Abstract
DNA hypermethylation of the p15INK4b tumor suppressor gene is commonly observed in acute myeloid leukemia (AML). Repressive histone modifications and their associated binding proteins have been implicated in the regulation of DNA methylation and the transcriptional repression of genes with DNA methylation. We have used high-density chromatin immunoprecipitation-on-chip to determine the histone modifications that normally regulate p15INK4b expression in AML cells and how these marks are altered in cells that have p15INK4b DNA methylation. In AML patient blasts without p15INK4b DNA methylation, a bivalent pattern of active (H3K4me3) and repressive (H3K27me3) modifications exist at the p15INK4b promoter. AML patient blasts with p15INK4b DNA methylation lose H3K4me3 at p15INK4b and become exclusively marked by H3K27me3. H3K27me3, as well as EZH2, extends throughout p14ARF and p16INK4a, indicating that polycomb repression of p15INK4b is a common feature in all AML blasts irrespective of the DNA methylation status of the gene. Reactivation of p15INK4b expression in AML cell lines and patient blasts using 5-aza-2′-deoxycytidine (decitabine) and trichostatin A increased H3K4me3 and maintained H3K27me3 enrichment at p15INK4b. These data indicate that AML cells with p15INK4b DNA methylation have an altered histone methylation pattern compared with unmethylated samples and that these changes are reversible by epigenetic drugs.
Introduction
Acute myeloid leukemia (AML) is characterized by a series of genetic and epigenetic alterations that disrupt the differentiation, proliferation, and survival of myeloid progenitor cells. One of the most prevalent epigenetic alterations in AML is the transcriptional silencing of the p15INK4b (CDKN2b) tumor suppressor gene by DNA hypermethylation, reported in up to 80% of AML patients.1,2 p15INK4b is a regulator of cell-cycle arrest in the G1 phase of the cell cycle through inhibition of cyclin-dependent kinase 4 (CDK4) and cyclin-dependent kinase 6 (CDK6).3 In myeloid cells, this role is evident during the maturation of myeloid progenitor cells, in which p15INK4b is up-regulated by cytokines in association with cell-cycle arrest.4,5 An additional role for p15INK4b in early-stage myelopoiesis has recently been revealed in p15INK4b knockout mice.6 In this system, loss of p15INK4b results in increased production of granulocyte-macrophage progenitors.
p15INK4b and neighboring tumor suppressor genes p14ARF and p16INK4a compose a 41-kb locus located on chromosome 9p21, which is critical for the regulation of cell survival and proliferation. Although different combinations of all 3 genes are altered in a wide spectrum of human neoplasias, in AML, DNA methylation of p15INK4b occurs independently of genetic or epigenetic changes at p14ARF and p16INK4a.1,2 The high incidence of p15INK4b DNA methylation in AML and its role in cell-cycle control make it an attractive target for reactivation by epigenetic therapies, including DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors.7,8
Histone modifications play an important role in the gene expression patterns at the INK4b-ARF-INK4a locus.9 The signatures of polycomb gene repression, Bmi-1 and enhancer of zeste-2 (EZH2) binding as well as the trimethylation of histone H3 lysine 27 (H3K27me3) modification, have been found at p14ARF and p16INK4a in fibroblasts,10 malignant rhabdoid tumor cell lines,11 and embryonic stem cells.12 In fibroblasts, polycomb binding at p14ARF and p16INK4a appears critical in maintaining gene repression before signals for oncogene-induced senescence.10,13 Compared with fibroblasts, CD34+ and CD133+ hematopoietic progenitor/stem cells have H3K27me3 spread across a broader region of the locus encompassing p15INK4b.10,14 This suggests that polycomb repression of p15INK4b is a common regulatory mechanism in some hematopoietic cell lineages. Overexpression of Bmi-1 or EZH2 has been observed in cancer, including AML, and may lead to abnormal repression of genes regulated by polycomb complexes.15 Thus, the disruption of pathways that control the distribution of histone modifications may represent a mechanism by which tumor suppressor genes are aberrantly repressed in cancer.
DNA methylation and histone modifications can regulate the distribution of each other and collaborate to solidify repression at target genes. DNA methylation of promoters can induce repressive histone modifications through recruitment of methyl-binding proteins in association with histone-modifying enzymes.16 Alternatively, H3K27me3 and H3K9me3 have been implicated in the initiation of DNA methylation. Heterochromatin protein-1 and EZH2, which bind H3K9me3 and H3K27me3, respectively, interact with DNMTs and may recruit them to CpG islands.16
Based on these observations, we were interested in examining the association of histone modifications with p15INK4b DNA methylation in AML. We used high-resolution chromatin immunoprecipitation (ChIP)–on-chip to view a large area surrounding and including p15INK4b. This approach allowed us to assay histone modifications not only at p15INK4b, but throughout the entire INK4b-ARF-INK4a region. We demonstrate here that AML cells with p15INK4b DNA methylation have lower enrichment of the activation-associated H3K4me3 modification at the p15INK4b promoter compared with unmethylated samples. Although low levels of H3K9me3 were observed in all AML samples, another repressive modification, H3K27me3, is found at p15INK4b as well as p14ARF and p16INK4a in both samples with p15INK4b DNA methylation as well as those in which the gene is unmethylated. Thus, in AML samples without p15INK4b DNA methylation, p15INK4b is in a bivalent H3K27me3/H3K4me3 pattern, which has been described for a subset of developmentally regulated genes in embryonic stem cells.16,17 The presence of H3K27me3 and the binding of EZH2 over this region suggest that polycomb repression is a common feature at p15INK4b in AML cells. Our data indicate that loss of H3K4me3 can be reversed using epigenetic therapies that reactivate p15INK4b expression. Interestingly, this reversal is associated with retention of H3K27me3 enrichment, therefore returning p15INK4b to a bivalent state.
Methods
Cell lines and AML patient samples
All cell lines were obtained from ATCC. HL-60, KG-1, KG-1a, and U-937 cells were grown in RPMI 1640 containing 10% fetal bovine serum (FBS) and penicillin-streptomycin (Invitrogen). Kasumi-1 cells were grown in RPMI 1640 with 20% FBS and penicillin-streptomycin. AML-193 cells were maintained in Iscove modified Dulbecco medium with 20% FBS, 2 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor, and 2 ng/mL recombinant human interleukin-3 (R&D Systems).
AML specimens (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were obtained from Johns Hopkins University School of Medicine as part of a protocol approved by the National Institutes of Health Institutional Review Board. Mononuclear cells from AML patients were isolated from whole blood or marrow using density gradient centrifugation with Ficoll-Hypaque (GE Healthcare), assessed for cytogenetics and AML subtype classification, and cryopreserved as previously described.18 AML patient samples chosen for this study possessed greater than 90% blasts. Bone marrow-derived CD34+ cells from healthy donors were obtained from the National Disease Research Interchange. AML blasts and CD34+ cells were thawed in DNaseI solution and grown for 12 hours in StemSpan Serum-Free Expansion Medium with StemSpan CC-100 cytokine cocktail (StemCell Technologies) to a final concentration of 100 ng/mL recombinant human Fms-like tyrosine kinase 3 ligand, 100 ng/mL recombinant human stem cell factor, 20 ng/mL recombinant human interleukin-3, and 20 ng/mL recombinant human interleukin-6.
Drug treatment
AML cell lines and AML patient blasts were treated for 72 hours with 5-aza-2′-deoxycytidine (5-aza-dC, decitabine, 2μM; dissolved in dimethyl sulfoxide [DMSO]; Sigma-Aldrich) with drug replacement every 24 hours. For trichostatin A (TSA; Sigma-Aldrich) experiments, TSA (250nM; dissolved in DMSO) was added for the final 12 hours of the 72-hour 5-aza-dC treatment period.
Bisulfite PCR, quantitative PCR, and reverse-transcriptase PCR
RNA from cell lines and patient samples was isolated using Trizol reagent (Invitrogen). cDNA was produced from 500 ng or 1 μg of total RNA on independent RNA isolates from each cell line using a cDNA reverse transcription kit (Applied Biosystems). Quantitative polymerase chain reaction (PCR) was performed in triplicate using one-tenth the cDNA reaction with predesigned gene expression assays (Applied Biosystems) for p15INK4b (Hs00793225), p14ARF (Hs99999189), p16INK4a (Hs00233365), β-actin (4333762), and glyceraldehyde-3-phosphate dehydrogenase (4333764). The sample cDNA copy numbers were calculated using plasmid DNA standards produced from PCR target regions cloned into pCR2.1 (Invitrogen). Relative quantitation was carried out by the comparative threshold cycle (Ct) method.
Bisulfite sequencing was conducted on genomic DNA (1 μg) using a published protocol.19 The PCR was carried out using primers p15BiHuF and p15BiHuR (supplemental Table 2). PCR products (811 bp) were cloned into pCR2.1, and at least 10 clones were sequenced using the BigDye Terminator, Version 1.1 Cycle Sequencing kit (Applied Biosystems). DNA methylation of p16INK4a and p14ARF was analyzed using previously described primers (supplemental Table 2).19,20
ChIP and tiling microarray analysis
ChIP was conducted as previously described.21 Cell lines were fixed in 0.8% formaldehyde for 6 minutes at 25°C. After lysis, samples were sonicated using a probe sonicator (Misonix) for 15 cycles (20 seconds per cycle/1 minute cooling) at a power level of 3. AML clinical samples and CD34+ cells were fixed in 1% formaldehyde for 10 minutes at 25°C and sonicated for 8 20-second cycles. Chromatin sheering was optimized to a size range of 200 to 600 bp. Chromatin (150-200 μg) was immunoprecipitated with antibodies H3K9me3 (Abcam, ab8898), H3K27me3 (Upstate, 07-449), H3K4me3 (Abcam, ab8580), H3K36me3 (Abcam, ab9050), H3K9/14Ac (Upstate, 06-599), EZH2 (Active Motif 39103), or rabbit IgG (Sigma-Aldrich, 15006). A 10% aliquot was removed as an input fraction. Control enrichment was measured by PCR using primers for RNA polymerase II large subunit (supplemental Table 2) for H3K4me3 and H3K36me3 antibodies or hsSat2 for H3K9me3 and H3K27me3 antibodies. ChIP DNA and input DNA were amplified as previously described (WGA2, Sigma-Aldrich).21 A total of 2 μg of amplified DNA was labeled with Cy3 (input) or Cy5 (IP) dUTP (PerkinElmer Life and Analytical Sciences) using the CGH labeling kit (Invitrogen).
Custom 4 × 44k or 8 × 15k tiling arrays (Agilent) contained probes spanning human chromosome 9 from base pairs 21770030 to 22999745 (University of California Santa Cruz genome browser-hg18). Probes, average of 60 bp in length, were predesigned or designed using eArray (Agilent), covering INK4b-ARF-INK4a at 20-bp resolution. Cross-hybridizing probes were excluded. A total of 3 μg of labeled ChIP and input DNA was cohybridized to the microarray for 40 hours at 65°C, washed, and scanned using an Agilent Scanner controlled by Agilent Scan Control 7.0 software. Data were extracted and with Feature Extraction 9.1 software and analyzed using ChIP Analytics 1.3 software (Agilent) correcting for background differences and normalizing for dye bias. Normalized and raw data files can be accessed at the GEO database GSE16730 and supplemental Table 4. To determine the reproducibility of experiments, KG-1 cell lines were repeated in 2 independent experiments (supplemental Figure 2) with a calculated linear regression R2 value of 0.95. ChIP enrichment across p15INK4b was validated with quantitative PCR for each AML cell line (supplemental Figure 3).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 software. An unpaired t test was performed on measurements of p15INK4b, p14ARF, and p16INK4a expression from AML samples from 3 experimental replicates. One-way analysis of variance followed by post hoc analysis using Dunnett multiple comparison test was used to measure differences in gene expression and ChIP enrichment in AML cell lines treated with 5-aza-dC and TSA. Each experiment was performed in 3 experimental replicates.
Results
H3K27me3 is enriched at the INK4b-ARF-INK4a locus in AML cell lines with p15INK4b DNA methylation
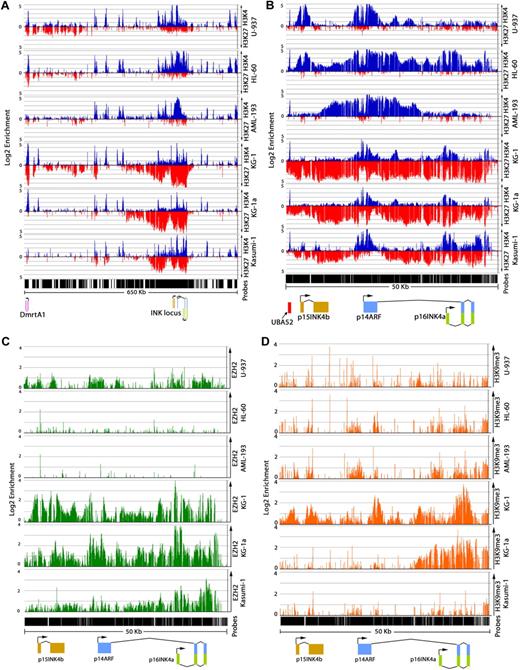
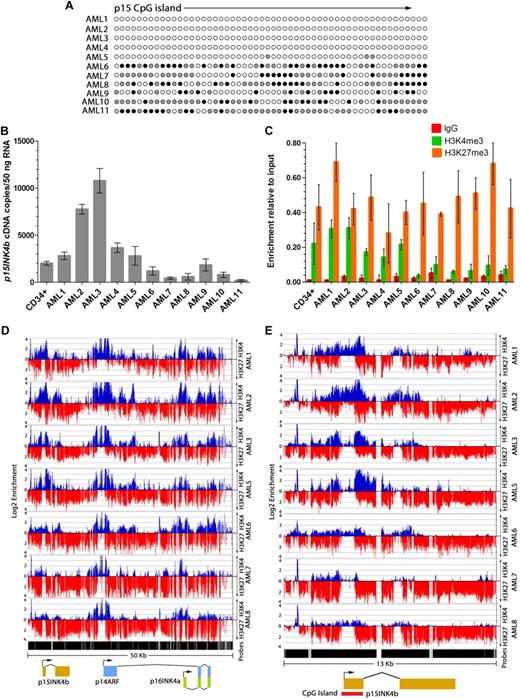
To assess the histone modifications associated with DNA methylation at p15INK4b, we carried out ChIP-on-chip experiments in AML-derived cell lines U-937, HL-60, AML-193, KG-1, KG-1a, and Kasumi-1. Consistent with previous reports, these cell lines display differences in the density of DNA methylation at the p15INK4b CpG island located at exon 1.22-24 p15INK4b is densely methylated in KG-1a, Kasumi-1, and AML-193 cells and possesses a variegated methylation pattern in KG-1 cells (Figure 1A). U-937 and HL-60 cells are devoid of p15INK4b DNA methylation. Regardless of the DNA methylation status, we found that expression of p15INK4b is low (less than single cDNA copy per cell) in all analyzed cell lines. However, cell lines with p15INK4b DNA methylation have lower levels of p15INK4b expression (30 cDNA copies/100 ng total RNA) compared with unmethylated cell lines (428 cDNA copies/100 ng total RNA; P < .01; unpaired t test; Figure 1B). With the exception of U-937 and AML-193 cells, expression of p16INK4a is also low in AML cell lines, even though they are free of p16INK4a DNA methylation23,25 (Figure 1B-C).
p15INK4b DNA methylation and expression in AML cell lines used in study. (A) Bisulfite sequencing of the p15INK4b exon 1 CpG island. Filled circles represent percentage of DNA methylation at CpG positions from 10 independently sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine). (B-C) Quantitative reverse-transcriptase (RT)–PCR analysis of p15INK4b, p14ARF, and p16INK4a expression in AML cell lines. Copies of cDNA per 100 ng of total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD.
, 10%-49% methylcytosine). (B-C) Quantitative reverse-transcriptase (RT)–PCR analysis of p15INK4b, p14ARF, and p16INK4a expression in AML cell lines. Copies of cDNA per 100 ng of total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD.
p15INK4b DNA methylation and expression in AML cell lines used in study. (A) Bisulfite sequencing of the p15INK4b exon 1 CpG island. Filled circles represent percentage of DNA methylation at CpG positions from 10 independently sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine). (B-C) Quantitative reverse-transcriptase (RT)–PCR analysis of p15INK4b, p14ARF, and p16INK4a expression in AML cell lines. Copies of cDNA per 100 ng of total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD.
, 10%-49% methylcytosine). (B-C) Quantitative reverse-transcriptase (RT)–PCR analysis of p15INK4b, p14ARF, and p16INK4a expression in AML cell lines. Copies of cDNA per 100 ng of total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD.
ChIP-on-chip experiments were performed using a tiling microarray, featuring probes spanning a 1.2-megabase region of human chromosome 9 (21770030-22999745). This array provides a high resolution (20 bp) of binding events at p15INK4b, p14ARF, and p16INK4a. Because of the reported association of H3K27me3 and H3K9me3 with proteins that interact with DNMTs, we initiated our study looking at the relationship of these histone modifications with p15INK4b DNA methylation. In AML cell lines with p15INK4b DNA methylation (KG-1, KG-1a, and Kasumi-1), we observed that H3K27me3 is enriched over a broad region upstream of p15INK4b (chromosome 9: 22017185-22067089) and over INK4b-ARF-INK4a (chromosome 9: 21955560-22001115; Figure 2A). H3K27me3 spans p15INK4b with high levels of enrichment over exon 2 (Figure 2B; supplemental Figure 3). We found a sharp boundary downstream of p16INK4a (chromosome 9: 21955560) where enrichment of H3K27me3 ends, suggesting that the spread of H3K27me3 outside the INK4b-ARF-INK4a region is blocked at this location. Interestingly, in CD4+ T cells, this region is bound by CCCTC-binding factor (CTCF), which has previously been identified at boundaries of active and inactive domains.26 EZH2, a member of the polycomb repressive complex 2, binds at INK4b-ARF-INK4a, overlapping the area of H3K27me3 in KG-1, KG-1a, and Kasumi-1 cells (Figure 2C). The highest level of EZH2 binding is located at p16INKa exon 1, whereas lower levels of enrichment extend into p15INK4b and p14ARF. In contrast to the AML cell lines with p15INK4b DNA methylation, U-937 and HL-60 cells exhibit low levels of H3K27me3 and EZH2 enrichment over INK4b-ARF-INK4a (Figure 2B-C). The AML-193 cell line, which has DNA methylation of p15INK4b, displayed a unique pattern of histone modifications compared with other AML cell lines and clinical samples (Figure 3D) analyzed in our study. In these cells, we measured no H3K27me3 or EZH2 enrichment over INK4b-ARF-INK4a (Figure 2B). This suggests that DNA methylation at p15INK4b is maintained independently of polycomb complexes in this cell line. The lack of H3K27me3 and EZH2 binding at the INK4b-ARF-INK4a region in AML-193 cells does not result from global changes in H3K27me3 or EZH2 protein levels compared with other AML cell lines (supplemental Figure 4).
Distribution of H3K4me3, H3K27me3, and EZH2 at INK4b-ARF-INK4a differs in AML cell lines with DNA methylation. ChIP-on-chip analysis of enrichment of H3K4me3 (blue), H3K27me3 (red) on chromosome 9: 21800000 to 22450000 (A) and chromosome 9: 21953000 to 22003000 (B) in AML cell lines. Top axis represents H3K4me3; and bottom axis, H3K27me3. Enrichment of EZH2 (green; C) and H3K9me3 (orange; D) at the INK4b-ARF-INK4a region in AML cell lines. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
Distribution of H3K4me3, H3K27me3, and EZH2 at INK4b-ARF-INK4a differs in AML cell lines with DNA methylation. ChIP-on-chip analysis of enrichment of H3K4me3 (blue), H3K27me3 (red) on chromosome 9: 21800000 to 22450000 (A) and chromosome 9: 21953000 to 22003000 (B) in AML cell lines. Top axis represents H3K4me3; and bottom axis, H3K27me3. Enrichment of EZH2 (green; C) and H3K9me3 (orange; D) at the INK4b-ARF-INK4a region in AML cell lines. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
AML patient blasts with p15INK4b DNA methylation display reduced H3K4me3 enrichment at the promoter. (A) Bisulfite sequencing of the p15INK4b exon 1 CpG island in AML patient samples. Filled circles represent the frequency of DNA methylation at CpG positions in sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine). (B) Quantitative RT-PCR analysis of p15INK4b expression in AML patient blasts. cDNA copies per 50 ng total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD. Quantitative PCR analysis (C) of H3K4me3 and H3K27me3 enrichment at the p15INK4b promoter. Data are averages of ΔΔCt compared with input DNA from 3 technical replicates with error bars of SD. ChIP-on-chip analysis of enrichment of H3K4me3 (blue) and H3K27me3 (red) on chromosome 9: 21953000 to 22003000 (D) and chromosome 9: 21990000 to 22003000 (E) in AML patient samples. Top axis represents H3K4me3; and bottom axis, H3K27me3. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
, 10%-49% methylcytosine). (B) Quantitative RT-PCR analysis of p15INK4b expression in AML patient blasts. cDNA copies per 50 ng total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD. Quantitative PCR analysis (C) of H3K4me3 and H3K27me3 enrichment at the p15INK4b promoter. Data are averages of ΔΔCt compared with input DNA from 3 technical replicates with error bars of SD. ChIP-on-chip analysis of enrichment of H3K4me3 (blue) and H3K27me3 (red) on chromosome 9: 21953000 to 22003000 (D) and chromosome 9: 21990000 to 22003000 (E) in AML patient samples. Top axis represents H3K4me3; and bottom axis, H3K27me3. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
AML patient blasts with p15INK4b DNA methylation display reduced H3K4me3 enrichment at the promoter. (A) Bisulfite sequencing of the p15INK4b exon 1 CpG island in AML patient samples. Filled circles represent the frequency of DNA methylation at CpG positions in sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine). (B) Quantitative RT-PCR analysis of p15INK4b expression in AML patient blasts. cDNA copies per 50 ng total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD. Quantitative PCR analysis (C) of H3K4me3 and H3K27me3 enrichment at the p15INK4b promoter. Data are averages of ΔΔCt compared with input DNA from 3 technical replicates with error bars of SD. ChIP-on-chip analysis of enrichment of H3K4me3 (blue) and H3K27me3 (red) on chromosome 9: 21953000 to 22003000 (D) and chromosome 9: 21990000 to 22003000 (E) in AML patient samples. Top axis represents H3K4me3; and bottom axis, H3K27me3. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
, 10%-49% methylcytosine). (B) Quantitative RT-PCR analysis of p15INK4b expression in AML patient blasts. cDNA copies per 50 ng total RNA were calculated from a plasmid DNA standard curve and represent the averages of 3 independent experiments with error bars of SD. Quantitative PCR analysis (C) of H3K4me3 and H3K27me3 enrichment at the p15INK4b promoter. Data are averages of ΔΔCt compared with input DNA from 3 technical replicates with error bars of SD. ChIP-on-chip analysis of enrichment of H3K4me3 (blue) and H3K27me3 (red) on chromosome 9: 21953000 to 22003000 (D) and chromosome 9: 21990000 to 22003000 (E) in AML patient samples. Top axis represents H3K4me3; and bottom axis, H3K27me3. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
Although there was a previous report demonstrating an association of H3K9me3 with p15INK4b DNA methylation in AML cells,27 we found low levels of H3K9me3 at p15INK4b irrespective of the p15INK4b DNA methylation status (Figure 2D). In addition, our data indicated that low levels of H3K9me3 also exist at p14ARF-p16INK4a in AML cell lines, with the exception of KG-1 and KG-1a cells. Because genome-wide studies have indicated that the H3K9me3 histone modification is located at both actively transcribed as well as repressed genes,21 its role in the regulation of promoter CpG islands remains uncertain.
H3K4me3 is lost at p15INK4b in AML cell lines with p15INK4b DNA methylation
The activation-associated H3K4me3 modification has been inversely correlated with CpG island DNA methylation in other cell types28,29 and shown to directly inhibit the recruitment of DNMT3L.30 Our data suggest that the loss of H3K4me3 in association with DNA methylation is a common phenomenon. We found that AML cells that have p15INK4b DNA methylation (KG-1, KG-1a, Kasumi-1, and AML-193) do not exhibit H3K4me3 at p15INK4b (Figure 2B). In contrast, cell lines lacking p15INK4b DNA methylation (U-937 and HL-60) have H3K4me3 enrichment surrounding p15INK4b (chromosome 9: 21997400-22000500; Figure 2B). H3K4me3 enrichment was also found at the promoter of p14ARF in U-937 and HL-60 cells and p16INK4a in HL-60, KG-1, and KG-1a cells that are free of DNA methylation (Figure 2B). AML-193 cells display a unique pattern of H3K4me3 enrichment, with a large region of H3K4me3 initiating near the end of p15INK4b and spanning p14ARF and p16INK4a (Figure 2B). The presence of this activation-associated mark is consistent with the moderate expression level of p14ARF and p16INK4a in these cells. All cell lines display a peak of H3K4me3 located 3 kb upstream of p15INK4b corresponding to a transcribed region related to human ubiquitin A-52 residue ribosomal protein fusion product 1 (UBA52; Figure 2B).
H3K9/14 acetylation and H3K36me3 have been associated with actively transcribed genes.26 In AML cell lines, we found that the enrichment pattern of H3K9/14Ac was similar to that of H3K4me3 (supplemental Figure 5). In all AML cell lines without p15INK4b DNA methylation, the H3K9/14Ac mark is located around the p15INK4b promoter. We also compared the distribution of H3K36me3 in AML cell lines with and without p15INK4b DNA methylation. In U-937 and HL-60 cells free of p15INK4b DNA methylation, H3K36me3 is distributed over the p15INK4b open reading frame (supplemental Figure 6). KG-1 cells, which have p15INK4b DNA methylation and lower levels of p15INK4b expression, displayed lower levels of H3K36me3. H3K36me3 is also decreased in CD34+ cells (supplemental Figure 6) in which p15INK4b is unmethylated but have low p15INK4b expression (Figure 3B). Therefore, it is probable that loss of H3K36me3 results from transcriptional repression of the gene rather than from the acquisition of DNA methylation.
AML patient blasts with p15INK4b DNA methylation are marked by H3K27me3 whereas unmethylated blasts are bivalent
Because long-term cultured AML cell lines may not recapitulate events observed in primary tissue, we compared the pattern of histone modifications associated with p15INK4b DNA methylation in human AML clinical samples with that in AML cell lines. Consistent with prior reports,31,32 we found that p15INK4b DNA methylation is highly variegated in blasts (Figure 3A; supplemental Figure 1). p14ARF and p16INK4a are unmethylated, with the exception of AML6, which has p14ARF DNA methylation (supplemental Figure 7). Expression of p15INK4b is low in all AML samples and in CD34+ cells. Six AML samples (AML6-11) with high levels of p15INK4b DNA methylation displayed a 6.6-fold decrease in p15INK4b expression compared with unmethylated samples (AML1-5, P < .01, unpaired t test; Figure 3B).
The enrichment patterns of H3K27me3 in AML clinical samples differed from AML cell lines. Notably, we observed that all AML samples have H3K27me3 at p15INK4b irrespective of the DNA methylation status of the gene (Figure 3C). Similar to AML cell lines with p15INK4b DNA methylation, H3K27me3 extends throughout INK4b-ARF-INK4a (Figure 3D-E). AML clinical samples have low levels of H3K9me3 enrichment at p15INK4b in both DNA methylated and unmethylated samples (supplemental Figure 8). Higher levels of H3K9me3 were detected over exons 2 and 3 of p16INK4a and p14ARF in 3 of the 4 analyzed samples, suggesting that this mark may regulate these genes.
The most consistent feature associated with p15INK4b DNA methylation in both cell lines and clinical samples was the loss of H3K4me3 enrichment around the p15INK4b promoter. Clinical samples with p15INK4b DNA methylation had a 3.1-fold lower enrichment of H3K4me3 compared with unmethylated samples (P < .01, unpaired t test; Figure 3C). Low levels of H3K4me3 enrichment are evident over the region surrounding the p15INK4b transcription start site in samples with p15INK4b DNA methylation (Figure 3D-E).
AML patient samples with an unmethylated p15INK4b CpG island have overlapping regions of H3K4me3 and H3K27me3 enrichment at p15INK4b (Figure 3E). H3K27me3 extends into p14ARF and p16INK4a, suggesting a coordinated repression of the INK4b-ARF-INK4a locus even in the absence of p15INK4b DNA methylation (Figure 3D). This observation in clinical samples differs from that in AML cell lines where samples without p15INK4b DNA methylation displayed H3K4me3 in the absence of H3K27me3 at INK4b-ARF-INK4a. The coexistence of H3K4me3 and H3K27me3 at p15INK4b is consistent with bivalent genes, which have been identified in embryonic stem cells, and correlates with the low level of p15INK4b expression in unmethylated AML samples.
Epigenetic therapies induce H3K4me3 at the p15INK4b promoter
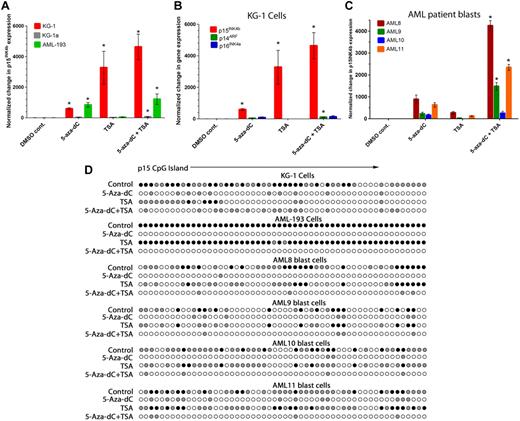
Because of the heterogeneity in the histone modification and DNA methylation patterns we observed between AML cell lines and patient blasts, we were interested in whether certain histone modifications and DNA methylation patterns affect the ability of epigenetic drugs to reactivate p15INK4b. For these studies, we examined the response of KG-1 and KG-1a cells with partial or full DNA methylation of p15INK4b, respectively, in the presence of H3K27me3 and AML-193 cells, which have full DNA methylation of p15INK4b, and no H3K27me3. Treatment of KG-1 and AML-193 cells with the DNMT inhibitor 5-aza-dC for 72 hours was sufficient to reactivate p15INK4b 592-fold in KG-1 and 838-fold in AML-193 over control-treated cells (P < .01, 1-way analysis of variance post hoc analysis; Figure 4A; supplemental Table 3). Reactivation of p15INK4b in KG-1 cells occurred independently of changes in p14ARF or p16INK4a expression (Figure 4B). Reactivation of p15INK4b in KG-1 and AML-193 cells correlated with a reduction in p15INK4b DNA methylation (Figure 4D; supplemental Figure 1) and fewer cycling cells (supplemental Figure 9). Similar to previous reports,24 no significant change in p15INK4b DNA methylation or gene expression was observed in KG-1a cells treated with 5-aza-dC. Surprisingly, KG-1 cells exhibited an even greater p15INK4b reactivation (6-fold) when treated with the histone deacetylase inhibitor TSA for 24 hours compared with 5-aza-dC (P < .01; Figure 4A). Reactivation with TSA was associated with a loss of p15INK4b DNA methylation (Figure 4D; supplemental Figure 1). This suggests that p15INK4b repression and DNA methylation in KG-1 cells may be maintained by the lack of histone acetylation at the gene or a protein altered in its expression by TSA treatment. This effect was unique to KG-1 cells as TSA was insufficient for p15INK4b reactivation or changes in DNA methylation in AML-193 or KG-1a cells. Although TSA has previously been reported to repress the expression of DNMTs,33,34 we observed only slight repression of DNMT3a and no alteration in DNMT 1 or 3b protein levels in TSA-treated KG-1 cells (supplemental Figure 10). Combined 5-aza-dC and TSA treatment caused loss of p15INK4b DNA methylation in KG-1 and AML-193 cells. p15INK4b levels after this treatment were comparable with TSA alone in KG-1 cells (P < .01) and 5-aza-dC alone in AML-193 cells (Figure 4A).
Reactivation of p15INK4b expression and loss of DNA methylation after treatment with DNMT or HDAC inhibitors. (A) Quantitative RT-PCR analysis of p15INK4b expression in KG-1, KG-1a, and AML-193 cells after reactivation with 5-aza-dC and/or TSA. (B) Quantitative RT-PCR for p15INK4b, p14ARF, and p16INK4a expression in KG-1cells after reactivation with 5-aza-dC and/or TSA. (A-B) *Increase in expression relative to control (P < .05, 1-way analysis of variance). (C) Quantitative RT-PCR for p15INK4b expression in AML patient blasts treated with 5-aza-dC or TSA. Quantitative RT-PCR data in both cell lines and clinical samples were normalized to control cells and calculated from ΔΔCt averages of 3 independent experiments with error bars of SD. *Significant increase in expression in cells treated with a combination of 5-aza-dC and TSA compared with cells treated with 5-aza-dC alone (P < .05, 1-way analysis of variance). (D) Bisulfite sequencing of the p15INK4b CpG island in individual clones from control and treated KG-1, AML-193, and AML8 blasts after treatment with 5-aza-dC and/or TSA. Filled circles represent percentage DNA methylation at individual CpG positions from 10 independently sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine).
, 10%-49% methylcytosine).
Reactivation of p15INK4b expression and loss of DNA methylation after treatment with DNMT or HDAC inhibitors. (A) Quantitative RT-PCR analysis of p15INK4b expression in KG-1, KG-1a, and AML-193 cells after reactivation with 5-aza-dC and/or TSA. (B) Quantitative RT-PCR for p15INK4b, p14ARF, and p16INK4a expression in KG-1cells after reactivation with 5-aza-dC and/or TSA. (A-B) *Increase in expression relative to control (P < .05, 1-way analysis of variance). (C) Quantitative RT-PCR for p15INK4b expression in AML patient blasts treated with 5-aza-dC or TSA. Quantitative RT-PCR data in both cell lines and clinical samples were normalized to control cells and calculated from ΔΔCt averages of 3 independent experiments with error bars of SD. *Significant increase in expression in cells treated with a combination of 5-aza-dC and TSA compared with cells treated with 5-aza-dC alone (P < .05, 1-way analysis of variance). (D) Bisulfite sequencing of the p15INK4b CpG island in individual clones from control and treated KG-1, AML-193, and AML8 blasts after treatment with 5-aza-dC and/or TSA. Filled circles represent percentage DNA methylation at individual CpG positions from 10 independently sequenced clones (● represents ≥ 50% methylcytosine;  , 10%-49% methylcytosine).
, 10%-49% methylcytosine).
Because the effects of these drugs in AML cell lines may not reflect the consequences of their use in the clinic on primary leukemia cells, we studied the effects of 5-aza-dC and TSA on 4 AML patient samples with p15INK4b DNA methylation. In all 4 samples, p15INK4b could be reactivated by 5-aza-dC (P < .01) on average of 475-fold. Reactivation of p15INK4b correlated with reduced DNA methylation (Figure 4D) and reduced cycling cells (supplemental Figure 9) in all 4 samples. Reactivation of p15INK4b was observed in TSA-treated samples AML8 and AML11 (P < .05); however, p15INK4b expression in these 2 samples was lower compared with 5-aza-dC–treated samples. p15INK4b DNA methylation is unaffected by TSA treatment (Figure 4D). Importantly, we observed a significant increase in p15INK4b expression in 3 of 4 AML samples with combined 5-aza-dC and TSA treatment compared with individual drug treatments (P < .01; Figure 4C). This indicates that the synergy between these drugs is more dramatic in clinical samples than in AML cell lines.
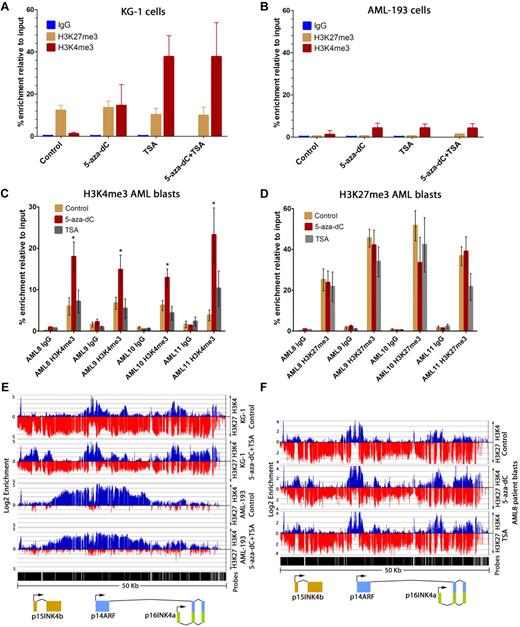
Histone modifications at p15INK4b were measured after drug treatment in KG-1 cells, AML-193 cells, and in AML patient samples. Reactivation of p15INK4b expression in KG-1 cells correlated with an increase in H3K4me3 at the promoter after 5-aza-dC (P < .01), TSA (P < .01), and 5-aza-dC and TSA treatments (P < .01; 1-way analysis of variance post hoc analysis; Figure 5A). H3K27me3 enrichment remained unaffected by these treatments (Figure 5A). Although p15INK4b reactivation correlated with loss of DNA methylation in AML-193 cells after 5-aza-dC treatment, we observed no change in H3K4me3 or H3K27me3 enrichment (Figure 5B).
Reactivation of p15INK4b using DNMT and HDAC inhibitors is associated with an increase in H3K4me3 and maintenance of H3K27me3 in KG-1 and AML blasts. ChIP-quantitative PCR measured enrichment of H3K4me3 and H3K27me3 at the p15INK4b promoter after reactivation in KG-1 (A) and AML-193 cells (B). The enrichment of H3K4me3 (C) and H3K27me3 (D) measured at the p15INK4b promoter by quantitative PCR in clinical samples AML8, AML9, AML10, and AML11 patient blasts treated with DMSO (control), 5-aza-dC, or TSA. (C) *Significant increase in H3K4me3 enrichment compared with control treatment (P < .05, 1-way analysis of variance). Enrichment was calculated from ΔΔCt averages of 3 independent experiments with error bars of SD. (E) ChIP-on-chip for enrichment of H3K4me3 (blue) and H3K27me3 (red) at INK4b-ARF-INK4a in DMSO control and 5-aza-dC plus TSA-treated KG-1 and AML-193 cells. (F) ChIP-on-chip for enrichment of H3K4me3 (blue) and H3K27me3 (red) at INK4b-ARF-INK4a in AML8 patient blasts treated with control DMSO, 5-aza-dC, or TSA. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
Reactivation of p15INK4b using DNMT and HDAC inhibitors is associated with an increase in H3K4me3 and maintenance of H3K27me3 in KG-1 and AML blasts. ChIP-quantitative PCR measured enrichment of H3K4me3 and H3K27me3 at the p15INK4b promoter after reactivation in KG-1 (A) and AML-193 cells (B). The enrichment of H3K4me3 (C) and H3K27me3 (D) measured at the p15INK4b promoter by quantitative PCR in clinical samples AML8, AML9, AML10, and AML11 patient blasts treated with DMSO (control), 5-aza-dC, or TSA. (C) *Significant increase in H3K4me3 enrichment compared with control treatment (P < .05, 1-way analysis of variance). Enrichment was calculated from ΔΔCt averages of 3 independent experiments with error bars of SD. (E) ChIP-on-chip for enrichment of H3K4me3 (blue) and H3K27me3 (red) at INK4b-ARF-INK4a in DMSO control and 5-aza-dC plus TSA-treated KG-1 and AML-193 cells. (F) ChIP-on-chip for enrichment of H3K4me3 (blue) and H3K27me3 (red) at INK4b-ARF-INK4a in AML8 patient blasts treated with control DMSO, 5-aza-dC, or TSA. Normalized enrichment data (IP/input) are plotted on a log 2 scale.
Similar to KG-1 cells, 5-aza-dC increased H4K4me3 at p15INK4b compared with control treatment (P < .01) in 4 AML clinical samples with p15INK4b DNA methylation (Figure 5C). However, in contrast to KG-1 cells, TSA alone did not induce significant H3K4me3 changes at p15INK4b compared with control treatment (Figure 5C). Microarray analysis of an AML clinical sample (AML8) treated with 5-aza-dC showed that p15INK4b reactivation correlated with an average 3-fold increase in H3K4me3 enrichment at probes around the p15INK4b transcription start site (chromosome 9: 21999095-219999315) compared with the control treatment (P < .05, 1-way analysis of variance; Figure 5F). No change in H3K4me3 was observed at p14ARF or p16INK4a after drug treatment in KG-1, AML-193, or in AML8 (Figure 5E-F). The maintenance of H3K27me3 at p15INK4b and p14ARF-p16INK4a after treatment in both KG-1 and in AML patient blasts indicates that p15INK4b is marked by H3K4me3 and H3K27me3 once reactivated (Figure 5D-F). Overall, these data show that epigenetic reactivation of p15INK4b can return the gene to the bivalent histone modification state observed in AML patient samples without p15INK4b DNA methylation.
Discussion
Because of their function in the regulation of gene expression, histone modifications represent an additional epigenetic pathway that may be altered in AML. In this study, we used high-density ChIP-on-chip to determine the histone modifications present at p15INK4b in AML cells and how these marks are altered in cells that have p15INK4b DNA methylation. An important observation in this study was that H3K4me3 occupancy is reduced around the p15INK4b promoter in AML cells with p15INK4b DNA methylation compared with samples without DNA methylation. This result is consistent with genome-wide studies in neuronal and fibroblast cells, which have indicated that many genes with DNA methylation display reduced H3K4me3.28,35 The loss of H3K4me3 at genes with DNA methylation could imply that H3K4me3 protects the p15INK4b CpG island from DNMTs. In support for this model, H3K4me3 has been shown to inhibit the binding of DNMT3L, thereby preventing the recruitment of de novo DNA methylation machinery to promoters.30 In addition, binding of the H3K4 methyltransferase MLL can protect CpG islands from acquiring DNA methylation.36 An alterative to this model is that DNA methylation at an earlier stage of cell development could alter the chromatin structure, preventing H3K4 histone methyltransferases from accessing the CpG island.37 This would suggest that DNA methylation serves to solidify repression at the promoter by preventing the gene from reacquiring activation-associated histone marks.
Another important observation in our study is that H3K27me3 enrichment spans p15INK4b and extends into p14ARF and p16INK4a in AML patient blasts irrespective of the DNA methylation status of p15INK4b. Although a previous report27 implicated changes in H3K9me3 in AML with p15INK4b DNA methylation, we found low levels of this mark at p15INK4b, indicating that H3K27me3 is the primary repressive mark at this gene. In AML clinical samples without p15INK4b DNA methylation, H3K27me3 enrichment overlaps an area of H3K4me3, creating a bivalent histone modification pattern at p15INK4b typically found at developmentally regulated genes in stem and progenitor cells.14,17,38,39 Genes in this conformation are maintained in a transcriptionally poised state in which expression is low. Accordingly, we find that expression of p15INK4b is low even in samples in which p15INK4b DNA is unmethylated. Because the region of H3K27me3 enrichment is also bound by EZH2, our results imply that polycomb-mediated repression is important in the regulation of p15INK4b as well as p14ARF and p16INK4a in AML cells. Polycomb proteins Bmi-1 and EZH2 have been identified as important regulators of p14ARF and p16INK4a in response to signals for cellular senescence in human and mouse fibroblasts.10,13 Interestingly, in these cell lineages, H3K27me3 and EZH2 appear to be distributed primarily at p14ARF and p16INK4a and not p15INK4b. We and others have found that hematopoietic progenitor cell lineages, CD34+ cells10 (supplemental Figure 11) and CD133+ cells,14 have H3K27me3 over INK4b-ARF-INK4a. Interestingly, these cells also exhibit low H3K4me3 at p15INK4b. Although CD34+ cells represent a mixed cell population, this would suggest that, in some early hematopoietic cells, polycomb repression may be a common feature of p15INK4b regulation. However, in CD4+ lymphoid cells, p15INK4b is marked by H3K4me3 in the absence of H3K27me3,26 thereby emphasizing the dynamic nature of histone modifications during cellular differentiation.
Although we did not observe p15INK4b DNA methylation in normal CD34+ cells (supplemental Figure 11), p15INK4b DNA methylation has been previously reported in these cells.40 During mouse neuronal cell differentiation, DNA methylation appears to be a normal process during lineage commitment used to solidify repression at promoters marked by H3K27me3.35 It remains unknown whether p15INK4b DNA methylation observed in AML represents a subset of myeloid lineage-committed cells in which this gene is normally terminally repressed or is an abnormal acquisition during transformation.
The histone modification pattern we observed in AML cell lines did not fully recapitulate the patterns we described in AML clinical samples. Cell lines free of p15INK4b DNA methylation (U-937 and HL-60) have resolved the bivalent histone modification state displaying high levels of H3K4me3 at the p15INK4b promoter in the absence of H3K27me3 throughout INK4b-ARF-INK4a. Although these cell lines may represent a unique subpopulation or differentiation state of AML cells, changes acquired during the establishment of these cell lines may explain the differences in our studies. Even though H3K4me3 in the absence of H3K27me3 is associated with gene activation,26 we found that p15INK4b expression was low in unmethylated cell lines. A genome-wide study identified a subset of genes with positive H3K4me3 enrichment that display low levels of expression, suggesting that H3K4me3 may be necessary but not sufficient for gene activation.41 Interestingly, p15INK4b is strongly up-regulated by monocytic differentiation in both U-937 and HL-609 cells, suggesting that additional activating events are necessary to achieve full expression potential.
Because EZH2 has been found to interact with DNMT1 and is overexpressed in certain AML subtypes,15 it is interesting to speculate that EZH2 directly regulates DNA methylation at p15INK4b. However, studies have suggested that DNA methylation can exist independently of polycomb marks. In mouse cells, Komashko et al21 found that 10% of genes with DNA methylation do not possess K3K9me3 or H3K27me3. In addition, DNA methylation can exist in cells without EZH2 expression,42 and not all genes with H3K27me3 exhibit DNA methylation.21 We found that p15INK4b DNA methylation exists in the absence of H3K27me3 or EZH2 in AML-193 cells. In addition, siRNA knockdown of EZH2 in KG-1 cells was unable to abrogate DNA methylation or induce expression of p15INK4b (T.A.P., unpublished results, November 2009). Although we cannot rule out a role for EZH2 in de novo DNA methylation, DNA methylation at p15INK4b may operate through polycomb independent or interdependent pathways. Recent studies have identified novel small RNAs, including microRNA-29b43 and p15-AS,44 as regulators of p15INK4b expression and DNA methylation.
We found that epigenetic reactivation of p15INK4b in KG-1 and AML patient cells restores p15INK4b to a bivalent state observed in unmethylated AML samples. Increases in H3K4me3 at gene promoters and maintenance of H3K27me3 have been previously observed after reactivation of genes with DNA methylation in the colon carcinoma cell line HCT116.45,46 It is notable that the reestablishment of the bivalent state at tumor suppressor genes would not be expected to fully reactivate these genes but would restore their ability to be reactivated after additional cellular stimuli that abrogate polycomb repression. In our experiments, we observed differences in the ability of p15INK4b to be reactivated by DNMT and HDAC inhibitors in AML cell lines and clinical samples. Our data indicate that gene reactivation in AML patient blasts, which have p15INK4b DNA methylation coexisting with repressive histone modifications, may benefit from the combined use of HDAC inhibitors with DNMT inhibitors. However, the lack of a synergistic effect of 5-aza-dC and TSA on p15INK4b reactivation in one AML patient sample suggests that heterogeneity may exist in leukemic cell response to these drugs. The unexpected reactivation and demethylation of p15INK4b in KG-1 cells by the treatment with the HDAC inhibitor TSA are consistent with previous reports for E-cadherin and RARβ2 in HEK 293 cells.47 Although TSA has been demonstrated to repress the expression of DNMTs,33,34 we have observed only slight repression of DNMT3a and no alteration of DNMT1 or 3b in KG-1 cells treated with 5-aza-dC or TSA (supplemental Figure 10). The short duration (12 hours) of TSA treatment in our experiments suggests that DNA methylation loss may have occurred through an active process.
Reactivation of epigenetically silenced tumor suppressor genes remains an attractive target for reducing the proliferation and survival of leukemia cells. Recent work suggests that, like DNA methylation, enzymes that regulate histone methylation can be manipulated to reverse abnormal gene repression in cancer.48 It remains of interest to describe the full assortment of alterations in histone modifications that occur in AML cells. However, before this can be achieved, the dynamics of how these histone marks normally regulate genes in myeloid cell lineages needs further analysis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Emily Prentice, Josh Chenoweth, Horatiu Muresan, and Rita Humeniuk for assistance with this project.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
National Institutes of Health
Authorship
Contribution: T.A.P. and L.W. designed research, analyzed data, and wrote the paper; J.B. performed some experiments; and D.S. generated critical reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Linda Wolff, NIH, Bldg 37, Rm 4124, 37 Convent Dr, MSC 4263, Bethesda, MD 20892-4263; e:-mail: lwolff@helix.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal