Abstract
It is generally conceded that selective combinations of transcription factors determine hematopoietic lineage commitment and differentiation. Here we show that in normal human hematopoiesis the transcription factor nuclear factor I-A (NFI-A) exhibits a marked lineage-specific expression pattern: it is upmodulated in the erythroid (E) lineage while fully suppressed in the granulopoietic (G) series. In unilineage E culture of hematopoietic progenitor cells (HPCs), NFI-A overexpression or knockdown accelerates or blocks erythropoiesis, respectively: notably, NFI-A overexpression restores E differentiation in the presence of low or minimal erythropoietin stimulus. Conversely, NFI-A ectopic expression in unilineage G culture induces a sharp inhibition of granulopoiesis. Finally, in bilineage E + G culture, NFI-A overexpression or suppression drives HPCs into the E or G differentiation pathways, respectively. These NFI-A actions are mediated, at least in part, by a dual and opposite transcriptional action: direct binding and activation or repression of the promoters of the β-globin and G-CSF receptor gene, respectively. Altogether, these results indicate that, in early hematopoiesis, the NFI-A expression level acts as a novel factor channeling HPCs into either the E or G lineage.
Introduction
Hematopoietic stem cells (HSCs), endowed with extensive self-renewal potential and multilineage differentiation capacity, sustain the hematopoietic system throughout life.1 HSCs generate undifferentiated hematopoietic progenitor cells (HPCs), which gradually lose their self-renewal ability. Multipotent common myeloid progenitors, the functional equivalent of colony-forming units of mixed type (CFU-mix) or CFU-GEMM, undergo progressive restriction of their lineage commitment, eventually separating into 2 major bipotent progenitor types: megakaryocyte-erythroid progenitor and granulocyte-monocyte progenitor. These progenitor types are responsible for producing unilineage progenitors for the erythroid (E), megakaryocytic (Mk), granulocytic (G), and monocytic (M) lineage, respectively, termed burst-forming unit (BFU-E) and CFU-E, BFU-Mk and CFU-Mk, CFU-G, and CFU-M. The unilineage HPCs, in turn, generate differentiated precursors, which eventually mature into terminal circulating blood cells.
Multiple hematopoietic genes, particularly transcription factors (TFs) and growth factor receptors, are promiscuously coexpressed in HSCs and HPCs, a phenomenon referred to as lineage priming.2 Their coexistence grants differentiation flexibility and allows lineage fate commitment at the multipotent stage. Key lineage-restricted factors are characterized by a low level of expression in the undifferentiated state but become dominant in their specific lineages, whose development is thereby promoted while alternative cell fates are antagonized.3
Whereas GATA-1 and PU.1 represent a paradigm of lineage programming TFs driving the fate of HSCs/HPCs,4–6 growing evidence indicates that other TFs play a pivotal role in the complex scenario of lineage specification.7,8 In this regard, the TF Nuclear Factor I-A (NFI-A) acts as a transcriptional activator or repressor in a cell type and promoter-specific context.9,10 NFI-A belongs to the NFI TF family consisting of 4 different genes that contain a highly conserved N-terminal DNA-binding/dimerization domain and a divergent C-terminal transactivation/repression domain, which confers unique functions to each member. NFI-A has a major role in brain development and shows a unique pattern of expression in the developing mouse beginning early on in the heart and brain with a broader tissue expression pattern as development progresses.11 Recent studies showed that NFI-A is a relevant target of the myeloid regulator miR-223 in promyelocytic leukemia cells12 and suppresses monocytic differentiation of HPCs,13 whereas nothing is known on its role in normal erythro-granulopoiesis.
Two lineage-associated genes, β-globin and G-CSFR, are key determinants for the erythroid and granulocytic pathways, respectively. The human β-globin locus consists of 5 genes, including the embryonic ϵ, the fetal Gγ, Aγ, the adult δ, and β, which are sequentially activated during ontogeny: the transcriptional control of hemoglobin from embryonic to adult life is only partially understood14,15 and involves complex DNA-protein and protein-protein interactions at cis-elements located in both proximal promoter regions and distal regulatory sequences.16 Granulocyte colony-stimulating factor (G-CSF), the main cytokine stimulator of the G pathway, binds to the G-CSF receptor (G-CSFR) to mediate activation of neutrophils and differentiation of their progenitors/precursors in the bone marrow.17–19 In this study, we describe a central role of NFI-A in the human erythro-granulopoietic lineage decision and differentiation. In HPCs, NFI-A upmodulation favors the development of the E lineage and represses the G pathway, in part by direct transcriptional activation of β-globin and repression of G-CSFR.
Methods
Cell culture
Cord blood (CB) was obtained from healthy, full-term pregnancies according to institutional guidelines and with the approval of the University of Rome “La Sapienza.” Informed consent was obtained in accordance with the Declaration of Helsinki. Human growth factors were purchased from PeproTech, human erythropoietin (Epo) was provided by Amgen.
CD34+ collection, isolation, and unilineage cultures were performed as previously described.20 In unilineage cultures, serum-free medium was supplemented with: Epo (3 U/mL), interleukin-3 (IL-3; 0.01 U/mL) and GM-CSF (0.001 ng/mL) for erythroid (E) and IL-3 (1 U/mL), GM-CSF (0.1 ng/mL), and G-CSF (500 U/mL) for granulocytic (G).
In bilineage E + G culture, serum-free medium was supplemented with IL-3 (1 U/mL), GM-CSF (0.1 ng/mL), Epo (0.6 U/mL), and G-CSF (500 U/mL).
Viable cells were counted every 2 to 3 days using Trypan Blue Staining (Sigma-Aldrich) and passaged at 2 × 105 cells/mL.
In clonogenic assay, 102 CD34+ cells were plated in duplicate in serum-free medium containing 0.9% methylcellulose MethoCult H4100 (StemCell Technologies). The following cytokines were used: for BFU-E assay, IL-3 (0.01 U/mL), GM-CSF (0.001 ng/mL), and Epo (3 U/mL). For CFU-G assay, IL-3 (1 U/mL), GM-CSF (0.1 ng/mL), and G-CSF (500 U/mL). For BFU-E/CFU-G assay, IL-3 (1 U/mL), GM-CSF (0.1 ng/mL), Epo (0.6 U/mL), and G-CSF (500 U/mL; supplemental data, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Morphologic analysis
Cells were harvested from day 3 to days 21 to 24, smeared on glass slides by cytospin centrifugation, stained with standard May-Grünwald-Giemsa, and observed by conventional light field microscopy, to evaluate morphology and cell differentiation through the erythroid and granulocytic pathways. Proerythroblasts, basophilic, polychromatophilic, and orthochromatic cells in the erythroid series, myeloblasts, promyelocytes, myelocytes, and mature cells (meta/band) in the granulocytic series were identified, and their percentage value was obtained. At least 500 cells were counted per slide.
Slides were viewed with a Nikon Eclipse E1000 microscope (Nikon) using Plan Fluor lenses at 20×/0.75 PH and 40×/1.30 PH oil with DPX mounting medium (Laboratory Supplies). Colonies were viewed using a Plan Fluor lense at 10×/0.30 PH. Images were acquired using Nikon digital camera model DXM1200 and were processed with Nikon ACT-1 software Version 2.63.
RNA analysis
Total RNA was isolated by Trizol (Invitrogen) following the manufacturer's instructions. First-strand cDNA was synthesized with SuperScript II (Invitrogen) using equivalent amounts of total RNA for each sample and oligo (dT) (Invitrogen) primers. mRNA quantification by real-time polymerase chain reaction (PCR) was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) and inventoried TaqMan gene expression assays (Applied Biosystems) with ΔCt values normalized using endogenous GAPDH as control.
ChIP assay
Formaldehyde was added to cultured cells (2 × 106) at a final concentration of 1% for 10 minutes at 37°C to cross-link DNA and protein complexes. Chromatin was sheared by sonication and immunoprecipitated with 3 μg of anti-NFI-A (Abcam). Genomic regions of approximately 200 bp containing the NFI-A consensus site were amplified by multiplex PCR21 in the presence of [α-32P] dATP using the primers listed in supplemental Table 1. PCR signals were quantified using Typhoon Trio Phosphoimager (Amersham Biosciences) and ImageQuant software (Molecular Dynamics). The relative occupancy of NFI-A present on the promoter (Prom) specific regions is normalized to the input (I) signal, following the algorithm: [(IP-BO)/I]PROM/[(IP-BO)/I]UR. IP represents chromatin immunoprecipitated by the NFI-A antibody. BO (beads only) represents nonspecific signal in absence of antibody. The coamplification of tubulin (mentioned as unrelated genomic region [UR]) within the same multiplex reaction serves as an internal control for background nonspecific chromatin. PCRs were performed within the linear range of amplification (supplemental data).
Lentiviral production and infection
Infectious particles were produced as previously described.22 Cells were infected by the spin-inoculation method. CD34+ cells were infected at day 1 of culture. At 48 hours after infection, the cells were monitored for GFP expression by flow cytometry using FACScan instrument (BD Biosciences), showing an average transduction efficiency ranging from 15% to 20% (for the HA-NFI-A construct) to 30% to 40% (for vector and siNFI-A). At this time, GFP+ cells were sorted using the FACSaria instrument (BD Biosciences) and placed in liquid phase culture or clonogenic medium (supplemental data).
Flow cytometry
Phycoerythrin (PE)-conjugated antihuman Glycophorin-A (GPA), CD34, CD15, CD14, CD11b, and CD114 antibodies were purchased from BD Biosciences Pharmingen. G-CSFR total protein expression was assessed by immunostaining using a PE-conjugated anti-CD114 antibody in cells fixed and permeabilized with 100 μL of Cytofix/Cytoperm (BD Biosciences Pharmingen). Immunophenotype and 7-amino-actinomycin D (Calbiochem) cell viability staining (final concentration of 5 μg/mL), performed according to the manufacturer's instructions were analyzed by flow cytometry using FACScan instrument (BD Biosciences).
Assay of globin-chain content
High performance liquid chromatography (HPLC) separation of globin chains was performed according to the previously published method15 (supplemental data).
Plasmids, constructs, luciferase assay
The lentiviral vector encoding HA-NFI-A was previously described.13 The siNFI-A vector was generated by cloning the siNFI-A sense 5′-AACCAGAGGTCAAGCAGAA-3′ into the pRRLcPPT.hPGK.EGFP.WPRE lentiviral vector (named vector; supplemental data). Supplemental data contain information on luciferase assay, and supplemental Table 1 contains information on PCR primers.
Immunoblotting
Total protein was extracted using CelLytic M Lysis Reagent (Sigma-Aldrich) following the manufacturer's protocol, supplemented with Protease Inhibitor Cocktail (Sigma-Aldrich). Total cell extracts were fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by electroblotting. Blots were probed with the primary and secondary antibodies listed in Supplemental data. Immunoreactivity was measured using the enhanced chemiluminescence method (Amersham Biosciences).
Statistical analysis
Comparison between multiple groups was made by one-way analysis of variance and the Tukey post test to analyze significance. Two-tailed t test analysis for significance was used to compare individual data to control values. Differences were considered significant at P values less than .05 or less than .01.
Results
NFI-A expression is upmodulated during erythropoiesis and shut off during granulopoiesis
Human CB CD34+ HPCs were induced into unilineage erythroid (E; Figure 1Ai-iii) or granulocytic (G; Figure 1Bi-iii) proliferation and differentiation/maturation in serum-free suspension cultures. The unilineage cultures recapitulate the in vivo differentiation/maturation of HPCs20,23–25 ; therefore, these assays allow analysis of sequential, discrete stages of differentiation/maturation through a single lineage.
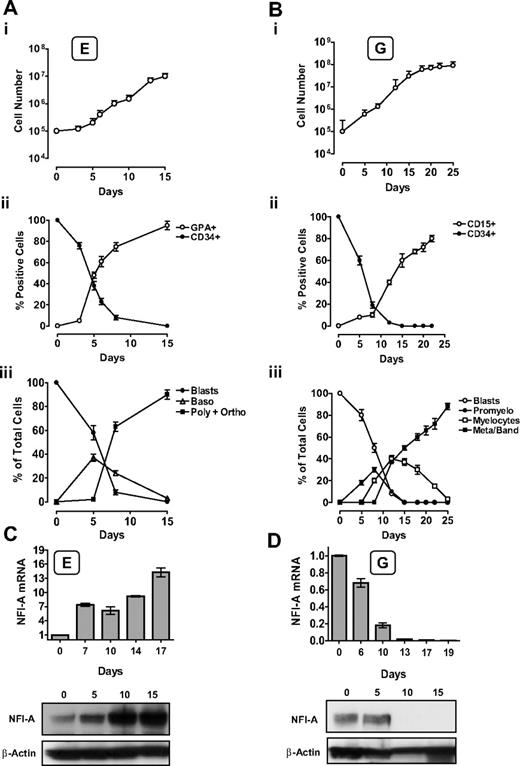
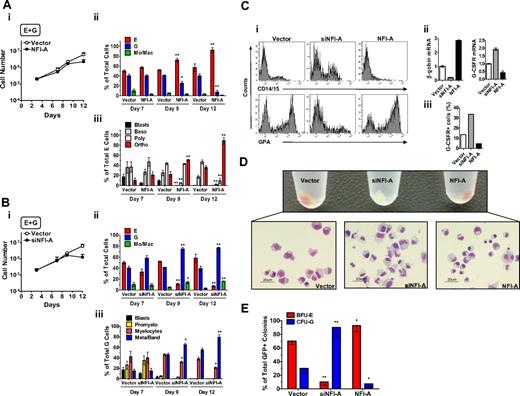
Unilineage erythroid (E) and granulocytic (G) culture of HPCs: NFI-A expression pattern during E and G differentiation/maturation. (A) Unilineage E culture of CB CD34+ HPCs. (i) Growth curve, (ii) CD34 and GPA surface marker expression, and (iii) percentage of blasts, proerythroblasts (Blasts), basophilic (Baso), polychromatophilic (Poly), and orthochromatic (Ortho) erythroblasts (mean ± SEM values; n = 7). (B) Unilineage G culture of CB CD34+ HPCs. (i) Growth curve, (ii) CD34 and CD15 surface marker expression, and (iii) percentage of blasts, promyelocytes (Promyelo), myelocytes, mature metamyelocytes, and band cells (Meta/band). Mean ± SEM values (n = 8). (C) NFI-A increases in E culture. (Top) Real-time PCR evaluation of NFI-A mRNA. Normalized mean ± SEM values (n = 3). (Bottom) Immunoblot of NFI-A protein. β-Actin was used as loading control. (D) NFI-A decrease in G culture. (Top) Real-time PCR evaluation of NFI-A mRNA. Normalized mean ± SEM values (n = 3). (Bottom) Immunoblot of NFI-A protein.
Unilineage erythroid (E) and granulocytic (G) culture of HPCs: NFI-A expression pattern during E and G differentiation/maturation. (A) Unilineage E culture of CB CD34+ HPCs. (i) Growth curve, (ii) CD34 and GPA surface marker expression, and (iii) percentage of blasts, proerythroblasts (Blasts), basophilic (Baso), polychromatophilic (Poly), and orthochromatic (Ortho) erythroblasts (mean ± SEM values; n = 7). (B) Unilineage G culture of CB CD34+ HPCs. (i) Growth curve, (ii) CD34 and CD15 surface marker expression, and (iii) percentage of blasts, promyelocytes (Promyelo), myelocytes, mature metamyelocytes, and band cells (Meta/band). Mean ± SEM values (n = 8). (C) NFI-A increases in E culture. (Top) Real-time PCR evaluation of NFI-A mRNA. Normalized mean ± SEM values (n = 3). (Bottom) Immunoblot of NFI-A protein. β-Actin was used as loading control. (D) NFI-A decrease in G culture. (Top) Real-time PCR evaluation of NFI-A mRNA. Normalized mean ± SEM values (n = 3). (Bottom) Immunoblot of NFI-A protein.
In the E culture, NFI-A was sharply upmodulated at both the RNA and protein levels (Figure 1C), in parallel with the increase of Glycophorin-A+ (GPA) cells and erythroblast differentiation/maturation (Figure 1Aii-iii). Conversely, in G culture, we observed a drastic decrease of NFI-A, reaching undetectable RNA and protein levels starting from days 10 to 13 (Figure 1D): this correlated with the increasing number of CD15+ cells and G differentiation/maturation at morphology level (Figure 1Bii-iii).
In line with these findings, NFI-A was expressed at a higher level in the erythroleukemic K562 cell line than the myeloblastic leukemia HL-60 cell line; moreover, NFI-A levels decreased in HL-60 cells undergoing G differentiation after treatment with retinoic acid (RA; supplemental Figure 1A).
NFI-A up-regulation is required for erythroid differentiation of HPCs
The dichotomy of NFI-A expression patterns in the E versus G lineage suggested that this TF may exert opposite functional actions in these cell differentiation pathways.
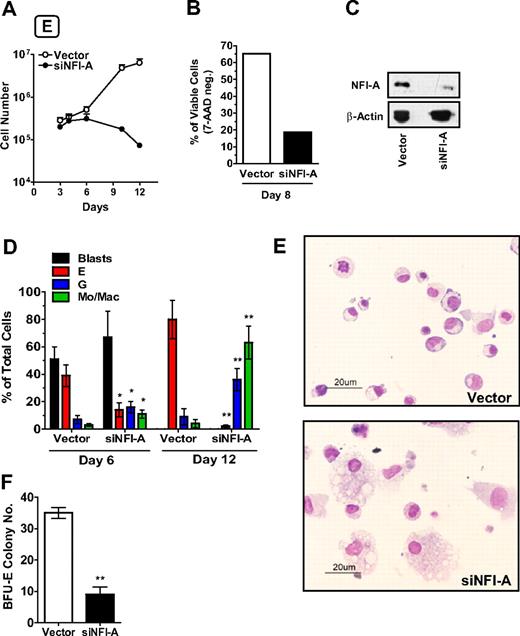
To verify this hypothesis, we first monitored the effects of NFI-A knockdown in HPCs seeded in E culture using a lentiviral construct encoding an shRNA-targeting NFI-A mRNA (named siNFI-A). To exclude possible off-target effects, siNFI-A was compared with a previously published shRNA against NFI-A12 (here named siNFI-A-2): both the constructs specifically knocked down NFI-A, and siNFI-A showed the maximum silencing efficiency (supplemental Figure 1B) without triggering an off-target interferon response (supplemental Figure 1C). In E culture, HPCs stably expressing siNFI-A showed an impaired growth capacity: by day 8, the majority of the cells were nonviable (Figure 2A-B), with no significant changes in the cell cycle status of the remaining viable cells (data not shown). NFI-A protein knockdown in siNFI-A expressing cells was confirmed by Western blot (Figure 2C). Moreover, E differentiation/maturation in siNFI-A cells was dramatically afflicted: at day 12, the residual cells were almost totally of granulocytic and monocytic type (Figure 2D-E). Similarly, clonogenic assays in E medium showed that siNFI-A transduction caused a drastic reduction of the BFU-E colony number (Figure 2F). These results indicate that NFI-A upmodulation is necessary to allow normal E differentiation/maturation of HPCs.
NFI-A knockdown impairs erythroid (E) differentiation of HPCs. (A) Growth curve of HPCs in unilineage E culture infected with control vector or siNFI-A (mean ± SEM values of 3 independent experiments). (B) The histogram shows the percentage of viable (or 7-amino-actinomycin D-negative) cells detected by flow cytometry (representative of 3). (C) Western blot analysis confirming NFI-A knockdown in siNFI-A expressing HPCs in unilineage E culture. (D) Wright-Giemsa staining of vector- and siNFI-A–transduced cells. Percentage of blasts, erythroid (E), granulocytic (G), and monocytic/macrophage (Mo/Mac) cells (mean ± SEM values of 3 independent experiments). (E) Morphology of vector and siNFI-A cells at day 12 of E culture (representative field, original magnification ×400). See “Morphologic analysis” for more image information. (F) Number of BFU-E colonies generated by vector- and siNFI-A–infected HPCs (mean ± SEM values; n = 3).
NFI-A knockdown impairs erythroid (E) differentiation of HPCs. (A) Growth curve of HPCs in unilineage E culture infected with control vector or siNFI-A (mean ± SEM values of 3 independent experiments). (B) The histogram shows the percentage of viable (or 7-amino-actinomycin D-negative) cells detected by flow cytometry (representative of 3). (C) Western blot analysis confirming NFI-A knockdown in siNFI-A expressing HPCs in unilineage E culture. (D) Wright-Giemsa staining of vector- and siNFI-A–transduced cells. Percentage of blasts, erythroid (E), granulocytic (G), and monocytic/macrophage (Mo/Mac) cells (mean ± SEM values of 3 independent experiments). (E) Morphology of vector and siNFI-A cells at day 12 of E culture (representative field, original magnification ×400). See “Morphologic analysis” for more image information. (F) Number of BFU-E colonies generated by vector- and siNFI-A–infected HPCs (mean ± SEM values; n = 3).
NFI-A accelerates erythroid differentiation of HPCs and it restores erythropoiesis in the presence of suboptimal or minimal erythropoietin (Epo) stimulus
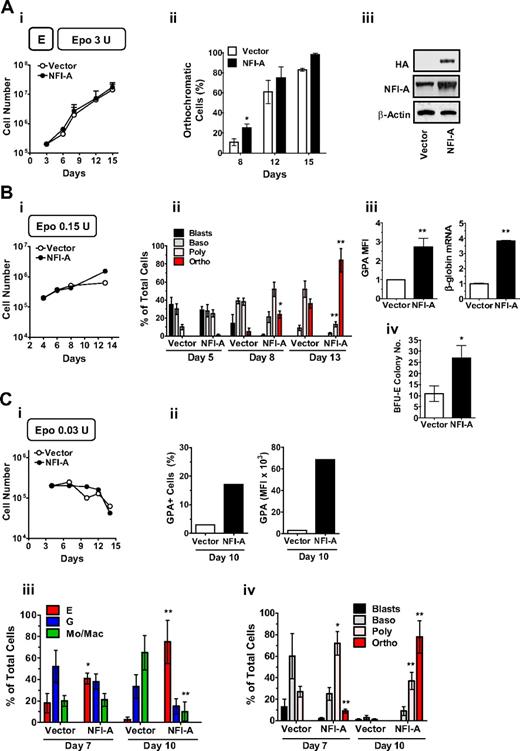
We then hypothesized that the enforced overexpression of NFI-A may favor E differentiation, compared with empty vector cells. Indeed, lentiviral NFI-A overexpression in HPCs seeded in E culture (Figure 3Ai) resulted in an accelerated differentiation and maturation, as demonstrated by the increase of orthochromatic erythroblasts at day 8 (Figure 3Aii). Western blot analysis using anti-HA and anti-NFI-A antibodies confirmed the expression of exogenous NFI-A (Figure 3Aiii). In addition, NFI-A ectopic expression in K562 caused an increase in benzidine-stained cells and GPA expression in both baseline conditions and Ara-C-induced E differentiation with no significant change in cell growth rates (supplemental Figure 2Ai-iii).
NFI-A overexpression favors differentiation and overcomes erythropoietin (Epo) dependence of erythroid (E) culture. (Ai) Growth curve of vector- and NFI-A–transduced HPCs in standard unilineage E culture (Epo 3 U/mL; mean ± SEM values; n = 3). (ii) Percentage of orthochromatic cells at sequential stages of E culture generated by vector- and NFI-A–expressing HPCs (mean ± SEM values; n = 3). (iii) Western blot showing ectopic expression of HA-tagged NFI-A in NFI-A–infected HPCs in E culture at day 8. (Bi) Growth curve (a representative experiment of 3 is shown) and (ii) morphologic evaluation (mean ± SEM values; n = 3) of E differentiation of vector- and NFI-A–transduced HPCs seeded in E culture with suboptimal amounts of Epo (0.15 U). (iii) Increase of GPA mean fluorescence intensity (MFI) (mean ± SEM values; n = 3) and real-time PCR showing β-globin mRNA expression (mean ± SEM values from 2 independent infections). (iv) Number of BFU-E colonies plated in Epo 0.15 clonogenic medium (mean ± SEM values of 2 paired experiments). (Ci) Growth curve of vector- and NFI-A–transduced HPCs in culture supplemented with minimal amounts of Epo (0.03 U). A representative experiment of 3 is shown. (ii) Percentage of GPA+ cells (left) and GPA MFI (right) at day 10. A representative experiment of 3 is shown. (iii-iv) Morphology analysis of vector- and NFI-A–infected HPCs (mean ± SEM values; n = 3).
NFI-A overexpression favors differentiation and overcomes erythropoietin (Epo) dependence of erythroid (E) culture. (Ai) Growth curve of vector- and NFI-A–transduced HPCs in standard unilineage E culture (Epo 3 U/mL; mean ± SEM values; n = 3). (ii) Percentage of orthochromatic cells at sequential stages of E culture generated by vector- and NFI-A–expressing HPCs (mean ± SEM values; n = 3). (iii) Western blot showing ectopic expression of HA-tagged NFI-A in NFI-A–infected HPCs in E culture at day 8. (Bi) Growth curve (a representative experiment of 3 is shown) and (ii) morphologic evaluation (mean ± SEM values; n = 3) of E differentiation of vector- and NFI-A–transduced HPCs seeded in E culture with suboptimal amounts of Epo (0.15 U). (iii) Increase of GPA mean fluorescence intensity (MFI) (mean ± SEM values; n = 3) and real-time PCR showing β-globin mRNA expression (mean ± SEM values from 2 independent infections). (iv) Number of BFU-E colonies plated in Epo 0.15 clonogenic medium (mean ± SEM values of 2 paired experiments). (Ci) Growth curve of vector- and NFI-A–transduced HPCs in culture supplemented with minimal amounts of Epo (0.03 U). A representative experiment of 3 is shown. (ii) Percentage of GPA+ cells (left) and GPA MFI (right) at day 10. A representative experiment of 3 is shown. (iii-iv) Morphology analysis of vector- and NFI-A–infected HPCs (mean ± SEM values; n = 3).
In standard E culture, saturating levels of Epo (3 U/mL) are required to induce optimal erythroblast expansion and differentiation/maturation.20,26 We hypothesized that NFI-A overexpression may restore erythropoiesis in the presence of a suboptimal or minimal Epo stimulus. To test this hypothesis, we supplemented E culture with a low or minimal Epo concentration, corresponding, respectively, to 1:20 (0.15 U) and 1:100 (0.03 U) of the saturating Epo level. Compared with control cells, NFI-A–transduced HPCs seeded in Epo 0.15 U medium displayed a similar proliferation rate (Figure 3Bi) while exhibiting enhanced differentiation/maturation as indicated by erythroblast morphology (Figure 3Bii), GPA expression, and β-globin mRNA level (Figure 3Biii). Interestingly, NFI-A enforced expression also increased BFU-E colony formation (Figure 3Biv). When Epo was lowered to 0.03 U, cell expansion was blocked (Figure 3Ci): importantly, erythroid cells were not present in control culture, whereas NFI-A overexpressing cells matured to the terminal stage, as shown by the increase of GPA+ cells (Figure 3Cii, supplemental Figure 2B) and morphology analysis (Figure 3Ciii-iv).
We conclude that NFI-A overexpression in HPCs promotes E differentiation and maturation while inducing a more mature E phenotype in K562 cells. Furthermore, NFI-A overexpression restores erythroblast development in E culture supplemented with a suboptimal or minimal Epo stimulus.
NFI-A downmodulation is necessary to permit granulocytic differentiation/maturation
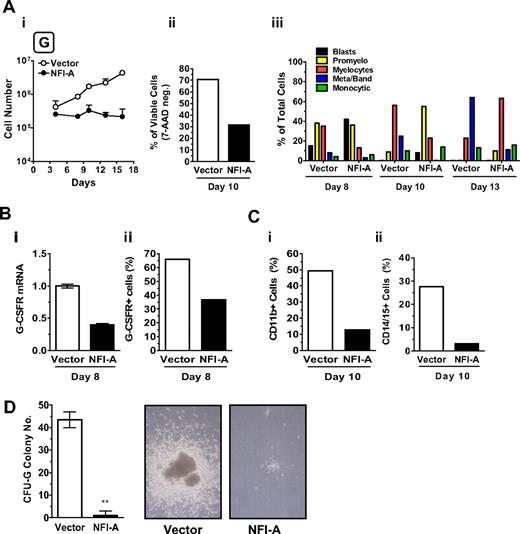
To examine the functional role of NFI-A in the G lineage, HPCs transduced with a lentivirus encoding NFI-A were seeded in G culture. Control cells were transduced with the empty vector. Cells expressing ectopic NFI-A had reduced viability (Figure 4Ai-ii) with no detectable differences in the cell cycle status (data not shown) and showed a delayed entry in the G pathway (Figure 4Aiii). NFI-A cells displayed a significant reduction of G-CSFR mRNA (Figure 4Bi) and total protein expression (Figure 4Bii, supplemental Figure 3A) in addition to a strong decrease in myeloid markers CD11b and CD14/15 (Figure 4Ci-ii). In clonogenic G assay, vector-transduced HPCs generated a significant number of CFU-G colonies of large size; whereas in cultures of NFI-A infected cells, almost all G clones were abortive (Figure 4D). In the HL-60 cell line, ectopic expression of NFI-A decreased the RA effect on G differentiation, as indicated by the lower level of differentiation markers (CD11b, CD15, and G-CSFR; supplemental Figure 3B-C), thus in line with previous observations on the promyelocytic leukemia NB4 cell line.12
Ectopic expression of NFI-A blocks granulocytic (G) differentiation of HPCs. (Ai) Growth curve of unilineage G cultures transduced with vector or NFI-A lentivirus (mean ± SEM values of 3 independent experiments). (ii) The histogram shows the percentage of viable (or 7-amino-actinomycin D-negative) cells detected by flow cytometry (representative of 3). (iii) Wright-Giemsa staining of infected cells, showing the percentage of cells at various stages of differentiation. A representative experiment of 3 is shown. (Bi) Real-time PCR analysis of G-CSFR mRNA and (ii) flow cytometry analysis of total G-CSFR protein expression in vector and NFI-A cells at day 8. (C) Flow cytometry analysis of (i) CD11b and (ii) CD14/CD15 myeloid markers at day 10. (D) (Left) number of CFU-G colonies generated by transduced HPCs (mean ± SEM values; n = 3). (Right) Representative microphotographs of G colony or cluster in vector or NFI-A culture, respectively, at day 14. Original magnification ×100. See “Morphologic analysis” for more image information.
Ectopic expression of NFI-A blocks granulocytic (G) differentiation of HPCs. (Ai) Growth curve of unilineage G cultures transduced with vector or NFI-A lentivirus (mean ± SEM values of 3 independent experiments). (ii) The histogram shows the percentage of viable (or 7-amino-actinomycin D-negative) cells detected by flow cytometry (representative of 3). (iii) Wright-Giemsa staining of infected cells, showing the percentage of cells at various stages of differentiation. A representative experiment of 3 is shown. (Bi) Real-time PCR analysis of G-CSFR mRNA and (ii) flow cytometry analysis of total G-CSFR protein expression in vector and NFI-A cells at day 8. (C) Flow cytometry analysis of (i) CD11b and (ii) CD14/CD15 myeloid markers at day 10. (D) (Left) number of CFU-G colonies generated by transduced HPCs (mean ± SEM values; n = 3). (Right) Representative microphotographs of G colony or cluster in vector or NFI-A culture, respectively, at day 14. Original magnification ×100. See “Morphologic analysis” for more image information.
NFI-A is a regulator of the erythro-granulocytic lineage decision
Based on the results thus far, we hypothesized that NFI-A levels may function in directing HPCs into the E or G differentiation pathway. To test the hypothesis, we developed a culture system allowing HPCs to undergo bilineage E + G differentiation/maturation (Figure 5). This culture system is stimulated by an appropriate mixture of E and G growth factors (including Epo 0.6 U/mL and G-CSF 500 U/mL, see “Cell culture” for more information): as a result, the proliferating cells are characterized by a balanced ratio of E and G cells undergoing differentiation and maturation up to the terminal stage while including a small monocytic contaminant.
NFI-A regulates erythroid (E) versus granulocytic (G) lineage differentiation in bilineage E + G culture (mean ± SEM values from 3 independent experiments). (A) Ectopic expression of NFI-A promotes E and inhibits G differentiation/maturation. (i) Growth curve, (ii) morphology analysis of the cellular composition at sequential culture times, and (iii) differentiation/maturation of the E population of vector- and NFI-A–infected HPCs. (B) NFI-A knockdown blocks E and promotes G differentiation/maturation. (i) Growth curve, (ii) morphology analysis at sequential culture times, and (iii) differentiation/maturation of the G population in culture of vector- and siNFI-A–infected HPCs. (C) Lineage-specific marker expression at early stage (day 9) of culture. (i) Representative flow cytometry analysis of vector-, siNFI-A–, and NFI-A–infected cells using the erythroid GPA and the myeloid CD14/CD15 markers, (ii) β-globin and G-CSFR mRNA detected by real-time PCR, and (iii) percentage of cells expressing G-CSFR detected by flow cytometry (day 7). (D) Macroscopic and morphologic changes of vector-, siNFI-A–, and NFI-A–infected cells at day 9 of E + G culture. (Top) Macroscopic view of cellular pellets centrifuged from vector culture (mixed population, erythroid red cells in the center and peripheral myeloid-white cells in the surrounding ring), siNFI-A culture (only myeloid cells), and NFI-A culture (predominance of red cells). (Bottom) Representative morphology fields (original magnification, ×400). See “Morphologic analysis” for more image information. (E) E + G clonogenic activity of HPCs transduced with vector, siNFI-A, or NFI-A viruses. Histograms represent the relative GFP+ BFU-E and CFU-G colony distribution. GFP+ colony numbers were: vector BFU-E 34.3 ± 7, CFU-G 14.3 ± 5; siNFI-A BFU-E 4.6 ± 1.5, CFU-G 31 ± 9; NFI-A BFU-E 29.3 ± 6, CFU-G 3.5 ± 1 (mean ± SEM values from 3 paired experiments).
NFI-A regulates erythroid (E) versus granulocytic (G) lineage differentiation in bilineage E + G culture (mean ± SEM values from 3 independent experiments). (A) Ectopic expression of NFI-A promotes E and inhibits G differentiation/maturation. (i) Growth curve, (ii) morphology analysis of the cellular composition at sequential culture times, and (iii) differentiation/maturation of the E population of vector- and NFI-A–infected HPCs. (B) NFI-A knockdown blocks E and promotes G differentiation/maturation. (i) Growth curve, (ii) morphology analysis at sequential culture times, and (iii) differentiation/maturation of the G population in culture of vector- and siNFI-A–infected HPCs. (C) Lineage-specific marker expression at early stage (day 9) of culture. (i) Representative flow cytometry analysis of vector-, siNFI-A–, and NFI-A–infected cells using the erythroid GPA and the myeloid CD14/CD15 markers, (ii) β-globin and G-CSFR mRNA detected by real-time PCR, and (iii) percentage of cells expressing G-CSFR detected by flow cytometry (day 7). (D) Macroscopic and morphologic changes of vector-, siNFI-A–, and NFI-A–infected cells at day 9 of E + G culture. (Top) Macroscopic view of cellular pellets centrifuged from vector culture (mixed population, erythroid red cells in the center and peripheral myeloid-white cells in the surrounding ring), siNFI-A culture (only myeloid cells), and NFI-A culture (predominance of red cells). (Bottom) Representative morphology fields (original magnification, ×400). See “Morphologic analysis” for more image information. (E) E + G clonogenic activity of HPCs transduced with vector, siNFI-A, or NFI-A viruses. Histograms represent the relative GFP+ BFU-E and CFU-G colony distribution. GFP+ colony numbers were: vector BFU-E 34.3 ± 7, CFU-G 14.3 ± 5; siNFI-A BFU-E 4.6 ± 1.5, CFU-G 31 ± 9; NFI-A BFU-E 29.3 ± 6, CFU-G 3.5 ± 1 (mean ± SEM values from 3 paired experiments).
HPCs subjected to NFI-A enforced expression or knockdown were seeded into the E + G bilineage culture. Here again, control cells were infected with empty vector. As shown in Figure 5Ai, NFI-A– and vector-transduced HPCs similarly proliferated and differentiated up to day 9. Thereafter, the E population significantly prevailed and matured more rapidly in NFI-A–overexpressing cells (Figure 5Aii-iii). HPCs infected with siNFI-A proliferated similarly to control cells up to day 9 but then showed a blockage of cell expansion (Figure 5Bi). Notably, the siNFI-A–transduced cells displayed a predominant G morphology coupled with a more rapid maturation to mature granulocytes (Figure 5Bii-iii). Analysis of lineage-specific surface markers (GPA for the E and an equimolar mix of CD14/CD15 for the G lineage) at day 9 confirmed the morphologic observations. NFI-A–infected cells express more GPA and no G markers, whereas siNFI-A–infected cells express with a greater intensity G markers and little or no GPA (Figure 5Ci). In addition, β-globin and G-CSFR mRNA (Figure 5Cii), along with G-CSFR protein expression analyzed by flow cytometry (Figure 5Ciii, supplemental Figure 4A) showed an opposite pattern of expression in the siNFI-A versus NFI-A cells compared with the vector cells confirming the lineage-specific prevalence. At day 9, macroscopic observation of the control cell pellet showed a central core of red E cells, surrounded by white G cells, whereas the siNFI-A pellet is entirely G-white and NFI-A overexpressing cells are almost entirely E-red, as confirmed by microscopic morphologic analysis of the same cell pellets (Figure 5D).
In a clonogenic E + G assay, GFP+ colonies were scored after 14 to 18 days (Figure 5E). NFI-A–overexpressing cells showed an increased proportion of BFU-E colonies and a dramatic reduction in CFU-G colonies compared with vector colonies. In contrast, siNFI-A–infected HPCs showed an opposite distribution of colony types, with a predominance of CFU-Gs.
Altogether, these experiments indicate that NFI-A plays a key role in the erythro-granulocytic lineage decision at the level of HPCs and possibly early precursors.
A functional relationship between NFI-A levels and 2 lineage-associated genes: β-globin and G-CSFR
We then attempted to explore the mechanism of NFI-A action by screening possible cis-acting elements interacting with NFI-A within promoter regions of E- and G-specific genes.
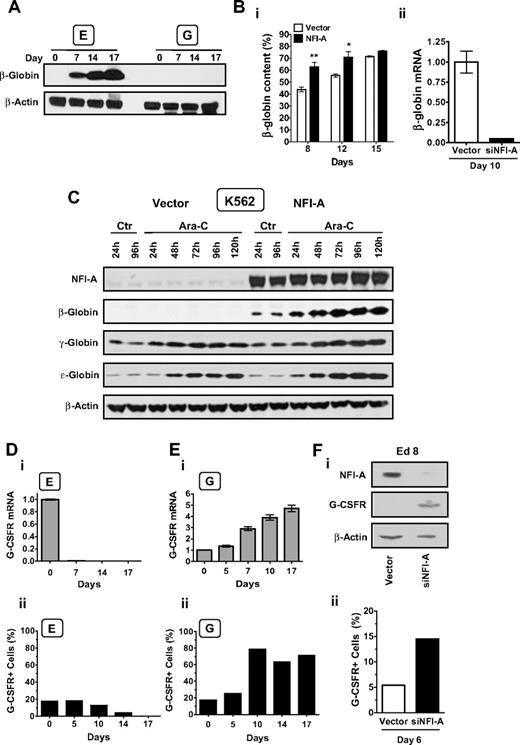
We selected the β-globin promoter as a target of NFI-A during E differentiation because binding sites were noted in the proximal promoter region.27 As expected, analysis of β-globin expression in E and G culture showed that the mRNA and protein level greatly increased in the E series (Figure 6A, supplemental Figure 5A) but was absent in the G lineage (Figure 6A). Using an HPLC assay, we observed that in E culture NFI-A–overexpressing cells showed an increased β-chain content at both early and intermediate days of differentiation (Figure 6Bi, supplemental Figure 5B). Conversely, real-time PCR showed a marked decrease of β-globin mRNA in siNFI-A cells (Figure 6Bii). Results in K562 cells were striking (Figure 6C, supplemental Figure 5C). In empty vector cells, β-globin was not detectable in both control cultures and after Ara-C-induced E differentiation, thus in line with previous reports.28,29 Conversely, β-globin mRNA and protein were strongly induced in NFI-A–overexpressing cells in either control or Ara-C-treated cultures. γ-Globin and ϵ-globin were equally present in control vector and NFI-A cells, and both increased in a time-dependent manner on Ara-C treatment. Altogether, these data show that ectopic expression of NFI-A specifically induces β-globin expression, not only in E culture of HPCs, but also in K562 cells, which normally do not express β-globin.
A functional relationship between NFI-A and 2 lineage-associated genes: β-globin and G-CSFR. (A) Immunoblot detection of β-globin at sequential stages of E and G culture. (B) (i) HPLC analysis of globin-chain content in unilineage E culture of HPCs transduced with empty vector or NFI-A. The β-globin chain content is expressed as the percentage of β-globin/non–α-globin chain content (ie, β/β + γ). Mean ± SEM values (n = 3). (ii) Real-time PCR of β-globin mRNA expression in siNFI-A–infected versus empty vector HPCs in E culture at day 10 (mean ± SEM values; n = 3). (C) Western blot analysis of β-globin, γ-globin, ϵ-globin, and NFI-A protein expression in control (Ctr) and Ara-C–treated vector- and NFI-A–transduced K562 cells. (Di) G-CSFR mRNA during E culture detected by real-time PCR (normalized mean ± SEM values; n = 3) and (ii) percentage of G-CSFR+ cells detected by flow cytometry that were permeabilized and stained with a PE-conjugated G-CSFR antibody. (Ei) G-CSFR mRNA during G culture, and (ii) percentage of G-CSFR+ cells detected by flow cytometry. (Fi) Western blot analysis showing G-CSFR reactivation after NFI-A knockdown in siNFI-A–infected HPCs in unilineage E culture at day 8. (ii) Surface expression of G-CSFR detected by flow cytometry in siNFI-A infected E cells at day 6.
A functional relationship between NFI-A and 2 lineage-associated genes: β-globin and G-CSFR. (A) Immunoblot detection of β-globin at sequential stages of E and G culture. (B) (i) HPLC analysis of globin-chain content in unilineage E culture of HPCs transduced with empty vector or NFI-A. The β-globin chain content is expressed as the percentage of β-globin/non–α-globin chain content (ie, β/β + γ). Mean ± SEM values (n = 3). (ii) Real-time PCR of β-globin mRNA expression in siNFI-A–infected versus empty vector HPCs in E culture at day 10 (mean ± SEM values; n = 3). (C) Western blot analysis of β-globin, γ-globin, ϵ-globin, and NFI-A protein expression in control (Ctr) and Ara-C–treated vector- and NFI-A–transduced K562 cells. (Di) G-CSFR mRNA during E culture detected by real-time PCR (normalized mean ± SEM values; n = 3) and (ii) percentage of G-CSFR+ cells detected by flow cytometry that were permeabilized and stained with a PE-conjugated G-CSFR antibody. (Ei) G-CSFR mRNA during G culture, and (ii) percentage of G-CSFR+ cells detected by flow cytometry. (Fi) Western blot analysis showing G-CSFR reactivation after NFI-A knockdown in siNFI-A–infected HPCs in unilineage E culture at day 8. (ii) Surface expression of G-CSFR detected by flow cytometry in siNFI-A infected E cells at day 6.
Next, we focused on the G-CSFR, which contains 2 conserved putative NFI sites. In E culture, the G-CSFR mRNA and protein levels (Figure 6Di-ii) dropped down to untraceable levels at terminal stages. Conversely, in G culture, the G-CSFR mRNA and protein level progressively increased through late maturation (Figure 6Ei-ii). In E cells, lentiviral knockdown of NFI-A was similarly associated with marked upmodulation of G-CSFR total protein and G-CSFR surface expression (Figure 6Fi-ii). These results were confirmed in HL-60 cells induced by RA treatment into G differentiation: NFI-A ectopic expression repressed the G-CSFR mRNA and surface protein expression. On the contrary, siNFI-A infection caused an induction of G-CSFR receptor protein level in HL-60 cells (supplemental Figure 3C-D).
NFI-A directly activates β-globin and represses G-CSFR proximal promoters, exhibiting a dual transcriptional activity in a lineage-specific context
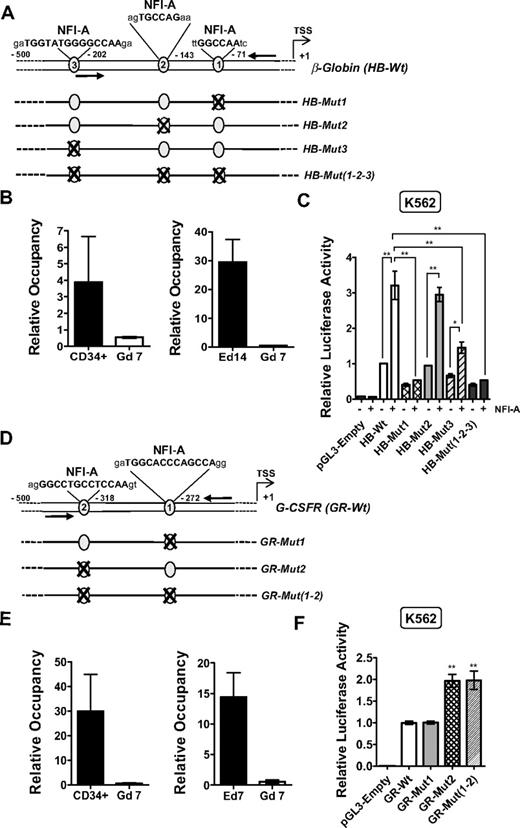
Because of the functional role of NFI-A in β-globin induction, we attempted to map the in vivo binding of endogenous NFI-A to the β-globin promoter using chromatin-immunoprecipitation (ChIP). Computational analysis by Chip-Mapper, Alibaba 2.1 and Mat-Inspector Professional software programs30–32 revealed 3 potential NFI-binding sites in the β-globin proximal promoter region (Figure 7A): specifically, phylogenetic footprinting showed site 1 to be embedded in a highly evolutionary conserved region (supplemental Figure 6A). Consequently, NFI-A chromatin occupancy in primary cells (physiologic binding) was determined using genomic DNA isolated from CD34+ HPCs, terminal E cells (E culture, day 14), and intermediate G cells (G culture, day 7) and immunoprecipitated using an NFI-A antibody. DNA was analyzed by multiplex PCR using primers specific for the promoter region containing the NFI-A–binding sites and a UR as an internal control. The linear range of amplification was confirmed by serial dilution of input DNA (supplemental Figure 7A). Figure 7B shows that NFI-A at these sites was present in CD34+ HPCs and greatly enriched in terminal E cells, characterized by elevated levels of both NFI-A and β-globin. Intermediate G cells, which lack NFI-A protein, were used as a negative control (Figure 7B, supplemental Figure 7Bi). We further extended ChIP analysis in K562 cells using real-time PCR in parallel with the multiplex PCR approach to asses NFI-A binding. ChIP was performed in vector- and NFI-A–transduced K562 cells in the absence or presence of Ara-C. As shown consistently by the 2 independent methods, only NFI-A–transduced cells show NFI-A binding to the β-globin promoter, and this occupancy increases in a time-dependent manner in the presence of Ara-C (supplemental Figure 8A,Ci-ii), complementing the trend observed in primary cells. As expected, real-time ChIP analysis in the HL-60 cell line shows no significant binding over the course of RA treatment (supplemental Figure 8E).
Antithetic effects of NFI-A on the β-globin and G-CSFR promoters in vivo. (A) Schematic representation of the β-globin proximal promoter. NFI-A DNA-binding site sequences are highlighted in bold characters. TSS indicates the transcriptional start site; arrows, the position of the primers used for the ChIP analysis. Mutant sites are numbered according to their vicinity to the TSS. (B) Relative quantification of NFI-A occupancy on the β-globin promoter. Chromatin was immunoprecipitated with an NFI-A antibody, and the bound DNA was analyzed by multiplex PCR using specific primers corresponding to the β-globin promoter region containing the NFI-A binding sites and an unrelated intergenic genomic region (UR) as an internal control. Histograms represent the mean ± SEM from 3 independent DNA preparations; PCR analyses were repeated at least 3 times. (C) Promoter assay showing a positive role of NFI-A in β-globin proximal promoter activation. Wild-type and mutant constructs were assayed for transcriptional activity relative to the endogenous NFI-A or in a cotransfection with an NFI-A–expressing plasmid. The firefly luciferase values were normalized to the Renilla luciferase values for each transfection, and the relative luciferase activity is represented as relative fold induction over the HB-Wt transfection (mean ± SEM values from 4 independent transfections). Each reading was repeated at least 2 times. (D) Schematic representation of the NFI-A binding sites on the G-CSFR promoter. (E) Relative quantification of NFI-A occupancy on the G-CSFR promoter. Chromatins were immunoprecipitated with an NFI-A antibody, and the bound DNA was analyzed by multiplex PCR using specific primers corresponding to the G-CSFR promoter region containing the NFI-A binding sites and an unrelated intergenic genomic region (UR) as an internal control. Histograms represent the mean ± SEM from 3 independent DNA preparations; multiplex PCR analysis was repeated at least 3 times. (F) Promoter assay showing NFI-A repressive activity on the G-CSFR promoter performed in K562 cells. Wild-type or mutant promoter constructs were transfected, and firefly luciferase activity was normalized to Renilla luciferase activity for each transfection. The relative luciferase activity is represented as relative fold induction over the GR-Wt transfection. Data represent mean ± SEM from 3 independent transfections. Each reading was repeated at least 2 times.
Antithetic effects of NFI-A on the β-globin and G-CSFR promoters in vivo. (A) Schematic representation of the β-globin proximal promoter. NFI-A DNA-binding site sequences are highlighted in bold characters. TSS indicates the transcriptional start site; arrows, the position of the primers used for the ChIP analysis. Mutant sites are numbered according to their vicinity to the TSS. (B) Relative quantification of NFI-A occupancy on the β-globin promoter. Chromatin was immunoprecipitated with an NFI-A antibody, and the bound DNA was analyzed by multiplex PCR using specific primers corresponding to the β-globin promoter region containing the NFI-A binding sites and an unrelated intergenic genomic region (UR) as an internal control. Histograms represent the mean ± SEM from 3 independent DNA preparations; PCR analyses were repeated at least 3 times. (C) Promoter assay showing a positive role of NFI-A in β-globin proximal promoter activation. Wild-type and mutant constructs were assayed for transcriptional activity relative to the endogenous NFI-A or in a cotransfection with an NFI-A–expressing plasmid. The firefly luciferase values were normalized to the Renilla luciferase values for each transfection, and the relative luciferase activity is represented as relative fold induction over the HB-Wt transfection (mean ± SEM values from 4 independent transfections). Each reading was repeated at least 2 times. (D) Schematic representation of the NFI-A binding sites on the G-CSFR promoter. (E) Relative quantification of NFI-A occupancy on the G-CSFR promoter. Chromatins were immunoprecipitated with an NFI-A antibody, and the bound DNA was analyzed by multiplex PCR using specific primers corresponding to the G-CSFR promoter region containing the NFI-A binding sites and an unrelated intergenic genomic region (UR) as an internal control. Histograms represent the mean ± SEM from 3 independent DNA preparations; multiplex PCR analysis was repeated at least 3 times. (F) Promoter assay showing NFI-A repressive activity on the G-CSFR promoter performed in K562 cells. Wild-type or mutant promoter constructs were transfected, and firefly luciferase activity was normalized to Renilla luciferase activity for each transfection. The relative luciferase activity is represented as relative fold induction over the GR-Wt transfection. Data represent mean ± SEM from 3 independent transfections. Each reading was repeated at least 2 times.
To assess the contribution of individual NFI-A sites in β-globin transcription, we designed promoter-luciferase fusion constructs containing the wild-type β-globin promoter and NFI-A site-specific mutants (Figure 7A). Figure 7C shows that cotransfection of HB-Wt with a plasmid encoding NFI-A in K562 cells led to an approximately 3-fold increase in transcription with respect to HB-Wt alone. In contrast, HB-Mut1 abolished transcriptional induction even in the presence of ectopic NFI-A. HB-Mut3 responded to NFI-A to a lesser extent, whereas HB-Mut2 behaved similarly to HB-Wt. The mutant construct, containing 3 mutated sites (HB-Mut1, Mut2, Mut3), exhibited no transcriptional induction on NFI-A ectopic expression, as observed for HB-Mut1. In conclusion, the results indicate that NFI-A binds to and activates the β-globin promoter through primarily site 1 and secondarily site 3.
The G-CSFR promoter contains 2 conserved putative NFI-binding sites (supplemental Figure 6B). As a result, we performed multiplex ChIP analysis of the promoter using primers including both sites (Figure 7D). Genomic DNA from CD34+ HPCs, intermediate E cells (E day 7), and intermediate G cells (G day 7) was immunoprecipitated using an NFI-A antibody. As shown in Figure 7E and supplemental Figure 7Bii, we found binding of NFI-A to the G-CSFR sites in CD34+ HPCs and E cells, but not in G cells. Binding of NFI-A to G-CSFR sites within NFI-A–transduced K562 cells was observed by multiplex and real-time PCR and increased with time on exposure to Ara-C (supplemental Figure 8B,Di-ii). On the contrary, in the HL-60 cell line, NFI-A is present on the G-CSFR promoter in both vector- and NFI-A–transduced control cells, and rapidly falls off in vector cells on RA treatment, whereas its occupancy is only slightly reduced in NFI-A–overexpressing cells on RA treatment (supplemental Figure 8F). To further demonstrate that NFI-A transcriptionally represses the G-CSFR promoter, we performed a promoter assay in K562 cells, using a wild-type promoter construct and site-specific mutants (Figure 7D). As shown in Figure 7F, the basal activity of the G-CSFR wild-type promoter construct (GR-Wt) was enhanced by mutating site 2 (GR-Mut2), as shown by a rescue of transcription of approximately 2-fold. The mutant construct bearing a mutation in site 1 (GR-Mut1) did not affect its transcriptional activity; conversely, the double-mutant construct (GR-Mut1, -Mut2) behaved similarly to GR-Mut2. In conclusion, we postulate that NFI-A binds and transcriptionally represses the G-CSFR promoter, specifically through site 2, whereas site 1 is inactive.
Discussion
Our studies indicate that NFI-A plays a major role in the control of the erythroid-granulocytic lineage decision at the level of HPCs. NFI-A accumulation during initial E differentiation results in progressive activation of β-globin gene transcription, coupled with repression of G-CSFR, thus channeling HPCs and early precursors into the E lineage and shutting off their G potential. Conversely, NFI-A suppression in early granulopoiesis activates G-CSFR transcription and impedes β-globin expression, thereby directing HPCs and early precursors into the G pathway.
As a model system, we used unilineage E and G cultures of human HPCs: these assays recapitulate the in vivo differentiation/maturation of these hematopoietic series, hence allowing analysis of lineage-specific cells at discrete sequential stages of erythropoiesis and granulopoiesis.20,23 The expression of NFI-A, although low in early HPC differentiation, was either sharply up-regulated or fully suppressed in later stages of E or G culture, respectively. A similar pattern has been observed for other TFs controlling lineage specification, such as GATA-1,33 PU.1,34 and C/EBPα35 : all are expressed at low levels in “primed” HSCs/HPCs, whereas their upmodulation in later stages is critical to promote lineage(s)-specific differentiation program(s) and to antagonize the expression of genes specific for the other series.
NFI-A accumulation is necessary for both E differentiation and suppression of the G potential. In E culture, siNFI-A–transduced HPCs were incapable of E differentiation but generated predominantly myeloid cells. This is presumably the result of the presence of small amounts of IL-3 and GM-CSF in the E medium: these 2 cytokines display a pro-survival activity on granulomonocytic cells.18,36 Furthermore, GM-CSF stimulates M-CSF release in monocytes,37 giving rise to a positive autocrine feedback loop. Conversely, HPCs transduced with NFI-A in G culture displayed a block in G differentiation.
NFI-A is not only necessary for E differentiation/maturation but is also capable of driving it. During normal erythropoiesis, Epo binds to the EpoR and activates signaling cascades, resulting in antiapoptotic effects and proliferation coupled with E differentiation/maturation.38 In suboptimal E culture, because of a low or minimal Epo stimulus, NFI-A overexpression was able to restore E differentiation/maturation: this suggests that NFI-A may directly target key molecules involved in the E gene program, independently of EpoR signaling.
Altogether, NFI-A upmodulation exerts sharp positive effects on erythropoiesis, whereas its downmodulation is permissive for granulopoiesis. Indeed, the results obtained in E + G liquid phase and clonogenic culture of HPCs indicate that NFI-A upmodulation or downmodulation governs the erythro-granulocytic lineage branchpoint, directing precursor cells into the E or G fate, respectively. Consequently, we hypothesized that NFI-A may exert these actions through transcriptional control of genes playing pivotal functions in the E or G program: indeed, a bioinformatic screen suggested that the β-globin and G-CSFR promoters were potential NFI-A targets.
NFI-A overexpression in E culture resulted in a significant increase of the β-globin content in erythroblasts. In the erythroleukemic K562 cells, incapable of synthesizing β-globin even on Ara-C induced E differentiation,28 ectopic expression of NFI-A induced a marked expression of β-globin. These results indicate a tight correlation between NFI-A levels and β-globin expression. Identifying new players involved in the transcriptional regulation of β-globin gene is potentially of clinical significance because mutations in the gene or promoter cis-elements are relevant in β-thalassemias. We observed that NFI-A specifically binds the β-globin proximal promoter, primarily at position −73 to −78, and activates transcription. This NFI-A–binding site overlaps with the CCAAT element, which also binds to other factors and is 1 of 3 elements required for maximal transcription of the β-globin promoter.39 Interestingly, a recently described case of β++-thalassemia (mild β-thalassemia intermedia) was linked to a mutation of −73 (A to T) within the conserved CCAAT box.40
NFI-A is a novel negative transcriptional regulator of G-CSFR expression, which plays a crucial role in the production and function of granulocytes.41 Knockdown of NFI-A in E culture led to activation of G-CSFR, whereas NFI-A transduction in G culture blocked the receptor expression. We found that NFI-A binds to a cis-element located between nucleotides −318 and −334 in the G-CSFR promoter and represses its transcription in reporter and functional assays. So far, only a few transcriptional regulators of this receptor have been discovered; some (eg, PU.1 and C/EBPα) have been extensively characterized,42,43 whereas the contribution of others (eg, AP-1 and AP-2, GF-1) still needs to be clarified.44 It is noteworthy that the G-CSFR is also expressed in human monocytes and directly modulates inflammatory cytokine secretion.45 In acute promyelocytic leukemias bearing leukemogenic fusion proteins, G-CSFR function is disrupted46 : NFI-A knockdown may provide an attractive therapeutic approach, possibly in combination with ATRA + G-CSF differentiation therapy.46
In ChIP experiments, we observed that chromatin occupancy by NFI-A at β-globin and G-CSFR sites increases with E differentiation. This is seemingly because of the rise of NFI-A concentration. In addition, other components may be involved, such as the chromatin status, interacting factors bound to nearby cis-acting elements, and the number and/or affinity of motif features.47 Further studies will be required to determine the mechanism of action of NFI-A on these promoters, as well as to identify other possible NFI-A targets playing a significant role in the E and G differentiation program.
Recent studies identified NFI-A as a major component of regulatory pathways controlling RA-induced leukemic G differentiation and monocyte development. In both cases, NFI-A expression was suppressed by lineage-specific microRNAs, the granulocytic miR-223, and the monocytic miR-424, respectively, controlled by the C/EBPα and PU.1 TFs.12,13 Whereas these studies indicated a role of NFI-A in myeloid differentiation, the mechanism of these actions was not elucidated, in that no NFI-A target gene was identified. Interestingly, miR-223 is sharply downmodulated during normal E differentiation (results not shown): hypothetically, this shutdown may add to the strong transcriptional induction of NFI-A expression.
TFs involved in hematopoietic lineage specification are part of cell-specific networks, participating in positive and negative cross-regulations at the transcriptional and posttranscriptional level.48 This report uncovers that NFI-A is a novel piece of this network: ongoing studies aim to further delineate the interlink of NFI-A with other known TFs and circuitries.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Cerio and A. D'Angiò for technical assistance, V. Michetti and M. Blasi for editorial assistance, and Dr Richard Gronostajski for sharing NFI reagents.
This work was supported by the Italy USA Oncology Program and the Biotechnology Program, Istituto Superiore di Sanità (C.P.); Associazione Italiana per la Ricerca sul Cancro (C.P., C.N., I.B.), University “La Sapienza,” and Ministry for University and Research (PRIN, C.N., I.B.); EU project SIROCCO (LSHG-CT-2006-037900), and ESF project “NuRNASu,” Centro di Eccellenza Biologia e Medicina Molecolare (I.B.). L.M.S. is supported by a fellowship from the University of Rome “La Sapienza.”
Authorship
Contribution: L.M.S. and A.S. designed and performed research, analyzed and interpreted data, and wrote the manuscript; E.P. performed research and analyzed data; M. Ballarino, O.M., S.S., N.F., G.C., M.L.D.M., and A.F. performed research; M. Biffoni performed flow cytometric analysis; G.M., M.G., and I.B. contributed vital new reagents or analytical tools; and C.P. and C.N. coordinated the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cesare Peschle, Istituto di Ricovero e Cura a Carattere Scientifico Multimedica, Via Gaudenzio Fantoli 16/15, Milan, 20138, Italy; e-mail: cesare.peschle@iss.it; or Clara Nervi, University of Rome “La Sapienza” & San Raffaele Biomedical Park Foundation, Via di Castel Romano 100, Rome, 00128, Italy; e-mail: clara.nervi@uniroma1.it.
References
Author notes
*L.M.S. and A.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal