Abstract
The t(14;18)(q32;q21) involving the immunoglobulin heavy chain locus (IGH) and the MALT1 gene is a recurrent abnormality in mucosa-associated lymphoid tissue (MALT) lymphomas. However, the nucleotide sequence of only one t(14;18)–positive MALT lymphoma has been reported so far. We here report the molecular characterization of the IGH-MALT1 fusion products in 5 new cases of t(14;18)–positive MALT lymphomas. Similar to the IGH-associated translocations in follicular and mantle cell lymphomas, the IGH-MALT1 junctions in MALT lymphoma showed all features of a recombination signal sequence–guided V(D)J-mediated translocation at the IGH locus. Furthermore, analogous to follicular and mantle cell lymphoma, templated nucleotides (T-nucleotides) were identified at the t(14;18)/IGH-MALT1 breakpoint junctions. On chromosome 18, we identified a novel major breakpoint region in MALT1 upstream of its coding region. Moreover, the presence of duplications of MALT1 nucleotides in one case suggests an underlying staggered DNA-break process not consistent with V(D)J-mediated recombination. The molecular characteristics of the t(14;18)/IGH-MALT1 resemble those found in the t(14;18)/IGH-BCL2 in follicular lymphoma and t(11;14)/CCND1-IGH in mantle cell lymphoma, suggesting that these translocations could be generated by common pathomechanisms involving illegitimate V(D)J-mediated recombination on IGH as well as new synthesis of T-nucleotides and nonhomologous end joining (NHEJ) or alternative NHEJ repair pathways on the IGH-translocation partner.
Introduction
Four recurrent translocations, t(1;14)(p22;q32), t(3;14)(p14.1;q32), t(11;18)(q21;q21), and t(14;18)(q32;q21), have been described in marginal zone B-cell lymphomas of the mucosa-associated lymphoid tissue (MALT) type.1-6 The 2 latter translocations involved the MALT1 gene and lead to an API2-MALT1 gene fusion in the t(11;18)(q21;q21) and an IGH-MALT1 fusion in the t(14;18)(q32;q21).1-3 The molecular analyses of the t(11;18)/API2-MALT1 have led to its complete characterization, facilitating the development of detection assays based on fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (PCR).
In contrast, less is known about the molecular genetic features of the t(14;18)/IGH-MALT1 in MALT lymphoma because only one case has been described molecularly, so far.7 The scarcity of data could be due to the relative low incidence of this translocation, because only up to 10% of MALT lymphomas are t(14;18)-positive, to the difficulty in obtaining densely infiltrated tumor tissue, and to the low proliferation rate of the neoplastic cells. In addition, t(14;18)/IGH-MALT1–positive MALT lymphomas originate at sites such as the liver, skin, ocular adnexa, or salivary glands.1,3,8
Nevertheless, the molecular characterization of t(14;18)/IGH-MALT1 is important to understand the underlying pathogenetic mechanisms and to develop detection methods for the diagnostic workup. In this context it is important to note that the t(14;18)/IGH-MALT1 in MALT lymphomas and the t(14;18)(q21;q32)/IGH-BCL2 in follicular lymphomas (FLs) are indistinguishable by conventional cytogenetic analysis.7
In other B-cell lymphomas, such as FL and mantle cell lymphoma (MCL), the analyses of the immunoglobulin heavy chain locus (IGH) breakpoint junctions have led to important insights in the mechanisms leading to the IGH-associated translocations in these lymphoma subtypes.9-13
The t(14;18)/IGH-BCL2 in FL and the t(11;14)/CCND1-IGH in MCL result from an illegitimate V(D)J recombination with insertions of templated nucleotides (T-nucleotides) at the junction sites.9,10 Moreover, the study of the breakpoints of translocations involving the IGH locus has been used to understand the cellular origin of B-cell lymphomas. The presence of DH segments carrying somatic mutations at the direct JH-BCL2 and CCND1-JH junctions has suggested that these translocations can also emerge in germinal/postgerminal center B cells and not only at early stages in the B-cell development.9,10,12
We herein report the molecular genetic characterization of the direct MALT1-JH and reciprocal DH-MALT1 junctions in 5 new MALT lymphomas harboring the t(14;18)/IGH-MALT1.
Methods
Clinical and cytogenetic data
Five cases with the diagnosis of an extranodal marginal zone B-cell lymphoma of the MALT type, the presence of the t(14;18)/IGH-MALT1 as shown by interphase FISH, and available frozen or paraffin-embedded tissue were collected from different genetic institutes (University Medical Center Hamburg-Eppendorf, Germany [n = 3]; Center Hospitalier Lyon Sud, France [n = 1]; University of Cambridge, United Kingdom [n = 1]) and included in the present study. The clinical and cytogenetic data of these cases are described in Table 1.
Clinical and cytogenetic data of patients with MALT lymphoma with t(14;18)/IGH-MALT1
| Case no. . | Sex/age . | Diagnosis . | Therapy/response . | Karyotype . |
|---|---|---|---|---|
| 1 | F/71 | Low-grade MALT lymphoma of the conjunctiva | RT/CR | No mitosis |
| 2 | M/53 | Low-grade MALT lymphoma of the lung | Lobectomy, CT (Chloramb)/CR | 49,XY,+3,+5,+12,t(14;18)(q32;q21)[4]/50,idem,+8[13]/46,XY[3] |
| 3 | M/57 | Low-grade MALT lymphoma of the orbit | NA | NA |
| 4 | F/65 | Low-grade MALT lymphoma of the skin | NA | NA |
| 5 | M/78 | Low-grade MALT lymphoma of the skin | NA | NA |
| 6* | M/76 | Low-grade MALT lymphoma of the orbit | RT/CR | 47,XY,+3,t(14;18)(q32;q21)[6]/46,XY[3] |
| Case no. . | Sex/age . | Diagnosis . | Therapy/response . | Karyotype . |
|---|---|---|---|---|
| 1 | F/71 | Low-grade MALT lymphoma of the conjunctiva | RT/CR | No mitosis |
| 2 | M/53 | Low-grade MALT lymphoma of the lung | Lobectomy, CT (Chloramb)/CR | 49,XY,+3,+5,+12,t(14;18)(q32;q21)[4]/50,idem,+8[13]/46,XY[3] |
| 3 | M/57 | Low-grade MALT lymphoma of the orbit | NA | NA |
| 4 | F/65 | Low-grade MALT lymphoma of the skin | NA | NA |
| 5 | M/78 | Low-grade MALT lymphoma of the skin | NA | NA |
| 6* | M/76 | Low-grade MALT lymphoma of the orbit | RT/CR | 47,XY,+3,t(14;18)(q32;q21)[6]/46,XY[3] |
The status for all cases was diagnosis at the time the sample was taken.
RT indicates radiation therapy; CR, complete remission; CT, chemotherapy; Chloramb, chlorambucil; and NA, not available.
Case 6 corresponds to case 1 of the study of Sánchez-Izquierdo et al.7
Interphase FISH
Four of 5 MALT lymphomas originating from the skin (n = 2), the lung (n = 1), and the conjunctiva (n = 1) were analyzed by interphase FISH as previously described with the use of probes for the MALT1 gene (PAC117B5 and PAC59N7) and the IGH locus (cosmid Cα1).1 One further MALT lymphoma of the orbit (case 3) was analyzed with the LSI MALT1 (18q21) Dual Color, Break Apart Rearrangement Probe, the LSI IGH (14q32) Dual Color, Break Apart Rearrangement Probe, and the LSI IGH/MALT1 t(14;18)(q32;q21) Dual Color, Dual Fusion Translocation Probe (all from Abbott Vysis).
Amplification of the direct and reciprocal IGH-MALT1 products on frozen tissue and statistical analysis
The direct IGH-MALT1 fusions (MALT1-JH) were confirmed in cases 1 and 2 by long-distance PCR with the use of the GeneAmp XL PCR system (Applied Biosystems) on patients' DNA with nested primers MALT1-R1a and MALT1-R1b in combination with JH6-R1a and JH5-R1a.
The reciprocal IGH-MALT1 fusion products (DH-MALT1) were amplified with the use of the AccuPrime Pfx DNA Polymerase (Invitrogen) and the MALT1-specific primer MALT1-F01a in combination with 1 of a set of 7 specific DH primers D1 to D7,9 followed by a nested PCR with MALT1-F01b with one of the nested D1N1 to D7N1 specific primers, as previously described.9
The products were amplified, sequenced twice in both directions, and analyzed with the use of the BLAST algorithm at National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast).
The presence of T-nucleotides at the IGH-MALT1 junctions was investigated by using the statistical approach described by Jäger et al.9 The de novo nucleotide additions found at the breakpoints were analyzed by using a binomial test. A P value of no more than .05 was selected as the point of high statistical significance.9,10
Amplification of the IGH-MALT1 products on paraffin-embedded tissue
For the amplification of the IGH-MALT1 fusion products in cases 3 to 5, DNA was isolated from formalin-fixed, paraffin-embedded sections, and PCR was performed with the use of the AccuPrime Pfx DNA Polymerase (Invitrogen). Pairs of nested MALT1 primers were designed to hybridize close to the MALT1 breakpoints detected in cases 1 and 2 and to enable the amplification of small-sized DNA fragments. For the detection of the MALT1-JH fusion products, the nested primers MALT1-R2a and MALT1-R2b (in cases 3 and 4) and the nested primers MALT1-R3a and MALT1-R3b (in case 5) were used combined with IGH primers JHEx-B and JHCo-B, described elsewhere.9
The products were amplified, sequenced, and analyzed as described above. For the amplification of the DH-MALT1 junctions different primer combinations were tested, but no fusion product could be amplified in these patients (cases 3-5).
Primer sequences
MALT1-JH fusion products were as follows: MALT1-R1a, 5′-CAGGGGCGGAAGAACAAATC-3′; MALT1-R1b, 5′-CCAATTAGACAGGTTTGCCGAG-3′; MALT1-R2a, 5′-CTTCTTAGAAAAGTCTGCCATGTA-3′; MALT1-R2b, 5′-AGAAAAGTCTGCCATGTACAAAG-3′; MALT1-R3a, 5′-GTCTTTACTGGAATCCTGC-3′; MALT1-R3b, 5′-TTACTGGAATCCTGCCTTT-3′; JH6-R1a, 5′-CATTCTTACCTGAGGAGACGGTG-3′; JH5-R1a, 5′-CTTTCTTTCCTGACCTCCAAAATG-3′.
DH-MALT1 fusion products were as follows: MALT1-F01a, 5′-ATGCCCATAAGAGAACCACAGG-3′; MALT1-F01b, 5′-GCGGGTAGTGACTGGATAGG-3′.
Results
We herein report the molecular characterization of the t(14;18)/IGH-MALT1 fusions in 5 new cases of MALT lymphomas: 2 originating from the skin, 1 from the conjunctiva, 1 from the lung, and 1 from the orbit. So far, molecular genetic data of one case representing a MALT lymphoma of the conjunctiva harboring the t(14;18)/IGH-MALT1 has been published.7
Cytogenetic data were available in one of our cases (case 2) and showed the t(14;18)/IGH-MALT1 accompanied by trisomies of chromosomes 3, 5, 8, and 12. In the case reported by Sánchez-Izquierdo et al,7 the t(14;18)/IGH-MALT1 occurred together with a trisomy 3 (case 6 in Table 2).
Sequences at the direct breakpoint junctions of the MALT1-JH fusion products
| Cases (organ)* . | Processing of ends† . | MALT1 sequence (18q21)‡ . | De novo nucleotide additions (D regions) . | JH sequence (14q32)‡ . | JH . | Processing of ends . |
|---|---|---|---|---|---|---|
| Germline | 27462TTTACTGGAATCCTGCCTTTTAATTTAAATTCAAC | ACTACTTTGACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | |||
| Germline | 27541CAAAGCGTTGCTTATGCAACCTCTCGTGTGCATAC | ACAACTGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH5 | |||
| Case 1 (conjunctiva) | +8 bp | 27541CAAAGCGTTGCTTATGCAACCTCTCGTGTGCAT | CCTAAGGGTTCGTGGGGCCGGGAGCCCAGGTC | GACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −8 bp |
| Case 2 (lung) | −4 bp | 27541CAAAGCGTTGCTTATGCAACCTtTCG | ACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −9 bp | |
| Case 3 (orbit) | Unknown | 27541CAAtGCGTTGCTTATGCAACCTCTC | AGTCGGTCTCGGCGCGCGA | TTGACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −6 bp |
| Case 4 (skin) | Unknown | 27541CAAAGCGTTGCTTATGCAACCTCT | ATGGTTTTGGC | CTGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH5 | −4 bp |
| Case 5 (skin) | Unknown | 27462TTTACTGGAATCCTGCCTTTTAATTT | CAGCGCGGCCGCGGCCCAGCCGGCCTATTA | AACCCaGGTCACCGTCTCCTCAGGT | JH5 | −28 bp |
| Case 6 (orbit) | Unknown | 27541CAAAGCGTTGCTTATGCA | GAGT | AACTgGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTtCTCAGGT | JH5 | −2 bp |
| Cases (organ)* . | Processing of ends† . | MALT1 sequence (18q21)‡ . | De novo nucleotide additions (D regions) . | JH sequence (14q32)‡ . | JH . | Processing of ends . |
|---|---|---|---|---|---|---|
| Germline | 27462TTTACTGGAATCCTGCCTTTTAATTTAAATTCAAC | ACTACTTTGACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | |||
| Germline | 27541CAAAGCGTTGCTTATGCAACCTCTCGTGTGCATAC | ACAACTGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH5 | |||
| Case 1 (conjunctiva) | +8 bp | 27541CAAAGCGTTGCTTATGCAACCTCTCGTGTGCAT | CCTAAGGGTTCGTGGGGCCGGGAGCCCAGGTC | GACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −8 bp |
| Case 2 (lung) | −4 bp | 27541CAAAGCGTTGCTTATGCAACCTtTCG | ACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −9 bp | |
| Case 3 (orbit) | Unknown | 27541CAAtGCGTTGCTTATGCAACCTCTC | AGTCGGTCTCGGCGCGCGA | TTGACTACTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH4 | −6 bp |
| Case 4 (skin) | Unknown | 27541CAAAGCGTTGCTTATGCAACCTCT | ATGGTTTTGGC | CTGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTCCTCAGGT | JH5 | −4 bp |
| Case 5 (skin) | Unknown | 27462TTTACTGGAATCCTGCCTTTTAATTT | CAGCGCGGCCGCGGCCCAGCCGGCCTATTA | AACCCaGGTCACCGTCTCCTCAGGT | JH5 | −28 bp |
| Case 6 (orbit) | Unknown | 27541CAAAGCGTTGCTTATGCA | GAGT | AACTgGGTTCGACCCCTGGGGCCAGGGAACCCTGGTCACCGTCTtCTCAGGT | JH5 | −2 bp |
Case 6 corresponds to case 1 of the study of Sánchez-Izquierdo et al.7
Status of coding ends at IGH locus or MALT1 end processing (−bp indicates deletion; +bp, addition).
Numbers in superscript in the MALT1 sequence indicate the position of the first nucleotide in BAC clone RP11-126O1 according to GenBank sequence AC104971. Mismatches to the germline are in lowercase letters. Point mutations are underlined and nucleotide insertions are specified above the line. These sequences are deposited at GenBank, accession nos. GQ406056, GQ406058, GQ406060, GQ406061, and GQ406062.
The presence of the t(14;18)/IGH-MALT1 fusion was confirmed by interphase FISH in all of our 5 cases. The number of MALT1/IGH-positive cells ranged from 6.6% to 70% (case 1, 50%; case 2, 54%; case 3, 70%; case 4, 7%; case 5, 8%).
In all 5 cases, we could confirm the presence of the IGH-MALT1 fusion by PCR. Even in DNA extracted from paraffin-embedded tissues (cases 3-5) and in cases with a low tumor cell infiltration (cases 4 and 5), we were able to amplify the IGH-MALT1 fusion products.
Characteristics of the breakpoints at the IGH locus
The sequencing of the PCR products showed a fusion of sequences upstream of the coding region of MALT1 to the JH segment of the IGH locus in all 5 cases. These results and the data of one other case published by Sánchez-Izquierdo et al7 are summarized in Table 2.
Our detailed analyses of the direct MALT1-JH breakpoints showed the typical features of a V(D)J-mediated recombination at the IGH locus. In our 5 cases and in the previously reported case, the breakage and rejoining occurred at the recombination signal sequence (RSS) adjacent to the join coding segments JH4 (cases 1, 2, and 3) and JH5 (cases 4, 5, and 6; Table 2). In these 6 cases, the t(14;18)/IGH-MALT1 occurred probably during the DH-to-JH joining before the complete VHDJH assembly.
In addition to the direct MALT1-JH products, we amplified and sequenced the reciprocal DH-MALT1 junctions in 2 patients (cases 1 and 2; Table 3; Figure 1). The sequencing showed a juxtaposition of MALT1 sequences to the diversity gene segments D2-15 in case 1 and D2-21 in case 2. Because the coding sequence of the DH segment present in case 1 consisted of a fragment of 2 nucleotides, the assignment to the D2-15 segment was performed according to the intronic sequence.
Sequences at the reciprocal breakpoint junctions of the DH-MALT1 fusion products
| Cases . | DH . | Processing of ends* . | DH sequence (14q32) . | De novo nucleotide additions† . | MALT1 sequence (18q21)‡ . | Processing of ends* . |
|---|---|---|---|---|---|---|
| Germline | D2-15 | AGGATATTGTAGTGGTGGTAGCTGCTACTCC | 27556GCAACCTCTCGTGTGCATACGAGTTAAATG27585 | |||
| Germline | D2-21 | AGCATATTGTGGTGGTGACTGCTATTCC | ||||
| Case 1 | D2-15 | −29 bp | CCAGTAGGGACAGGAGGATTTTGTGGGGGCTCGTGTCACTGTGAG | CAGAACCCGT | GTGTGCATACGAGTTAAATG27585 | +8 bp |
| Case 2 | D2-21 | −13 bp | AGCATATTGTGGTGG | AAACT | CATACGAGTTAAATG27585 | −4 bp |
| Cases . | DH . | Processing of ends* . | DH sequence (14q32) . | De novo nucleotide additions† . | MALT1 sequence (18q21)‡ . | Processing of ends* . |
|---|---|---|---|---|---|---|
| Germline | D2-15 | AGGATATTGTAGTGGTGGTAGCTGCTACTCC | 27556GCAACCTCTCGTGTGCATACGAGTTAAATG27585 | |||
| Germline | D2-21 | AGCATATTGTGGTGGTGACTGCTATTCC | ||||
| Case 1 | D2-15 | −29 bp | CCAGTAGGGACAGGAGGATTTTGTGGGGGCTCGTGTCACTGTGAG | CAGAACCCGT | GTGTGCATACGAGTTAAATG27585 | +8 bp |
| Case 2 | D2-21 | −13 bp | AGCATATTGTGGTGG | AAACT | CATACGAGTTAAATG27585 | −4 bp |
Status of coding ends at IGH locus or MALT1 end processing (−bp indicates deletion; +bp, addition).
Numbers in superscript in the MALT1 sequence indicate the position of the first nucleotide in BAC clone RP11-126O1 according to GenBank sequence AC104971.
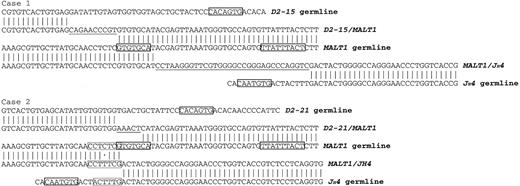
Comparison of the DNA sequences at the direct MALT1-JH and reciprocal DH-MALT1 breakpoint junctions and the IGH and MALT1 germline. The IGH-MALT1 fusion products of the cases 1 and 2 are aligned with germline sequences of IGH (GenBank accession no. X97051.1) and MALT1 (GenBank accession no. AC104971). Nucleotide homologies to the germline are represented by vertical lines and mismatches by dots. De novo nucleotide additions are underlined. Solid boxes indicate RSS heptamers in the JH and DH segment germlines and a cryptic RSS heptamer and nonamer in the MALT1 germline. Dashed line boxes in case 2 indicate the 6-bp microhomologous sequence flanking the translocation breakpoint.
Comparison of the DNA sequences at the direct MALT1-JH and reciprocal DH-MALT1 breakpoint junctions and the IGH and MALT1 germline. The IGH-MALT1 fusion products of the cases 1 and 2 are aligned with germline sequences of IGH (GenBank accession no. X97051.1) and MALT1 (GenBank accession no. AC104971). Nucleotide homologies to the germline are represented by vertical lines and mismatches by dots. De novo nucleotide additions are underlined. Solid boxes indicate RSS heptamers in the JH and DH segment germlines and a cryptic RSS heptamer and nonamer in the MALT1 germline. Dashed line boxes in case 2 indicate the 6-bp microhomologous sequence flanking the translocation breakpoint.
Further features at the MALT1-JH and DH-MALT1 junctions indicate that the V(D)J recombination machinery is responsible for the breaks at the IGH locus in the t(14;18)/IGH-MALT1. In the processing steps during the V(D)J recombination, antigen variability is introduced by nucleotide deletions at the coding ends and addition of nucleotides at the point of joining that were not present in the germline.14 Certainly, nucleotide deletions and/or de novo nucleotide additions (N-nucleotides) were also observed in the direct MALT1-JH junctions of all cases (Table 2). Similar to the direct breakpoints, the reciprocal DH-MALT1 breakpoints in cases 1 and 2 showed also deletions and additions of residues, resembling the processing of coding ends of V(D)J recombination products (Table 3).
T-nucleotides are present in the t(14;18)/IGH-MALT1 fusion products
T-nucleotides have recently been identified in the translocations t(14;18)/IGH-BCL2 in FL and t(11;14)/CCND1-IGH in MCL and consist of short copies of at least 5 nucleotides of the adjacent flanking sequences inserted in the breakpoints.9,10 Therefore, we investigated our cases with de novo nucleotide additions of at least 5 nucleotides at the breakpoints (cases 1-5; Tables 2–3) and examined their sequences for the presence of T-nucleotides. Interestingly, we identified T-nucleotides with highly significant homologies to the join coding segments involved in the translocation in all 5 cases (Table 4).
Templated nucleotide additions identified at the breakpoint junctions of the t(14;18)/IGH-MALT1
| Case no. . | De novo nucleotide additions* . | Breakpoint . | Template . | P . |
|---|---|---|---|---|
| 1 | TGGGGCCaGGGAgCCCaGGTCGAACCCgT | MALT1-JH | JH4 | |
| DH-MALT1 | < .001 | |||
| 2 | AAACT | DH-MALT1 | JH4 | .041 |
| 3 | GCCAGaGaaCCgCtGcgC | MALT1-JH | JH4 | < .001 |
| 4 | AcTGGTTttGgC | MALT1-JH | JH5 | .047 |
| 5 | CGACCCctGG | MALT1-JH | JH5 | < .001 |
| Case no. . | De novo nucleotide additions* . | Breakpoint . | Template . | P . |
|---|---|---|---|---|
| 1 | TGGGGCCaGGGAgCCCaGGTCGAACCCgT | MALT1-JH | JH4 | |
| DH-MALT1 | < .001 | |||
| 2 | AAACT | DH-MALT1 | JH4 | .041 |
| 3 | GCCAGaGaaCCgCtGcgC | MALT1-JH | JH4 | < .001 |
| 4 | AcTGGTTttGgC | MALT1-JH | JH5 | .047 |
| 5 | CGACCCctGG | MALT1-JH | JH5 | < .001 |
Mismatches to the germline are in lowercase letters. Point mutations are underlined, nucleotide insertions are specified above the line, and deletions are specified below the line. The sequence shown is the one of the copy.
In case 1, for example, T-nucleotides were found both in the direct and reciprocal fusion products. The JH4 segment involved in the IGH-MALT1 fusion served as template. In the direct junction, the homology sequence consisted of a fragment 21 basepairs (bp) in length with 3 mismatches. Eight of these 21 bp with 1 mismatch were also present in the reciprocal junction (Table 4). Taken together, in our cases and the published case, T-nucleotides were observed in 5 of 6 cases.
Characteristics of the breakpoints at the MALT1 gene
At the MALT1 locus, the analysis of the direct MALT1-JH sequences showed that all 5 breaks were located in the 5′ noncoding region of MALT1 on chromosome 18 (Table 2). The same was found for the published case.7 In all cases, the breakpoints were clustered in a 87-bp region between nucleotides 27.489 and 27.573, according to GenBank sequence AC104971 (BAC clone RP11-126O1, which contains the 5′ end of MALT1).
This is the first report that describes the presence of a putative major breakpoint region (MBR) for the t(14;18)/IGH-MALT1 in the 5′ noncoding region of the MALT1 gene.
The analysis of the reciprocal DH-MALT1 junctions could be performed in 2 of our cases. One case (case 2) showed a deletion of 4 nucleotides of MALT1. The other case (case 1) showed a duplication of 8 MALT1 nucleotides with the sequence GTGTGCAT found in the direct and the reciprocal IGH-MALT1 fusion (Table 3; Figure 1).
We identified one cryptic RSS sequence in the putative MALT1 MBR. This MALT1 RSS-like sequence fulfilled almost all requisites for its recognition by the recombination activating gene-1 (RAG-1)/RAG-2. It consisted of a heptamer with the essential trinucleotide CAC at the first 3 positions and an A residue at the fourth position, a 23-bp spacer, and a nonamer with a core of 3 consecutive A residues in the positions 4, 5, and 6 flanked by 2 residues other than A (Figure 1).15 However, this cryptic RSS was present in the 3′ to 5′ opposite orientation.
JH and DH utilization and nucleotide mutations at the breakpoints of the t(14;18)/IGH-MALT1
We have further investigated whether MALT lymphomas show a preferential use of determined JH and DH segments and/or the presence of somatic mutations as it has been described in FL and MCL.9,10 We observed that exclusively the JH4 and JH5 gene segments were used in the t(14;18)/IGH-MALT1 (Table 2). The rearranged DH genes were determined in 2 cases and showed a biased usage of the D2 family of the DH segments, D2-15 and D2-21 (Table 3).
We found mismatches to the germline close to the breakpoints in the sequences of the IGH and MALT1 genes in 4 patients (Table 2).
We detected IGH mutations in 2 cases with a C-to-T transition and an insertion of one G residue into the JH5 segment (case 6) and a T-to-A transversion into the JH5 segment (case 5) (Table 2).
MALT1 mutations were seen in 2 further cases with an A-to-T transversion (cases 2 and 4). In case 2, the origin of the sequence CTTTCG found at the direct breakpoint is ambiguous and can either be assigned to the MALT1 sequence CTCTCG with a C-to-T transition or to the IGH sequence CTTTG with a C insertion (Figure 1).
Discussion
The t(14;18)(q32;q21) involving the IGH and the MALT1 genes is a recurrent translocation in MALT lymphomas. However, since its identification 6 years ago, the molecular characteristics of this translocation have not been elucidated. This may be due to the relatively low incidence of this translocation and to the difficulties to obtain a sufficient amount of tumor tissue from these rare extranodal lymphomas.1,3,8
In this study, we performed a sequence analysis of the IGH-MALT1 junctions in 5 MALT lymphomas harboring the t(14;18)/IGH-MALT1. The results of our investigation strongly indicate that a failure in the V(D)J-mediated recombination process is responsible for the generation of breaks at the IGH locus in the t(14;18)/IGH-MALT1: the breakpoints in IGH were located at a RSS, and coding end processing, such as nucleotide deletions and addition of N-nucleotides, was detected in all cases.14,16 Similar findings have been published for other translocations associated with V(D)J recombination, such as the t(14;18)/IGH-BCL2 in FL and the t(11;14)/CCND1-IGH in MCL.9,10,17
On chromosome 18, all the breakpoints clustered in a 87-bp region in the 5′ noncoding region of MALT1. This clustering of breakpoints on chromosome 18 suggests the presence of a MBR in the MALT1 gene, as it occurs for BCL2 in t(14;18)/IGH-BCL2 and for CCND1 in the t(11;14)/CCND1-IGH.18,19
Interestingly, we observed one cryptic RSS sequence in the putative MALT1 MBR. However, several observations indicate that a RSS-guided V(D)J recombination is not responsible for the breaks at the MALT1 locus. First, the essential trinucleotide 5′CAC3′ of the heptamer and the nonamer sequence of this cryptic RSS were present in the 3′ to 5′ opposite orientation.15 Second, one of the breakpoints in MALT1 is located 75-bp downstream of this pseudo signal. Third, the IGH-MALT1 fusion sequence in one case showed a duplication of 8 nucleotides of MALT1, which were present both at the direct and reciprocal fusion products.
The presence of duplications is not consistent with the V(D)J recombination mechanisms. Duplications of sequences are supposed to be generated by a staggered double-stranded DNA break mechanism that is independent of the V(D)J recombination. Similar duplications have also been reported in the BCL2 and CCND1 genes in the IGH-BCL2 and CCND1-IGH translocations.9-11,20
Therefore, in the absence of any convincing recombination signals in the MALT1 locus, the definitive mechanisms of DNA recognition and cleavage leading to breaks on chromosome 18 remain to be elucidated.
In FL, most breaks on chromosome 18 cluster in a region of BCL2 known as MBR without a functional RSS sequence.13,17,18,21 Recent reports have shown that the RAGs recognize a specific “non-B DNA” conformation of the BCL2-MBR even in the absence of an RSS sequence.11,21,22 Therefore, the RAGs are supposed to be responsible for both cleavage at the IGH and the BCL2 loci by recognition of 2 different motifs, a sequence-dependent RSS signal at IGH and a non-B DNA structure at BCL2.22 It can be speculated if similar mechanisms play a role in the generation of breaks in MALT1 in the t(14;18)/IGH-MALT1.
Another novel finding of our study is the presence of T-nucleotides at the IGH-MALT1 junctions, which we have detected in all of our 5 analyzed cases. T-nucleotides have so far been reported in the translocations t(14;18)/IGH-BCL2 and t(11;14)/CCND1-IGH and consist of insertions of short copies of the surrounding JH, DH, and BCL2, or CCDN1 sequences at the breakpoints.9,10
The ascertainment of T-nucleotides has provided new insights into the mechanisms of IGH-associated translocations, in which processes other than illegitimate V(D)J-mediated recombination might be involved.23 For instance, T-nucleotides exhibit such a high homology to their template sequences that their synthesis by the template-independent terminal deoxynucleotidyl transferase of the V(D)J machinery is unlikely. Accordingly, T-nucleotides have not been described in normal V(D)J recombination products and RSS-guided V(D)J-mediated translocations.20,23 Moreover, the template sequences do not always border on the breakpoints, and mismatches with the germline have been observed, indicating that T-nucleotides are generated by an error-prone and template-dependent DNA polymerase.
T-nucleotides are not exclusively present in chromosomal translocations, but they have also been identified at the junctions of end-joining events in Drosophila. However the molecular mechanism that create T-nucleotides remains to be determined.24 On the basis of studies from flies and other organisms, McVey and Lee25 speculate that T-nucleotides might be generated by the action of an error-prone polymerase in an attempt to create microhomologies when flanking microhomologous sequences are inadequate. Furthermore, the investigators suggest that a nonhomologous end joining (NHEJ)–independent mechanism named microhomology-mediated end joining might be responsible for repairing of these DNA double-strand breaks in human neoplasias.25 In line with this hypothesis, all IGH-MALT1 junctions but one analyzed in this study displayed nucleotides insertions. The only IGH-MALT1 junction that did not show this feature contained a 6-bp imperfect microhomology sequence at the translocation breakpoint of MALT1-IGH.
The cell of origin of MALT lymphomas is still debated and probably belongs to more than one stage of B-cell differentiation.26 Interestingly, 4 MALT lymphomas analyzed here showed a total of 5 somatic mutations in the rearranged MALT1 and IGH genes; however, all mutations but one were found within the 20 bp of the IGH-MALT1 breakpoint junctions. The only mutation located distal from the breakpoint, namely a C-to-T transition described in the case published by Sánchez-Izquierdo et al,7 did not show the target motifs for a somatic hypermutation.27 Therefore, the mutations detected in our study could result from error-prone repair by end-joining events rather than from somatic hypermutation.28
In conclusion, our observations indicate that the pathomechanism underlying the t(14;18)/IGH-MALT1 in MALT lymphomas is probably based on an illegitimate V(D)J-mediated recombination at the IGH locus on chromosome 14 and a staggered double-stranded DNA break at the MALT1 locus on chromosome 18. Furthermore, we could identify a putative MBR proximal to the 5′ noncoding region of MALT1. The molecular characteristics of the t(14;18)/IGH-MALT1 resemble those found in the t(14;18)/IGH-BCL2 in FL and t(11;14)/CCND1-IGH in MCL, suggesting that these translocations could be generated by a common pathomechanism involving new synthesis of T-nucleotides and NHEJ or alternative NHEJ repair pathways.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by Deutsche Krebshilfe grant 106092 (J.D.).
Authorship
Contribution: E.M.M.P. designed the research, performed FISH experiments and molecular genetic analyses, analyzed the data, and wrote the manuscript; E.C.-B., S.G., F.B., and G. Salles performed cytogenetic analysis and contributed patients' data; H.Y. and M.-Q.D. performed FISH experiments and contributed patients' data; G. Schilling and N.A.-K. performed FISH experiments and molecular genetic analyses; E.V. performed statistical analyses; I.W. contributed patients' data; C.B. supervised research and wrote the manuscript; and J.D. designed and supervised the research, and wrote the manuscript. All authors have read and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eva Maria Murga Penas, Department of Hematology and Oncology, BMT with Section of Pneumology, Hubertus Wald Tumorzentrum-University Cancer Center Hamburg (UCCH), University Medical Center Hamburg-Eppendorf, Martinistr 52, 20246 Hamburg, Germany; e-mail: murga@uke.uni-hamburg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal