells. (D) Flow cytometry plot of samples shown in panel C. (E) Graph of the percentage of CD85j+ CD4 T cells in cells cotransfected with GFP. Samples were divided into quintiles based on GFP expression, and means + SDs from 3 transfections were calculated for each quintile. (F) Relative LILRB1 mRNA (exon 8) in cotransfected cells. cDNA was treated with DpnI before real-time PCR to digest plasmid DNA; n = 3 transfections. (G) LILRB1 5′-UTR sequences that include (BC015731) or exclude (AF283985) exon 1 were cloned upstream of the GFP ORF of pmaxGFP. Data showing GFP MFI relative to an unaltered GFP control vector are presented as mean + SD from 3 transfections.
Exon 1 sequences repress CD85j expression. Human PBMCs were transfected with expression constructs containing variousLILRB1 cDNAs and/or GFP. CD85j and GFP expression in CD4 T cells was analyzed 24 hours after transfection by flow cytometry. (A) Histograms and bar graph comparing CD85j expression by full-length (BC015731) cDNA, 5′-UTR plus coding region, and coding region alone. Results are representative of 6 experiments. MFI indicates mean fluorescence intensity. (B) Histograms and bar graph comparing various portions of the LILRB1 5′-UTR plus coding region and coding region alone. Δex1(AF84) and Δex1(AF85) contain the LILRB1 5′-UTR from AF283984 and AF283985 sequences, respectively. Δex1Δex2 begins with exon 3 and continues through the LILRB1 coding region. (C-F) A plasmid expressing GFP was cotransfected along with a LILRB1 5′-UTR plus coding region or coding region alone plasmid. Results are representative of 6 experiments. (C) Representative histograms showing CD85j and GFP expression in cotransfected CD4 T cells. (D) Flow cytometry plot of samples shown in panel C. (E) Graph of the percentage of CD85j+ CD4 T cells in cells cotransfected with GFP. Samples were divided into quintiles based on GFP expression, and means + SDs from 3 transfections were calculated for each quintile. (F) Relative LILRB1 mRNA (exon 8) in cotransfected cells. cDNA was treated with DpnI before real-time PCR to digest plasmid DNA; n = 3 transfections. (G) LILRB1 5′-UTR sequences that include (BC015731) or exclude (AF283985) exon 1 were cloned upstream of the GFP ORF of pmaxGFP. Data showing GFP MFI relative to an unaltered GFP control vector are presented as mean + SD from 3 transfections.
To address the hypothesis that distinct 5′-UTRs in B cells and monocytes account for their differences in protein expression, CD85j protein expression from transcripts containing these distinct 5′-UTRs were compared. We generated a series of vectors with LILRB1 coding region cDNA linked to various portions of the 5′-UTR. These vectors include the full 5′-UTR (containing exon 1) and 5′-UTRs from AF283984 and AF283985,23 both of which begin within exon 2, and a truncated 5′-UTR that begins with exon 3. Analysis of cells transfected with these vectors shows that exon 1 is responsible for the diminished protein expression conferred by the full-length LILRB1 5′-UTR (Figure 4B). This exon 1 effect was reliably observed by transfection of cells from many different donors; even transfecting double the amount of exon 1–containing DNA failed to reach CD85j levels seen with vectors lacking exon 1 (data not shown). Cotransfection with a separate GFP vector suggests that cells receiving the full LILRB1 5′-UTR plus coding region construct are as efficiently transfected as cells receiving the LILRB1 coding region construct (Figure 4C). In addition, in cells cotransfected with the 5′-UTR plus coding region construct, only those cells with high GFP expression, reflecting high delivery of plasmid DNA, exhibited high CD85j positivity (Figure 4D-E). In contrast, cells receiving LILRB1 coding region constructs begin to express CD85j even before GFP is detectible. qRT-PCR analysis from these cells suggests that cotransfected cells transcribe similar levels of LILRB1 mRNA despite the differences in protein expression (Figure 4F). Furthermore, the poor protein expression conferred by the LILRB1 5′-UTR can be transferred to another protein (GFP) and, in this context, is also exon 1 dependent (Figure 4G).
The sequence conferring translational repression of CD85j is mapped to 30 nt of exon 1
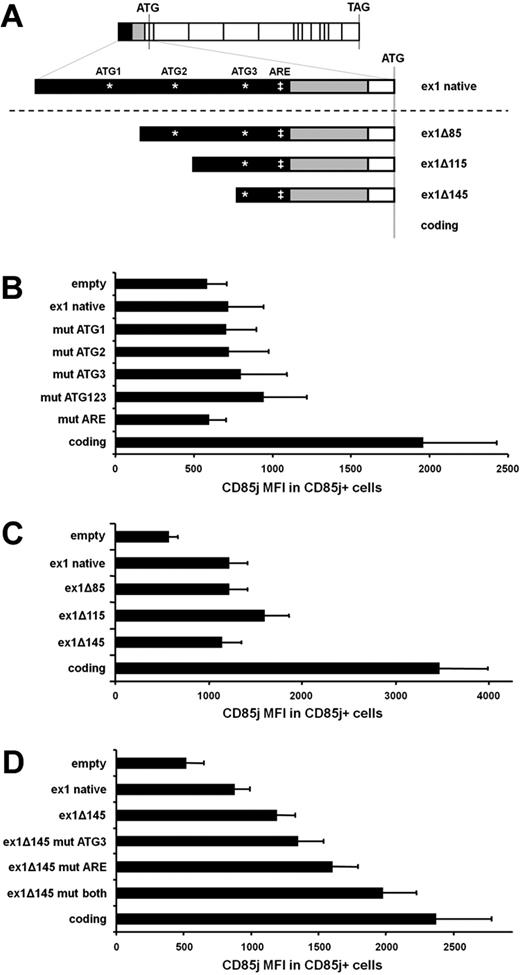
To further define the sequences within LILRB1 exon 1 that may be preventing full protein expression, we made a series of 5′-UTR plus coding region constructs containing alterations in the exon 1 sequence (Figure 5A). First, we examined the possibility that ATG sequences within exon 1 might act as false start codons, thereby interfering with ribosomal binding to the true start codon within exon 3. By site-directed mutagenesis, we destroyed each of the 3 ATG sequences within exon 1 by changing the T to an A. When constructs containing just one of these changes were transfected, CD85j expression did not improve compared with constructs containing the unaltered exon 1 (Figure 5B). To prevent all false translation initiations within exon 1, a construct was made in which all ATGs were destroyed. Transfection with this construct yielded only slightly higher CD85j expression (Figure 5B).
The distal 30 nt of LILRB1 exon 1 accounts for the poor protein expression by LILRB1 exon 1–containing constructs. (A) Schematic of constructs used to isolate the region within LILRB1 exon 1 responsible for poor protein expression. (B-D) Constructs used for transfections contained the LILRB1 coding region preceded by the portions of the LILRB1 5′-UTR indicated here. Sites in exon 1 that, in some constructs (B,D), were mutated are indicated by an asterisk (*) for ATG sequences and a double-dagger (‡) for the ARE sequence. (B) Graph comparing LILRB1 5′-UTR plus coding region constructs. Mutations were introduced into an LILRB1 5′-UTR construct containing the full-length exon 1. For mut ATG1, ATG2, ATG3, and ATG123, the 3 potential start codons found in LILRB1 exon 1 were changed to AAG. For mut ARE, the ATTTA sequence was changed to ATCTA; n = 4 transfections. (C) Graph comparing LILRB1 5′UTR plus coding region constructs. Progressively truncated LILRB1 exon 1 constructs were compared with an LILRB1 coding region construct; n = 3 transfections. (D) Graph comparing LILRB1 5′-UTR plus coding region constructs. The ATG and ARE sites in the ex1Δ145 construct were mutated, and CD85j expression was compared with the unmutated construct and an LILRB1 coding region construct; n = 5 transfections.
The distal 30 nt of LILRB1 exon 1 accounts for the poor protein expression by LILRB1 exon 1–containing constructs. (A) Schematic of constructs used to isolate the region within LILRB1 exon 1 responsible for poor protein expression. (B-D) Constructs used for transfections contained the LILRB1 coding region preceded by the portions of the LILRB1 5′-UTR indicated here. Sites in exon 1 that, in some constructs (B,D), were mutated are indicated by an asterisk (*) for ATG sequences and a double-dagger (‡) for the ARE sequence. (B) Graph comparing LILRB1 5′-UTR plus coding region constructs. Mutations were introduced into an LILRB1 5′-UTR construct containing the full-length exon 1. For mut ATG1, ATG2, ATG3, and ATG123, the 3 potential start codons found in LILRB1 exon 1 were changed to AAG. For mut ARE, the ATTTA sequence was changed to ATCTA; n = 4 transfections. (C) Graph comparing LILRB1 5′UTR plus coding region constructs. Progressively truncated LILRB1 exon 1 constructs were compared with an LILRB1 coding region construct; n = 3 transfections. (D) Graph comparing LILRB1 5′-UTR plus coding region constructs. The ATG and ARE sites in the ex1Δ145 construct were mutated, and CD85j expression was compared with the unmutated construct and an LILRB1 coding region construct; n = 5 transfections.
Next, we made a series of constructs with progressive truncation of exon 1. These constructs showed that as little as 30 nt of exon 1 sequence can prevent the full CD85j expression seen when cells are transfected with coding region constructs (Figure 5C). This 30-nt sequence contains 1 of the 3 ATGs found in exon 1 as well as the sequence ATTTA, a motif found in so-called AU-rich elements (ARE) that is known to mediate translational repression in other genes. We generated constructs lacking one or both of these sequences and tested CD85j expression after transfection. Similar to our findings with ATG mutants within the entire exon 1, disruption of the ATG within ex1Δ145 resulted in a slight enhancement of CD85j expression (Figure 5D). Disruption of the ARE sequence (ATTTA > ATCTA) yielded an even higher CD85j expression. When both the ATG and the ARE sequences were destroyed, CD85j expression nearly matched the strong expression seen when transfecting the LILRB1 coding region alone. However, when we disrupted the ARE sequence alone within the complete exon 1, we found no enhancement of CD85j expression (Figure 5B). The observed effect in the truncated constructs is in contrast to the negligible enhancement of the same mutations in the context of the entire exon 1, suggesting that false start codons and/or the ATTTA sequence only play an indirect role in the poor CD85j protein expression.
Discussion
Within the hematopoietic system, CD85j is expressed to various degrees by most cell types. In this report, we provide evidence that CD85j expression is regulated in a lineage-specific manner, and we identify a novel promoter used by lymphocytes, but not monocytes, that lies 13 kb upstream of the monocyte promoter and the main LILRB1 exon cluster on human chromosome 19. Use of the lymphocyte promoter results in an additional exon within the 5′-UTR that is absent in transcripts originating from the monocyte promoter. We show that transcripts containing this first exon do not efficiently translate CD85j protein compared with transcripts beginning with exon 2. Promoter choice, combined with translational repression, accounts for cell-specific differences in CD85j expression that are responsible for the context-dependent differences in CD85j function.
Our report suggests CD85j expression is regulated in a lineage-specific manner, whereby lymphocytes strongly favor CD85j expression from the upstream promoter and monocytes exclusively use the downstream promoter. For CD8 T cells, CD85j is well positioned to interfere with TCR activation by competing for CD8 binding to MHC class I25 and recruiting phosphatases to the synapse. For NK cells, CD85j is one of many inhibitory receptors that may be expressed to detect MHC class I on other cells.26 In these settings, the upstream promoter may be more amenable to the transcriptional apparatus needed to restrict CD85j expression to a subset of cells and the translational inefficiency of the resulting transcript may allow tighter regulation of CD85j protein levels.
Although B cells, CD8 T cells, and NK cells share a lineage and use the same LILRB1 promoter, B cells and monocytes use CD85j in a more similar functional context. First, whereas only a tightly defined subset of CD8 T cells expresses CD85j, it is ubiquitous on mature B cells and monocytes. Second, the defining operations of neither B cells nor monocytes directly involve MHC class I interactions. Naive B cells survey their surroundings by expressing many copies of a single rearranged surface immunoglobulin and are activated through combined signals delivered by (1) the antigen-crosslinked B-cell receptor (surface immunoglobulin plus Igα and Igβ signaling domains) and (2) a CD4 T cell–engaging MHC class II/antigen peptide complexes on the B-cell surface.27 Peripheral blood monocytes use surface receptors to sense chemokines and endothelial cell changes signaling inflammation,28 and, after extravasation into the tissue, additional interactions and chemical mediators drive their differentiation into phagocytic effector cells such as macrophages and DCs.29 B cell– and monocyte-activating signals require kinase activity that can be influenced by CD85j-recruited phosphatases.7,30-33 The MHC class I that CD85j encounters during these events exists either on the B cell or monocyte itself (cis) or on the surface of surrounding cells (trans), but not within the primary activation interface. CD85j functioning in a dispersed distribution on B cells and monocytes probably increases the threshold required to deliver activating signals by shifting the intracellular kinase/phosphatase balance rather than directly interfering with activating signals. Indeed, CD85j ligation on monocytes during in vitro DC generation dramatically affects the phenotype of resulting DCs that lack many characteristic surface markers and respond poorly to lipopolysaccharide stimulation.15
Although all mature B cells and monocytes express CD85j, a consequence of distinct promoter usage by B cells and monocytes is higher CD85j protein levels in monocytes. It is probable that the distinct roles these cell types play are best served by different CD85j levels. Monocytes are innate immune cells and lack the antigen specificity that defines B cells. When activated in the periphery, their effector functions act broadly and destructively toward surrounding tissues. An elevated activation threshold provided by high CD85j expression reserves highly damaging responses to all but the strongest inflammatory scenarios. However, mature B cells result from a meticulous process of receptor gene rearrangement and negative selection to ensure each B cell is functional and self-tolerant.34 These cells continuously circulate, awaiting antigen encounter. CD85j levels on B cells may help to establish a balance such that responses to self-antigens are avoided while allowing the subtle survival signals necessary to continue circulating.35 Interestingly, our finding that circulating transitional B cells lack CD85j expression suggests CD85j does not interfere with the signals required to establish the naive B-cell repertoire but becomes available to influence survival and activation signals on maturation.
Before this report, LILRB1 was assumed to have a single promoter, upstream of the main exon cluster. Using reporter constructs in cell lines, Nakajima et al24 demonstrated that this exon 2–proximal LILRB1 promoter is highly active in the monocyte-like THP-1 cell line and depends on PU.1 and Sp1 transcription factors. However, promoter activity in Jurkat cells, a T cell-like line, was weak compared with THP-1 cells. These results are consistent with our findings that primary T cells and monocytes use distinct LILRB1 promoters. Moreover, our findings from promoter reporter assays are reciprocal to those of Nakajima et al.24 Namely, exon 1-proximal promoter sequences are strongly active in T cells but not monocytes.
The upstream LILRB1 promoter used by lymphocytes serves as an alternative to the promoter used by monocytes. It is estimated that 30% to 50% of all human genes have alternative, and often distant, promoters.36,37 Some alternative promoters may be more active than others in a certain cell type and the hierarchy of promoter activities may differ among cell types.38,39 In other cases, distinct promoters yield unique 5′-UTRs that affect the character and/or quantity of the translated protein. When additional exons transcribed from alternative promoters contain an ATG sequence, it is possible the resulting protein will contain an altered N-terminus or even a completely new protein.40 Conversely, distinct 5′-UTRs may not alter the protein product but, rather, may affect transcript stability or translational efficiency.41 Our findings suggest use of the distant upstream LILRB1 promoter is cell type specific and results in inefficient CD85j protein expression without altering the resulting amino acid sequence. Similar to genes such as CDKN2C,42 we find the upstream LILRB1 promoter yields a 5′-UTR that profoundly effects protein expression in primary cells. Cells using the upstream LILRB1 promoter (such as B cells) express far less CD85j protein compared with cells using the downstream promoter (such as monocytes) despite similar mRNA levels.
We isolated the region responsible for poor CD85j protein expression by lymphocytes to the last 30 nt of exon 1. This region contains the last of 3 ATGs found in exon 1 and an ARE, a motif known to mediate translational repression by recruitment of RNA-binding proteins.43 Although AREs typically contain several repeats of this motif, they can function as a single pentamer.44 We mutated the ATG and ARE motif within the 30-nt sequence and found an improvement in CD85j expression. Expression was strongest when both mutations were present in the ex1Δ145 construct. Mutation of these elements in the context of the full exon 1 had no or only a modest effect on CD85j levels, suggesting that no single element is responsible for exon 1–mediated translational repression of CD85j expression and that the truncated exon 1 construct provides a context, such as a unique mRNA conformation, in which mutations are more potent than when the full exon 1 is present.
This study addressed steady-state CD85j expression in primary cells. Future studies might focus on how CD85j is acquired or lost during activation and differentiation states. For example, CD85j levels are known to increase during in vitro differentiation from monocytes into DCs.15 Conversely, DCs are known to down-regulate CD85j on activation.45 As suggested by the results in Figure 1, B cells acquire CD85j expression during the differentiation steps leading from transitional to mature B cells. Perhaps the most interesting scenario is the de novo acquisition of CD85j expression by CD8 T cells with advancing age, a phenomenon that can be mimicked by repeated stimulation cycles in vitro.16 This has important implications, given the pivotal role CD8 T cells play in infections, cancer, and autoimmunity, each of which disproportionately affect the elderly. It remains to be seen whether a given cell type can simultaneously activate and/or switch between, the 2 LILRB1 promoters or if expression is chiefly controlled by increased transcriptional activity at a single promoter. The disparity between translational efficiencies between the 2 promoters implies that even a small shift could profoundly affect CD85j protein levels. Potential therapeutic interventions, such as turning CD85j expression on in tumor cells or off in T cells, will require a thorough understanding of the mechanisms governing LILRB1 promoter choice and activity in a variety of cell types and settings.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Institutes of Health (grants RO1AR42527, RO1AR41974, RO1AI44142, U19AI57266, RO1EY11916, RO1AG15043, and T32GM008169-23, C.M.W. and J.J.G.; grant T32GM008169-23, D.L.L.) and by the American Foundation for Aging Research (D.L.L).
National Institutes of Health
Authorship
Contribution: D.L.L. designed and performed the research, analyzed data, and wrote the paper; and C.M.W. and J.J.G designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jörg J. Goronzy, Department of Medicine, Division of Immunology and Rheumatology, Stanford University School of Medicine, CCSR Bldg, Rm 2215, 269 Campus Dr West, Stanford, CA 94305-5166; e-mail: jgoronzy@stanford.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal