In this issue of Blood, Wei and colleagues investigate the overlapping roles of Ets1 and Ets2 in the regulation of endothelial cell function and survival during embryonic angiogenesis.1
Ets1 and Ets2 are known to regulate endothelial-restricted gene expression and endothelial function. For example, Ets1 has been shown to regulate the expression of several genes that regulate endothelial function and angiogenesis including VEGF, VEGF-R2, and Tie2.2 However, homozygous inactivation of Ets1 is associated with defects in T-cell function, but no defects in vascular development or angiogenesis.3 Similarly, while Ets2 is a critical regulator of trophoblast function during extraembryonic development, it is dispensable for development of the embryo proper.4 Mice with a homozygous hypomorphic mutant of Ets2, in which the conserved threonine 72 phosphorylation site is mutated, Ets2T72A/T72A mice, are viable and appear normal.5 In an elegant series of experiments, Wei et al uncover the overlapping roles of Ets1 and Ets2 during vascular development. First, they cross Ets1−/− mice with Ets2T72A/T72A to derive double mutant Ets1−/−; Ets2T72A/T72A mice. The double mutants are embryonal lethal and begin to develop vascular abnormalities at day E10.5, including a marked reduction in vascular complexity, defective branching, with the remaining vessels being dilated. To further validate that the phenotype is specifically related to reduced function of Ets2 in endothelial cells, double mutants were generated where targeted deletion of Ets2 was achieved by crossing floxed Ets2 mice with Tie2-Cre transgenic mice. When these mice were crossed with Ets1−/− mice to generate Tie2-Cre, Ets1−6/−, and Ets2fl/fl mice, they did not survive beyond E14.5 and exhibited the same phenotype as the Ets1−/− and Ets2T72A/T72A mice. Wei et al also demonstrate a 4-fold increase in the rate of apoptosis of endothelial cells in the double mutant mice that is at least in part explained by concomitant reductions in the expression of the antiapoptotic genes Bcl-XL and cIAP2.
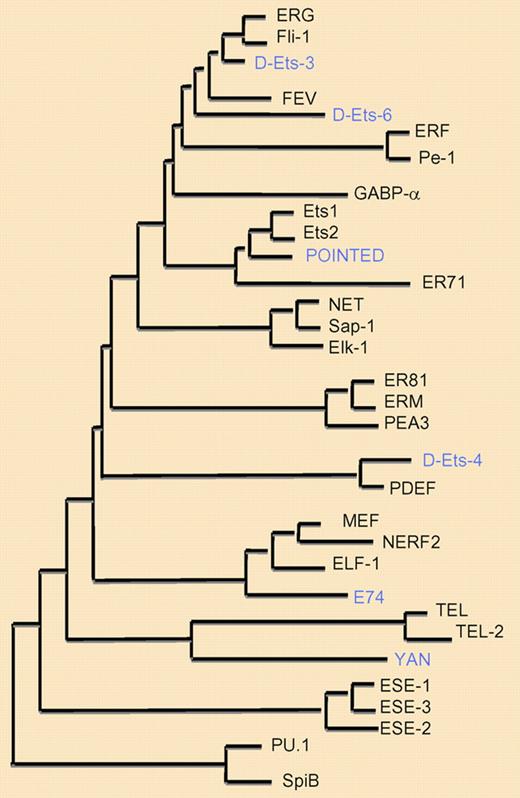
Phylogenetic tree of ETS family members. Phylogenetic tree demonstrating the evolutionary relationship between the different ETS factor family members, based on the relative conservation of the Ets domain, linking members with closely homologous amino acid sequences. Only human (black) and drosophila (blue) ETS members are shown.
Phylogenetic tree of ETS family members. Phylogenetic tree demonstrating the evolutionary relationship between the different ETS factor family members, based on the relative conservation of the Ets domain, linking members with closely homologous amino acid sequences. Only human (black) and drosophila (blue) ETS members are shown.
Ets factors can be subcategorized into subfamilies based on DNA and protein sequence homology (see figure). For example, Elf-1, Nerf-2, and Mef are highly related and belong to one subfamily. Similarly, Fli1 and ERG constitute another subfamily, as do Ets1 and Ets2. The subfamily members often have overlapping functions. From an evolutionary standpoint, this built-in redundancy may be protective against the effects of genetic mutations of the individual family members for critical developmental processes such as vasculogenesis. The study by Wei et al underscores the potential need to simultaneously disrupt the function of 2 or more highly related factors to uncover their overlapping functions in specific cell types, organs, or tissues.
From a therapeutic standpoint, recent studies support an important role for ETS factors as critical transcriptional regulators of tumor angiogenesis. Local administration of a dominant-negative form of Ets-1, encoding the DNA binding domain, significantly blocked tumor angiogenesis and tumor growth.6 Similarly, systemic delivery of membrane permeable peptides encoding the terminal portion the ETS domain of Elf-1 markedly inhibits the growth of B16 melanoma tumors by blocking tumor angiogenesis. By targeting the DNA binding domain, both of these therapeutic agents very likely target subfamilies of ETS factors rather than being entirely selective for a particular ETS factor.7 Finally, with recent advances in drug discovery, the identification and/or development of small molecule inhibitors of subfamilies of ETS factors may now be possible by taking advantage of structural similarities among the subfamilies in the DNA binding domain or other functionally conserved regions of the proteins.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal