After DNA replication, sister chromatids must be untangled, or decatenated, before mitosis so that chromatids do not tear during anaphase. Topoisomerase IIα (Topo IIα) is the major decatenating enzyme. Topo IIα inhibitors prevent decatenation, causing cells to arrest during mitosis. Here we report that acute myeloid leukemia cells fail to arrest at the mitotic decatenation checkpoint, and their progression through this checkpoint is regulated by the DNA repair component Metnase (also termed SETMAR). Metnase contains a SET histone methylase and transposase nuclease domain, and is a component of the nonhomologous end-joining DNA double-strand break repair pathway. Metnase interacts with Topo IIα and enhances its decatenation activity. Here we show that multiple types of acute leukemia cells have an attenuated mitotic arrest when decatenation is inhibited and that in an acute myeloid leukemia (AML) cell line this is mediated by Metnase. Of further importance, Metnase permits continued proliferation of these AML cells even in the presence of the clinical Topo IIα inhibitor VP-16. In vitro, purified Metnase prevents VP-16 inhibition of Topo IIα decatenation of tangled DNA. Thus, Metnase expression levels may predict AML resistance to Topo IIα inhibitors, and Metnase is a potential therapeutic target for small molecule interference.
Introduction
DNA double-strand breaks (DSBs) can result from normal cellular events, such as recombination during immune receptor rearrangement, recovery of stalled replication forks, replication of nicked templates, or failed decatenation.1 Such DSBs can result in translocations or deletions, and this genomic instability can lead to malignancy. Of the normal cell processes producing DSBs, the least well understood is chromosome decatenation. Sister chromatids become intertwined, or catenated, when chromosomes are replicated during DNA synthesis. When decatenation fails, chromatids can tear during anaphase, producing DSBs. Catenation status is actively monitored at 2 points in the cell cycle. One decatenation checkpoint blocks progression from G2 to M2, and another blocks progression from metaphase to anaphase during mitosis.2,,,–6 These checkpoints are conserved in most organisms, including yeast.7,8 Previously, it was reported that bladder and lung cancer cells lack decatenation checkpoints and proceed through mitosis even when decatenation is inhibited.9,10 It is possible that this failure of arrest at the decatenation checkpoints could be a general feature of malignancy, including acute leukemia. The critical decatenating enzyme is Topoisomerase IIα (Topo IIα).2,,,–6,11 Topo IIα inhibitors can trigger decatenation checkpoint arrest in normal cells at either of the 2 decatenation checkpoints. Recently, we identified and characterized a novel DNA repair protein called Metnase (also SETMAR). Metnase has an amino terminal SET histone methylase domain and a carboxy terminal transposase/nuclease domain. We previously reported that Metnase promotes nonhomologous end-joining of DSBs and methylates histone 3 lysine 36.12 Metnase appears to be localized to DSBs by interaction with Pso4, a poorly characterized DNA repair component.13 Metnase may promote DSB repair by interacting with DNA ligase IV, the final component of the nonhomologous end-joining pathway.14 Metnase is present only in primates and exhibits only partial transposase activity.12,14,,,,–19 We also found that Metnase has endonuclease activity specific for supercoiled DNA and improves Topo IIα-mediated decatenation in vitro and in vivo in noncancerous human cells.16,20 In this study, we hypothesized that Metnase mediates decatenation in acute leukemia cells. If there is a failure of decatenation arrest in acute leukemia cells when Topo IIα is inhibited, then perhaps Metnase mediated the continued progression of those cells through the decatenation checkpoints
We report here that both acute myeloid and acute lymphoblastic leukemia (AML and ALL) cells fail to arrest in mitosis when Topo IIα is inhibited. In AML, we show that Metnase levels mediate progression through mitosis by enhancing Topo IIα function. Most importantly, we found that Metnase mediates cell proliferation through and protects Topo IIα from VP-16, a Topo IIα inhibitor that is widely used in cancer treatment, including AML salvage therapy. This implies that Metnase is a critical mediator of leukemia resistance to the cytotoxic effects of Topo IIα inhibitors.
Methods
Western analysis of Metnase and Topo IIα expression
All leukemia cell lines were grown in RPMI media (HyClone) fully supplemented with 1% antimycotic/antibiotic (Cellgro), and 10% fetal bovine serum (Atlanta Biologicals). HEK-293T cells were cultured in fully supplemented Dulbecco modified Eagle medium (HyClone). Human CD34+ hematopoietic progenitors were purchased from the National Heart, Lung, and Blood Institute Programs of Excellence in Gene Therapy program at the Fred Hutchison Cancer Center (Seattle, WA), and cultured in StemSpan H3000 media (StemCell Technologies) supplemented with StemSpan CC100 (StemCell Technologies) for 5 days, incubated with 10 ng/mL granulocyte-macrophage colony-stimulating factor (Leukine; Bayer Healthcare Pharmaceuticals) for 24 hours as described previously.21 Western blot analysis was performed on total protein lysates using antisera specific for Metnase (1:4000; Lampire),12 Topo IIα (1:2000; Topogen),20 and finally β-actin (1:10 000; Sigma-Aldrich) as the loading control.
Manipulating Metnase levels
THP-1 cells were stably transfected with a control shRNA plasmid, pRS-shGFP, which expressed an shRNA against green fluorescent protein, or a combination of 3 shRNAs against Metnase, targeting nucleotides 1198 to 1226, 1270 to 1298, and 1800 to 1828 (from NM 006515) expressed from pRS (Origene). Stable transfectants were selected with 1 μg/mL puromycin and then cloned by limiting dilution. Metnase levels were assessed using reverse-transcribed polymerase chain reaction (RT-PCR) and Western blotting as we described.12,22 Each experiment using shRNA repressed Metnase was performed in at least 2 independent clones, with nearly identical results.
Metnase/Topo IIα coimmunoprecipitation
Coimmunoprecipitation of Metnase and Topo IIα (anti–Topo IIα from Topogen) was performed as follows: cells were lysed with radio immunoprecipitation assay (RIPA) buffer, and protein extracts precleared using 40 μL protein A/G beads. A total of 2 mg protein extract was used for each immunoprecipitation; 2 μL anti-V5 (Invitrogen), 2 μL anti-Metnase, or 2.5 μL anti–Topo IIα (Topogen) antibodies was used in a total volume of 500 μL. Lysates were then incubated overnight in 500 μL RIPA buffer with appropriate antibody at 4°C in the presence of 1U DNase I. A total of 30 μL of protein A/G beads was then added and incubated for 2 hours at 4°C. The beads were washed 3 times with RIPA buffer and twice with phosphate-buffered saline. The beads were then boiled, samples were run on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, and analyzed by Western blot using appropriate antisera. The input represents 1% of the total lysate for all experiments.
Mitotic arrest analysis
ICRF-193 inhibits Topo IIα without producing DNA damage, producing cell cycle arrest because of failed decatenation, and not from DNA damage. Three techniques were used to measure mitotic arrest after ICRF-193 exposure. Each of these techniques is complementary to the others. Immunofluorescence of tubulin condensation into a mitotic spindle and 4′-6-diamidino-2-phenylindole (DAPI)–stained chromosome alignment in a metaphase plate measured cells in metaphase as we described.20 Immunofluorescence of Mitotic Protein Monoclonal 2 (MPM-2), a mitotic marker,23 measured cells in mitosis. Flow cytometry using propidium iodide measured cells in G2/M phase of the cell cycle as we described.24 Briefly, unsynchronized cells were treated with 10, 25, or 100 μM ICRF-193 (MP Biomedicals) or equivalent volume of dimethyl sulfoxide (DMSO; vehicle control) for the required time points, harvested, cytospun, fixed with ice-cold methanol, and stained with anti-tubulin antibody conjugated to fluorescein isothiocyanate (FITC; Abcam) and/or anti–MPM-2 conjugated to Texas Red (Upstate Biotechnology) for 2 hours at room temperature. Cells were counterstained with DAPI to visualize nuclear morphology. Interphase and metaphase cells were counted by immunofluorescence microscopy using a Zeiss Confocal fluorescence microscope with 63× objectives equipped with laser and filter settings for DAPI, FITC, and Texas Red. The increase in cells in mitosis with ICRF-193 over the vehicle controls reflects mitotic decatenation arrest.3,,,–7 Each immunofluorescence experiment was performed in triplicate, and more than 350 cells were counted per condition. Statistical analyses were performed using Student t tests, and P values are as indicated.
Analysis of apoptosis
Apoptosis was analyzed by flow cytometric measurement of annexin-V expression in vector control or shRNA Metnase-transduced THP-1 cells treated with 0.5 μM VP-16 for 24 hours. The annexin V–FITC Apoptosis Detection Kit I (BD Biosciences PharMingen) was used.
Kinetoplast DNA decatenation
Purified recombinant Topo IIα (GE Healthcare) and catenated kinetoplast DNA (kDNA; Topogen) were used according to the manufacturer's instructions. Recombinant Metnase was purified as we described.12 kDNA (200 ng/μL) was incubated in the manufacturer's buffer with increasing concentrations of Metnase and/or Topo IIα for 1 hour at 37°C per the manufacturer's instructions.20 VP-16 or its buffer was added to reactions at a final concentration of 0.5 μM. kDNA decatenation was then visualized by 1% agarose gel electrophoresis in ethidium bromide and quantified by densitometry using ImageJ software (http://rsbweb.nih.gov/ij/).
Results
Metnase and Topo IIα interact in acute leukemia cells
We first examined whether the DNA repair component Metnase was expressed in 4 human acute leukemia cells and normal human CD34+ hematopoietic progenitors using Western blot analysis. We also measured the presence of Topo IIα in these lines. Normal CD34+ hematopoietic progenitors and the leukemia lines expressed both Metnase and Topo IIα at various levels (Figure 1A). Metnase can have 2 bands on Western analysis as it can be phosphorylated, as seen in the leukemia cell lines. HEK-293T cells express no Metnase, based on their transformation by SV-40 T antigen, and thus serve as a control.
Topo IIα and Metnase are expressed and interact in blood cells. (A) Western blot of multiple acute leukemia cell lines and normal CD34+ hematopoietic progenitors to show expression of Metnase and Topo IIα. HEK-293T cells do not express Metnase by virtue of their transformation by T antigen. They serve as a control for these studies. (B) Metnase and Topo IIα coimmunoprecipitated with each other from THP-1 AML cells in the presence of DNase I, indicating that their interaction is independent of DNA. (C) Metnase shRNA reduces Metnase mRNA in THP-1 AML cells as shown by RT-PCR. (D) Metnase shRNA reduces the expression of Metnase protein in THP-1 cells as shown by Western analysis.
Topo IIα and Metnase are expressed and interact in blood cells. (A) Western blot of multiple acute leukemia cell lines and normal CD34+ hematopoietic progenitors to show expression of Metnase and Topo IIα. HEK-293T cells do not express Metnase by virtue of their transformation by T antigen. They serve as a control for these studies. (B) Metnase and Topo IIα coimmunoprecipitated with each other from THP-1 AML cells in the presence of DNase I, indicating that their interaction is independent of DNA. (C) Metnase shRNA reduces Metnase mRNA in THP-1 AML cells as shown by RT-PCR. (D) Metnase shRNA reduces the expression of Metnase protein in THP-1 cells as shown by Western analysis.
We next tested whether Metnase and Topo IIα could interact in the AML cell line THP-1 by coimmunoprecipitation. We found that Metnase was present in immunoprecipitates of Topo IIα and that Topo IIα was present in Metnase immunoprecipitates (Figure 1B). The interaction of Metnase with Topo IIα was not dependent on DNA, as the inclusion of DNase in the immunoprecipitate buffer did not affect the results.
Defective mitotic decatenation arrest in acute leukemia cells
The bisdioxopiperazines, including ICRF-193, inhibit Topo IIα after DNA religation and therefore do not produce significant DNA DSBs. The obscuring effects of DNA damage on the cell cycle are minimized, and the mitotic arrest induced by these drugs primarily results from the failure of chromosomal decatenation.2,,,–6 Thus, ICRF-193 can be used to assess whether cells progress through mitosis despite the inhibition of decatenation. Because bladder and lung cancer cells do not demonstrate decatenation arrest after ICRF-193 exposure,9,10 we hypothesized that acute leukemia cells would also fail to arrest in mitosis when Topo IIα is inhibited with ICRF-193. We tested this by exposing several acute leukemia cell lines to ICRF-193 and measuring the increase in the fraction of cells in mitosis by 3 complementary methods. We used flow cytometry to measure the increase in the fraction of cells in the G2/M phase of the cell cycle. We used immunofluorescence assessment of tubulin condensation and DAPI nuclear morphology to measure cells in metaphase because mitotic spindle formation and chromosome alignment can be seen with this technique. We also used immunofluorescence to assess the presence of MPM-2, which measures total mitotic cells. The increase in the fraction of cells in mitosis in the presence of ICRF-193 with vehicle controls subtracted indicates the fraction of cells arrested because of decatenation failure.3,,–6 We examined 2 AML lines, THP-1 and KG-1, and 2 ALL lines 697 and REH, and compared these with normal human CD34+ hematopoietic progenitors and HEK-293T cells. The HEK-293T cells arrest appropriately at mitosis when Topo IIα is inhibited with ICRF-193 and served as a control.20
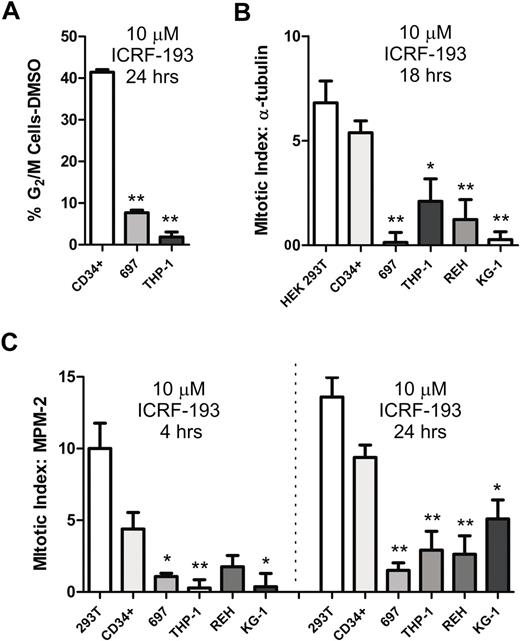
We first analyzed the ability of ICRF-193 to arrest CD34+ cells, 697 ALL and THP-1 AML cells in G2/M using flow cytometry (Figure 2A). We found that ICRF-193 increased the percentage of CD34+ cells in G2/M by 41%, whereas it increased the percentage of 697 cells by only 7.7% and THP-1 cells by 1.8%. Thus, there is a 5-fold increase in CD34+ cells in G2/M compared with 697 cells, and 22-fold increase compared with THP-1 cells, indicating that CD34+ cells are arrested significantly more efficiently by ICRF-193 inhibition of Topo IIα than acute leukemia cells.
Leukemia cells show reduced mitotic arrest when decatenation is inhibited. (A) Normal human CD34+ hematopoietic progenitors, 697 ALL, and THP-1 AML cells were treated with 10 μM ICRF-193 for 24 hours and then evaluated for the increase in G2/M cell-cycle fraction by propidium iodide staining for DNA content. (B) Mitotic arrest was induced by 10 μM ICRF-193 for 18 hours in CD34+ cells, 697 ALL and REH ALL cells, and THP-1 and KG-1 AML cells. HEK-293T cells, which do not express Metnase, served as a positive control for mitotic arrest after Topo IIα decatenation inhibition. Metaphase cells were imaged by tubulin immunofluorescence and DAPI nuclear morphology and quantified as a percentage of the total cell population with DMSO control subtracted. (C) Mitotic arrest induced by 10 μM ICRF-193 for 4 or 24 hours in CD34+ cells, 697 ALL and REH ALL cells, and THP-1 and KG-1 AML cells. HEK-293T cells, which do not express Metnase, served as a positive control for mitotic arrest after Topo IIα decatenation inhibition. Cells in mitosis were imaged by MPM-2 immunofluorescence and quantified as a percentage of the total population with DMSO control subtracted. The acute leukemia cell lines all have decreased mitotic arrest after Topo IIα inhibition with ICRF-193 compared with CD34+ cells. All experiments represent the average of at least 3 independent experiments, ± SEM. *Student t test (P < .05); **Student t test (P < .01).
Leukemia cells show reduced mitotic arrest when decatenation is inhibited. (A) Normal human CD34+ hematopoietic progenitors, 697 ALL, and THP-1 AML cells were treated with 10 μM ICRF-193 for 24 hours and then evaluated for the increase in G2/M cell-cycle fraction by propidium iodide staining for DNA content. (B) Mitotic arrest was induced by 10 μM ICRF-193 for 18 hours in CD34+ cells, 697 ALL and REH ALL cells, and THP-1 and KG-1 AML cells. HEK-293T cells, which do not express Metnase, served as a positive control for mitotic arrest after Topo IIα decatenation inhibition. Metaphase cells were imaged by tubulin immunofluorescence and DAPI nuclear morphology and quantified as a percentage of the total cell population with DMSO control subtracted. (C) Mitotic arrest induced by 10 μM ICRF-193 for 4 or 24 hours in CD34+ cells, 697 ALL and REH ALL cells, and THP-1 and KG-1 AML cells. HEK-293T cells, which do not express Metnase, served as a positive control for mitotic arrest after Topo IIα decatenation inhibition. Cells in mitosis were imaged by MPM-2 immunofluorescence and quantified as a percentage of the total population with DMSO control subtracted. The acute leukemia cell lines all have decreased mitotic arrest after Topo IIα inhibition with ICRF-193 compared with CD34+ cells. All experiments represent the average of at least 3 independent experiments, ± SEM. *Student t test (P < .05); **Student t test (P < .01).
Next, we analyzed the fractional increase of CD34+ and acute leukemia cells in metaphase after exposure to ICRF-193 using immunofluorescence of the condensation of tubulin into a mitotic spindle and alignment of chromosomes along a metaphase plate (Figures 3–4). After exposure to ICRF-193, there were 5.4% more CD34+ cells in metaphase, 0.1% more 697 cells, 2.1% more THP-1 cells, 1.2% more REH cells, and 0.3% more KG-1 cells (Figure 2B). It is not surprising that there is a lower percentage increase of cells using this method, as metaphase cells are only a small fraction of G2/M cells. Using this method, there were 2.6-fold more CD34+ cells in metaphase compared with THP-1 cells, 4-fold more compared with REH cells, 20-fold more compared with KG-1 cells, and 38-fold more compared with 697 cells. Each of these differences is statistically significant using the Student t test as indicated in Figure 2B. Thus, ICRF-193 arrested significantly more CD34+ cells in metaphase compared with all 4 acute leukemia lines. The fractional increase of CD34+ cells in metaphase after ICRF-193 is similar, although slightly lower, to that seen in HEK-293T cells, which have an intact mitotic decatenation arrest.20
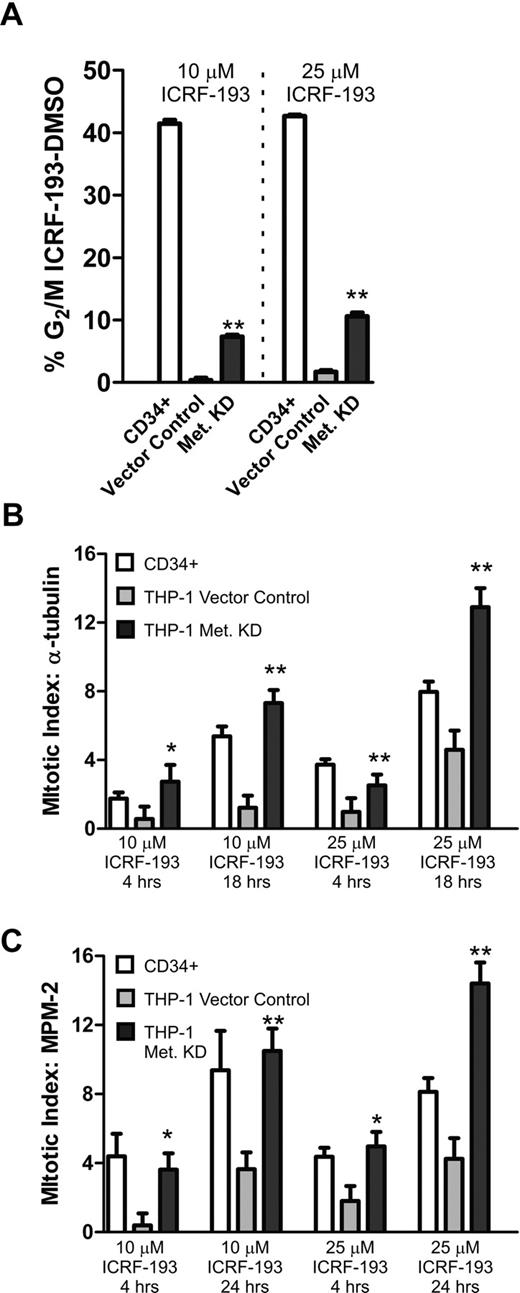
The reduction of Metnase restores mitotic arrest in THP-1 cells. (A) CD34+ progenitors and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector were treated with 10 or 25 μM ICRF-193 for 24 hours and then evaluated for the increase in G2/M cell-cycle fraction by propidium iodide DNA staining. (B) Mitotic arrest was induced by 10 or 25 μM ICRF-193 for 4 or 18 hours in CD34+ cells, and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector. Metaphase cells were imaged by tubulin immunofluorescence and DAPI nuclear morphology, and quantified as a percentage of the total cell population. (C) Mitotic arrest induced by 10 or 25 μM ICRF-193 for 4 or 24 hours in CD34+ cells, and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector. Cells in mitosis were imaged by MPM-2 immunofluorescence and quantified as a percentage of the total population. Repressing Metnase increased the mitotic arrest of the THP-1 cells after ICRF-193 exposure so that they approximated the arrest of CD34+ cells. All experiments represent the average of at least 3 independent experiments, ± SEM. *Student t test (P < .05); **Student t test (P < .01).
The reduction of Metnase restores mitotic arrest in THP-1 cells. (A) CD34+ progenitors and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector were treated with 10 or 25 μM ICRF-193 for 24 hours and then evaluated for the increase in G2/M cell-cycle fraction by propidium iodide DNA staining. (B) Mitotic arrest was induced by 10 or 25 μM ICRF-193 for 4 or 18 hours in CD34+ cells, and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector. Metaphase cells were imaged by tubulin immunofluorescence and DAPI nuclear morphology, and quantified as a percentage of the total cell population. (C) Mitotic arrest induced by 10 or 25 μM ICRF-193 for 4 or 24 hours in CD34+ cells, and THP-1 cells stably transduced with a vector control or the Metnase shRNA vector. Cells in mitosis were imaged by MPM-2 immunofluorescence and quantified as a percentage of the total population. Repressing Metnase increased the mitotic arrest of the THP-1 cells after ICRF-193 exposure so that they approximated the arrest of CD34+ cells. All experiments represent the average of at least 3 independent experiments, ± SEM. *Student t test (P < .05); **Student t test (P < .01).
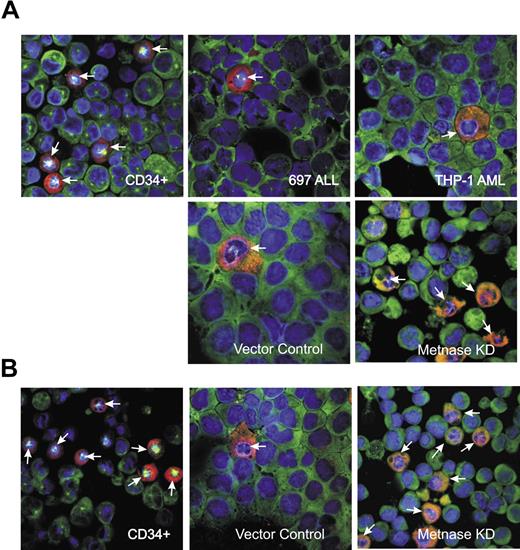
Photomicrographs showing that leukemia cells show a reduced mitotic arrest after ICRF-193 and restoration of mitotic arrest with reduced Metnase. (A) Images showing both tubulin (green) and MPM-2 (red) staining, as well as counterstaining with DAPI, after 10 μM ICRF treatment for 24 hours. There are more cells arrested in mitosis after ICRF-193 in the CD34+ cells than in the THP-1 AML cells. When Metnase is repressed in the THP-1 cells, the fraction of cells in mitosis after ICRF-193 exposure becomes similar to that seen with CD34+ cells. (B) Similar to panel A, but treated with 25 μM ICRF for 24 hours.
Photomicrographs showing that leukemia cells show a reduced mitotic arrest after ICRF-193 and restoration of mitotic arrest with reduced Metnase. (A) Images showing both tubulin (green) and MPM-2 (red) staining, as well as counterstaining with DAPI, after 10 μM ICRF treatment for 24 hours. There are more cells arrested in mitosis after ICRF-193 in the CD34+ cells than in the THP-1 AML cells. When Metnase is repressed in the THP-1 cells, the fraction of cells in mitosis after ICRF-193 exposure becomes similar to that seen with CD34+ cells. (B) Similar to panel A, but treated with 25 μM ICRF for 24 hours.
We next analyzed the fractional increase of CD34+ and acute leukemia cells in mitosis generally after exposure to ICRF-193 using immunofluorescence of MPM-2 expression (Figure 2C). After exposure to ICRF-193 for 24 hours, there were 9.4% more CD34+ cells in metaphase, 1.5% more 697 cells, 2.9% more THP-1 cells, 2.6% more REH cells, and 5.1% more KG-1 cells. In this assay, there were 1.8-fold more CD34+ cells in mitosis compared with KG-1 cells, 3.2-fold compared with THP-1 cells, 3.2-fold compared with REH cells, and 6.2-fold compared with 697 cells. Each of these differences is statistically significant using the Student t test as indicated in Figure 2C. The CD34+ cells arrest in mitosis in response to ICRF-193 somewhat less efficiently than the decatenation control HEK-293T cells. It should be noted that there was a progressive increase in mitotic arrest, ruling out checkpoint bypass, as measured by MPM-2 after ICRF-193 at 2, 4, 8, and 24 hours (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition, increasing ICRF-193 concentrations to very high levels did not quantitatively alter the decreased mitotic arrest of the acute leukemia cells compared with the CD34+ cells (supplemental Figure 2). Thus, acute leukemia cells do not become sensitive to ICRF-193 with increasing time or concentration. Representative photomicrographs are shown in Figure 4. In summary, each of these 3 assays demonstrated that, after Topo IIα inhibition with ICRF-193, human acute leukemia cells arrested in mitosis significantly less than normal CD34+ progenitors.
Reducing Metnase levels restores mitotic decatenation arrest in the AML line THP-1
In Figure 2, we found that acute leukemia cells did not arrest in mitosis when Topo IIα was inhibited. We had previously found that Metnase enhanced Topo IIα activity.20 Therefore, we next investigated whether reducing Metnase protein levels in the AML cell line THP-1 would affect this progression through mitosis despite Topo IIα inhibition. Metnase protein levels were reduced by at least 90% in THP-1 cells stably expressing Metnase shRNAs, as shown by RT-PCR and Western analysis (Figure 1C-D). We then examined these THP-1 cells with reduced Metnase levels for their ability to progress through mitosis when Topo IIα was inhibited using the same 3 assays. Using flow cytometry to assess the fraction of cells in G2/M after exposure to ICRF-193, we found that the repression of Metnase decreased progression through mitosis, but not to the same extent as seen in CD34+ cells. Vector control-transduced THP-1 cells (with normal Metnase levels) had 1.7% cells in G2/M, whereas CD34+ cells had 42% cells in G2/M. Repressing Metnase levels significantly increased THP-1 cells in G2/M by 6-fold, to 10.6% (Figure 3A). Next, using immunofluorescence to measure condensed tubulin to assess cells in metaphase after ICRF-193 exposure for 18 hours, we found that repressing Metnase increased arrested cells from 1.2% in the vector control to 7.3%, a 6-fold increase. Of note, there were 5.4% CD34+ cells arrested in metaphase in this assay. Thus, repression of Metnase completely restored metaphase arrest to ICRF-193 (Figure 3B). Finally, we assessed mitotic arrest using MPM-2 immunofluorescence (Figure 3C). We found that repressing Metnase increased mitotic arrest after ICRF-193 exposure for 24 hours from 3.6% to 10.4% in vector controls, compared with 9.4% in CD34+ cells. Representative photomicrographs are shown in Figure 4. In summary, these data demonstrated that Metnase is required for progression through mitosis when decatenation is inhibited by ICRF-193.
Metnase promotes THP-1 cell proliferation in the presence of the Topo IIα inhibitor VP-16
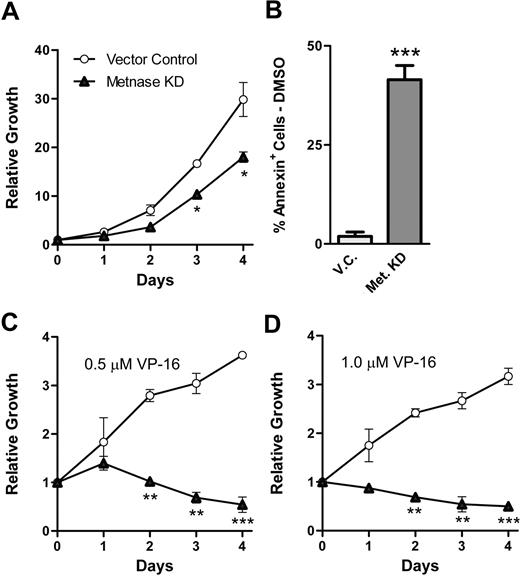
Because Metnase mediated resistance to the Topo IIα inhibitor ICRF-193, we asked whether it also mediated resistance to the clinically relevant Topo IIα inhibitor VP-16.25 Therefore, we examined whether THP-1 clones with reduced levels of Metnase were more sensitive to the antiproliferative effects of VP-16. THP-1 cells with repressed Metnase proliferated at a slightly decreased rate than vector control cells (Figure 5A), perhaps because of decreased endogenous decatenation rates. We next examined the proliferation of THP-1 cells with repressed Metnase in the presence of VP-16, compared with vector control cells. We found that these AML cells showed a markedly decreased ability to proliferate in the presence of VP-16 (Figure 5C-D); but when Metnase was decreased in these cells, they did not proliferate at all. Indeed, they were actively killed as indicated by the negative slope in Figure 5C and D. Finally, we tested whether repressing Metnase increased apoptosis in the THP-1 cells using flow cytometric analysis of the expression of annexin (Figure 5D). We found that repressing Metnase increased apoptosis in the THP-1 cells by 22-fold. In summary, these data indicate that Metnase not only mediates resistance to the non–DNA-damaging Topo IIα inhibitor ICRF-193 but also mediates resistance to the DNA damaging, clinically relevant Topo IIα inhibitor VP-16.
Metnase protects cells from the cytotoxic effects of the clinically relevant Topo IIα inhibitor VP-16. (A) Proliferation was measured in THP-1 cells expressing Metnase shRNA or vector control using growth curves. (B) THP-1 cells expressing Metnase shRNA or a vector control were subjected to 24 hours of 0.5 μM VP-16 and then analyzed for apoptosis using annexin-V expression. (C-D) THP-1 cells transfected with control vector proliferate in the presence of 0.5 μM (C) or 1.0 μM (D) VP-16, but THP-1 cells expressing Metnase shRNA do not. Data are averages (± SEM) of triplicate experiments performed twice. *Student t test (P < .05); **Student t test (P < .01); ***Student t test (P < .005).
Metnase protects cells from the cytotoxic effects of the clinically relevant Topo IIα inhibitor VP-16. (A) Proliferation was measured in THP-1 cells expressing Metnase shRNA or vector control using growth curves. (B) THP-1 cells expressing Metnase shRNA or a vector control were subjected to 24 hours of 0.5 μM VP-16 and then analyzed for apoptosis using annexin-V expression. (C-D) THP-1 cells transfected with control vector proliferate in the presence of 0.5 μM (C) or 1.0 μM (D) VP-16, but THP-1 cells expressing Metnase shRNA do not. Data are averages (± SEM) of triplicate experiments performed twice. *Student t test (P < .05); **Student t test (P < .01); ***Student t test (P < .005).
Metnase directly mediates Topo IIα resistance to inhibitors
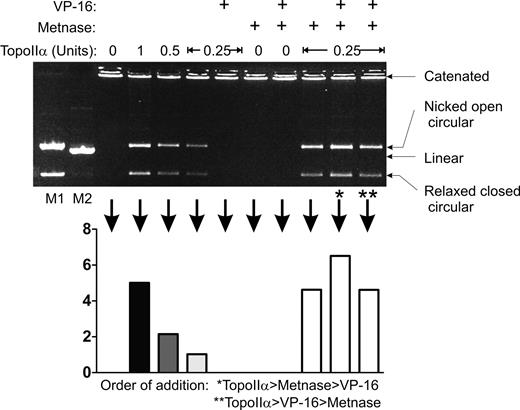
In previous studies, we found that Metnase mediated resistance to 2 Topo IIα inhibitors, ICRF-193, and VP-16. However, these studies were performed in intact cells. Even though Metnase coimmunoprecipitated Topo IIα, it is possible that they are in a complex, and there could be an intermediary factor translating the effect of Metnase to Topo IIα. Thus, it was not clear whether the ability of Metnase to enhance Topo IIα function was a direct effect or whether Metnase enhanced Topo IIα via another nuclear factor. To address this question, we analyzed in vitro decatenation of kDNA-interlocked mini-circles using purified Topo IIα and Metnase (Figure 6). Purified Metnase enhances the ability of pure Topo IIα to decatenate kDNA in vitro by 3-fold as we reported previously.20 As expected, VP-16 completely blocks the ability of Topo IIα to decatenate kDNA. Significantly, Metnase fully restores Topo IIα decatenation activity in the presence of VP-16, whether Metnase is added before or after VP-16. These data demonstrate that Metnase directly interacts with Topo IIα to enhance decatenation and does not act through an intermediary.
Metnase mediates resistance to the Topo IIα inhibitor VP-16 in vitro. In vitro analysis of Topo IIα decatenation of kDNA was performed in the presence of Metnase and/or VP-16. The relaxed closed circle DNA is the fully decatenated species. kDNA decatenation (graph) was quantified by densitometric scans of the gel image. Pure Metnase (200 ng/μL) enhanced pure Topo IIα decatenation when added before or after VP-16.
Metnase mediates resistance to the Topo IIα inhibitor VP-16 in vitro. In vitro analysis of Topo IIα decatenation of kDNA was performed in the presence of Metnase and/or VP-16. The relaxed closed circle DNA is the fully decatenated species. kDNA decatenation (graph) was quantified by densitometric scans of the gel image. Pure Metnase (200 ng/μL) enhanced pure Topo IIα decatenation when added before or after VP-16.
Discussion
Decatenation is a common yet poorly understood cause of DSBs that can lead to chromosomal translocations.1,2,8 Topo IIα is the key decatenating enzyme, but beyond that little is known about the biochemical mechanism of decatenation. When Topo IIα is inhibited by ICRF-193, which does not cause DNA damage and does not activate the DNA damage checkpoints, cells should normally arrest before mitosis, in a decatenation checkpoint.3,,,,–8 However, bladder and lung cancer cells progressed through mitosis, even when Topo IIα was inhibited by ICRF-193.9,10 Here we found that both ALL and AML cells also did not arrest in mitosis when ICRF-193 was present, even with increasing concentrations and increasing exposure times. This progression through mitosis when Topo IIα was inhibited was not shared by normal human CD34+ hematopoietic progenitors. In case the failure of mitotic arrest by the acute leukemia cells was an artifact of the assay, we examined this phenomenon using 3 distinct assays: flow cytometry of G2/M accumulation, expression of MPM-2 to define mitosis, and mitotic spindle formation by tubulin with chromosome alignment to define metaphase. Each of these complementary assays gave the same qualitative result.
This progression through mitosis by acute leukemia cells, even when Topo IIα is inhibited, has several interesting implications. First, the lack of decatenation arrest seen in a third type of cancer may indicate that this is a general feature of malignancy. The finding that normal human CD34+ progenitors did not share this characteristic makes it seem more probable that this indeed is a characteristic of transformation, at least in leukemia. If so, it may contribute to the general genomic instability seen in all cancer. Such a decatenation failure could even be an initiating event in malignant transformation, contributing to the chromosomal events that lead to oncogenesis.1 Or it could be a result of transformation, contributing to unregulated proliferation by allowing malignant cells to more rapidly progress through mitosis.9,10 This resistance to decatenation checkpoint arrest may also contribute to the clonal evolution of drug resistance; not just resistance to Topo IIα inhibitors, but by promoting genomic instability, decatenation failure would increase the evolution of selecting mutations.
Although the signaling that regulates decatenation is quickly evolving, the physical mechanism by which decatenation takes place is less well understood.9,10 We recently found that the DNA repair component Metnase enhances Topo IIα decatenation activity.20 Therefore, we examined whether the enhancement of Topo IIα by Metnase might be one mechanism by which acute leukemia cells bypass the mitotic decatenation arrest. We found that repressing Metnase levels using shRNA in the AML cell line THP-1 partially restores the total G2/M arrest, yet completely restored the ability of these cells to arrest in mitosis, specifically, when Topo IIα is inhibited by ICRF-193. Thus, to some extent, Metnase mediates resistance to ICRF-193; and when Metnase levels are reduced, these cells become sensitive to mitotic arrest in the presence of ICRF-193.
Repressing Metnase levels in the THP-1 cells also restored their sensitivity to the clinically relevant Topo IIα inhibitor VP-16. Thus, Metnase appears to mediate resistance to Topo IIα inhibitors generally. The restoration of sensitivity to VP-16 by the repression of Metnase raises an intriguing question. THP-1 cells normally proliferate in the presence of low doses of VP-16. VP-16 causes DNA DSBs by binding to and locking Topo IIα in the cleavage complex stage, before DNA strand religation and causing subsequent DNA DSBs. The question raised by the data here is whether the THP-1 cells with repressed Metnase died in the presence of VP-16 from catastrophic mitosis after failed decatenation or because of unrepaired DNA DSBs. Metnase is a component of the DNA DSB repair pathway, so it is conceivable that these cells could die of DNA damage-induced apoptosis. The alternative is that the lack of Metnase in these cells allowed free access of VP-16 to Topo IIα, and these cells underwent a catastrophic mitotic event. Given the in vitro data in Figure 6, where Metnase allows Topo IIα to function even in the presence of VP-16, a situation where the rest of the components of the nonhomologous end-joining pathway of DSB repair are not present, the latter hypothesis is more attractive.
The in vitro kDNA decatenation experiments showed that pure Metnase improved the decatenation ability of Topo IIα without an intermediary, and the coimmunoprecipitation experiments indicate that Metnase and Topo IIα interact. These data seem to imply that Metnase directly protects Topo IIα from its inhibitor in cells. There are several possible mechanisms by which it could do this. Metnase could have a higher affinity for Topo IIα than its inhibitors and sterically block ICRF-193 and VP-16 from interaction with Topo IIα. Metnase protected Topo IIα, even after addition of VP-16 in the in vitro kDNA assays, which could mean that it displaces VP-16 from Topo IIα. Alternatively, it could enhance the function of the small amount of Topo IIα that remains free from VP-16. The finding that Metnase directly enhances Topo IIα decatenation seems to imply cells proficient for Metnase never engage the decatenation checkpoint-induced cell cycle arrest because Topo IIα-mediated decatenation is not inhibited. As such, it functions as a positive regulator of Topo IIα and perhaps plays a greater role in the physical decatenation process than in the decatenation checkpoint signaling pathway. Cells may arrest in mitosis when Metnase is repressed not just because the decatenation checkpoint is activated, but also because they physically cannot separate their chromosomes to progress through metaphase. It is also possible that the acute leukemia cells failed to arrest in mitosis when Topo IIα inhibitors are present because Metnase protected Topo IIα from inhibition, and the checkpoint signaling cascade was never activated.
The data in this study demonstrate that Metnase protection of Topo IIα is a novel mechanism of resistance of cancer cells to VP-16. Probably the best investigated mechanism of VP-16 resistance is active efflux of the drug via the ABC transporters.26,,–29 Loss of the checkpoint proteins p21 or p53 can also result in VP-16 resistance.30,31 The role in which the enzymatic DNA repair components play in clinical VP-16 resistance is less well investigated. A rare report showed that increased levels of the recombination repair component RAD51 and the nonhomologous end-joining repair component DNA protein kinase catalytic subunit correlated with VP-16 resistance in lung cancer cells.32 Interestingly, there are several different tumor types that show intrinsic or acquired resistant to VP-16,26,33,–35 and it is possible that Metnase mediates this resistance and could be used as a clinical marker for treatment outcome.
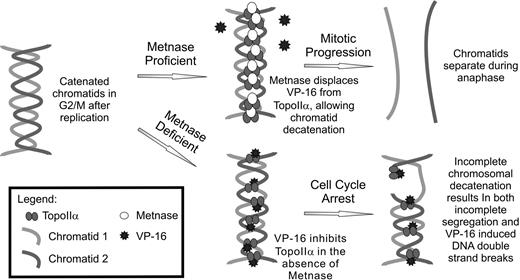
These data give rise to a model of mitotic progression of leukemia cells when VP-16 is present (Figure 7). Cells with appropriate levels of functional Metnase prevent Topo IIα inactivation by VP-16 (or ICRF-193) and can decatenate sister chromatids. Such cells can progress through anaphase because the chromatids are no longer entangled. This model is important for at least 3 reasons. First, acute leukemia cells fail to arrest in mitosis when Topo IIα inhibitors are present, compared with normal CD34+ cells. It is possible that this may be a general characteristic of malignant transformation for all types of cancer. Second, Metnase enhancement of Topo IIα decatenation in the presence of VP-16 sheds light on the molecular mechanism of decatenation in the cancer cell. It demonstrates that Metnase is a novel enhancer of Topo IIα biochemical function, allowing cancer cells to progress rapidly through mitosis by preventing difficulties with the mechanical separation of sister chromatids. Third, decreasing Metnase levels in AML cells restores their sensitivity to VP-16. Therefore, Metnase levels may predict clinical responsiveness to Topo IIα inhibitors. In this context, Metnase is an excellent candidate for small molecule inhibition to enhance the efficacy of Topo IIα inhibitors in AML therapy, particularly salvage chemotherapy.25
Model of Metnase mediating resistance to Topo IIα inhibitors in AML. Topo IIα inhibitors are common in the clinical management of AML during salvage therapy. Metnase expression has been shown here to mediate resistance to VP-16. When Metnase is present in AML cells, it is able to interact with Topo IIα, enhancing its function and results in proper chromosomal decatenation during metaphase. In the case where Metnase is deficient, the presence of VP-16 reduces Topo IIα activity and cells are not able to decatenate sister chromatids, which results in metaphase arrest.
Model of Metnase mediating resistance to Topo IIα inhibitors in AML. Topo IIα inhibitors are common in the clinical management of AML during salvage therapy. Metnase expression has been shown here to mediate resistance to VP-16. When Metnase is present in AML cells, it is able to interact with Topo IIα, enhancing its function and results in proper chromosomal decatenation during metaphase. In the case where Metnase is deficient, the presence of VP-16 reduces Topo IIα activity and cells are not able to decatenate sister chromatids, which results in metaphase arrest.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (NIH, Bethesda, MD; R01 CA100862, J.A.N.; an Activities to Promote Research Collaborations supplement to CA100862, J.A.N., R.H.; R01 CA102283, R.H.; and R01 HL075783, R.H.), and the Leukemia & Lymphoma Society (White Plains, NY; SCOR 7388-06, R.H., C.L.W.). Images were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility, which received support from NCRR 1 S10 RR14668, NSF MCB9982161, NCRR P20 RR11830, NCI P30 CA118100, NCRR S10 RR19287, NCRR S10 RR016918, the University of New Mexico Health Sciences Center, and the University of New Mexico Cancer Center. Data were generated in the Flow Cytometry Shared Resource Center supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center.
National Institutes of Health
Authorship
Contribution: J.W. performed research and wrote the paper; E.A.W. performed research and edited the paper; S.S. performed research; S.-H.L. provided reagents, edited the paper, and analyzed data; E.L. analyzed data; C.L.W. analyzed data and edited the paper; J.A.N. designed experiments, analyzed data, and edited the paper; and R.H. conceived the project, designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The present address of Dr Nickoloff is Department of Environmental and Radiological Health Sciences, Colorado State University, Ft Collins, CO.
Correspondence: Robert Hromas, Cancer Research and Treatment Center, University of New Mexico Health Science Center, 900 Camino de Salud, Albuquerque, NM 87131; e-mail: rhromas@salud.unm.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal