Abstract
In chronic lymphocytic leukemia (CLL), spontaneous regressions are an exceptional phenomenon, whose biologic features are unknown. We describe 9 CLL patients who underwent a spontaneous clinical regression over an 11-year follow-up, despite a residual neoplastic clone detected by flow cytometry. CD38 and ZAP-70 were negative in all cases. Immunoglobulin heavy chain variable region (IgVH) genes, mutated in all 7 evaluable patients, were restricted to the VH3 family in 6, with the usage of VH3-30 gene in 2. The light chain variable region genes were mutated in 6 of 8 cases, with the use of Vκ4-1 gene in 3. Microarray analysis of CLL cells showed a distinctive genomic profile with an overrepresentation of BCR-related and ribosomal genes, regulators of signal transduction and transcription. The number of activated T lymphocytes expressing IFN-γ, TNF-α, and IL-4 was similar between CLL in spontaneous regression and healthy persons. In conclusion, spontaneous clinical regressions can occur in CLL despite the persistence of the neoplastic clone, and the biologic features include negative CD38 and ZAP-70, mutated VH3-30 and Vκ4-1 genes. The peculiar gene profile suggests that BCR signaling may play an important role in this scenario as the most significant feature of the leukemic clone in regression.
Introduction
Chronic lymphocytic leukemia (CLL) is a B-cell disorder with a variable natural history and prognosis. More than one-half of the patients at diagnosis show an early stage of the disease. The median overall survival for this group of patients is approximately 10 years.1,2 They normally undergo clinical observation up to disease progression and treatment requirement. The interval from diagnosis to progression is extremely variable in duration, lasting from a few months to decades. Different biologic parameters recently identified allow clinicians to predict at diagnosis the time to progression and survival in this group of patients.3,4
We have previously reported on the unique biologic profile displayed by a group of patients with highly stable disease for more than 10 years,5,6 characterized by a typical morphology and immunophenotype, a normal T-cell subset distribution and normal serum immunoglobulin (Ig) levels, lack of CD38 expression, absence of poor prognosis genetic markers or p53 abnormalities, and a mutated profile of the Ig heavy chain variable region (VH) genes, with no Ig VH3-21 involvement.5 One-fourth of highly stable CLL used the VH3-72 gene and, in some cases, expressed VH and light chain variable region (VL) sequences with homologous third complementarity-determining region (CDR3s), suggesting recognition of a common antigen.6
The occurrence of a spontaneous regression of CLL in the absence of any previous treatment is exceptional. In the literature, there are only a few papers in which this phenomenon has been reported.7-11 The clinical features of these cases have been described, but there is no information on the biologic markers of their disease or on the pathogenesis of this event. Particularly, it is not known whether there are distinctive features that can identify among patients with good prognosis those who may experience over time a spontaneous regression of their disease. Moreover, it is interesting to note that such an event can occur also in patients with trisomy 12.9
In the present study, we describe 9 patients with CLL followed at our institution, in which the occurrence of a spontaneous clinical regression has been observed over a period of 3 to 28 years from diagnosis and who underwent an extended biologic evaluation of the disease.
Methods
Patients and samples
Spontaneous regression was defined as the achievement of a clinical regression of the disease (ie, absence of lymphadenopathy, splenomegaly, or constitutional symptoms; peripheral lymphocytosis less than 4 × 109/L; no anemia; no thrombocytopenia) in the absence of any previous treatment. Because no bone marrow (BM) assessment of CLL infiltration was performed, we could not evaluate the occurrence of a clinical remission.
All patients, 3 men and 6 women, were diagnosed and followed at the Institute of Hematology, “Sapienza” University of Rome. At the time of diagnosis, the median age was 53 years (range, 41-73 years). The proportion of circulating lymphocytes was 68% (range, 64%-90%) of the white blood cells, with a median lymphocyte count of 11.8 × 109/L (range, 8.7-27.6 × 109/L), 78% of which was represented by CLL cells (range, 65%-82%). Clinical stage according to the classifications of Binet et al1 and Rai et al2 was A/0 in 7 and A/II in 2 cases, who presented a mild splenomegaly (longitudinal diameter, 13 cm). After diagnosis, patients were seen at our institution every 3 months during the first year, every 4 months during the second year, every 6 months during the third year, and subsequently once a year.
Over the years, the disease remained stable, and no treatment requirement occurred. A progressive and gradual decrease in the lymphocyte count was observed over time. Median time from the highest lymphocyte count reached to the first lymphocyte count below 4 × 109/L was 9 years (range, 3-24 years). Thus, after a median follow-up of 11 years (range, 3-28 years) from diagnosis, a clinical spontaneous regression of the disease occurred. This phenomenon has remained stable, and the study was performed after a median follow-up of 16 years (range, 3-29 years) from diagnosis; at this time, the median lymphocyte count was 3.16 × 109/L (range, 1.3-4.0 × 109/L) and the median percentage of lymphocytes was 43% (30%-53%; Figure 1). The hemoglobin level and platelet counts remained within the normal range. No enlarged nodes or splenomegaly were detected by physical examination and abdominal ultrasound scans. The clinical features of the patients are summarized in Table 1.
Lymphocyte counts over time. The variation over time of the absolute number of lymphocytes (×109/L) is shown (m indicates month; y, year).
Lymphocyte counts over time. The variation over time of the absolute number of lymphocytes (×109/L) is shown (m indicates month; y, year).
Patients' clinical features
| Case . | Age at diagnosis, y/sex . | Stage at diagnosis . | Lymphocyte count, ×109/L . | Stage at study . | Medication intake . | Hepatitis B or C . | Years (from diagnosis to study) . | ||
|---|---|---|---|---|---|---|---|---|---|
| At diagnosis . | Highest . | At study . | |||||||
| 1 | 64/F | A0 | 9.8 | 17.6 | 1.39 | A0 | Ramipril, nitroglycerin skin patches, phenytoin (since 2005), anticholesterol drug | Negative | 13 |
| 2 | 51/F | A0 | 11.6 | 31.3 | 2.1 | A0 | Atorvastatin, antihypertensive, aspirin | Negative | 21 |
| 3 | 50/M | A2 | 16.2 | 16.2 | 3.16 | A0 | Antihypertensive/alcohol | Negative | 19 |
| 4 | 63/M | A0 | 10.6 | 13.6 | 3.5 | A0 | Bisoprolol, pantoprazole, perindopril, ticlopidine, simvastatin, pentoxifylline | Negative | 13 |
| 5 | 73/F | A2 | 15.7 | 15.7 | 3.86 | A0 | None | NA | 3 |
| 6 | 53/F | A0 | 27.6 | 31.0 | 4.0 | A0 | Losartan, hydrochlorothiazide | NA | 29 |
| 7 | 53/F | A0 | 11.7 | 12.9 | 2.5 | A0 | Carvedilol, ramipril, aspirin, atorvastatin, omega-3 fatty acids, acarbose, insulin | NA | 21 |
| 8 | 41/F | A0 | 12.3 | 26.0 | 1.4 | A0 | Hormonal replacement | NA | 16 |
| 9 | 66/M | A0 | 8.7 | 14.6 | 4.0 | A0 | Antihypertensive, aspirin | Negative | 13 |
| Case . | Age at diagnosis, y/sex . | Stage at diagnosis . | Lymphocyte count, ×109/L . | Stage at study . | Medication intake . | Hepatitis B or C . | Years (from diagnosis to study) . | ||
|---|---|---|---|---|---|---|---|---|---|
| At diagnosis . | Highest . | At study . | |||||||
| 1 | 64/F | A0 | 9.8 | 17.6 | 1.39 | A0 | Ramipril, nitroglycerin skin patches, phenytoin (since 2005), anticholesterol drug | Negative | 13 |
| 2 | 51/F | A0 | 11.6 | 31.3 | 2.1 | A0 | Atorvastatin, antihypertensive, aspirin | Negative | 21 |
| 3 | 50/M | A2 | 16.2 | 16.2 | 3.16 | A0 | Antihypertensive/alcohol | Negative | 19 |
| 4 | 63/M | A0 | 10.6 | 13.6 | 3.5 | A0 | Bisoprolol, pantoprazole, perindopril, ticlopidine, simvastatin, pentoxifylline | Negative | 13 |
| 5 | 73/F | A2 | 15.7 | 15.7 | 3.86 | A0 | None | NA | 3 |
| 6 | 53/F | A0 | 27.6 | 31.0 | 4.0 | A0 | Losartan, hydrochlorothiazide | NA | 29 |
| 7 | 53/F | A0 | 11.7 | 12.9 | 2.5 | A0 | Carvedilol, ramipril, aspirin, atorvastatin, omega-3 fatty acids, acarbose, insulin | NA | 21 |
| 8 | 41/F | A0 | 12.3 | 26.0 | 1.4 | A0 | Hormonal replacement | NA | 16 |
| 9 | 66/M | A0 | 8.7 | 14.6 | 4.0 | A0 | Antihypertensive, aspirin | Negative | 13 |
NA indicates not available.
All patients but one (no. 9) underwent peripheral blood (PB) sampling after giving their informed consent according to the Declaration of Helsinki for the blood collection and biologic analyses included in the present study, which was approved by the Institutional Review Board of the Department of Cellular Biotechnologies and Hematology, Sapienza University, Rome. The regression persisted thereafter, because all patients have remained in persistent regression after an updated follow-up of 16 years (range, 5-31 years) with the exception of case no. 9, who showed a lymphocyte count of 5.2 × 109/L at the last visit.
PB cell count, morphology, and immunophenotype
The PB counts were analyzed with the use of the H3 and ADVIA instruments (Bayer). The morphologic observation was carried out on May-Grünwald/Giemsa–stained PB films.
PB cells from fresh whole blood samples were characterized with 4-color immunostaining with B- and T-cell markers by flow cytometry. The panel included the following monoclonal antibodies (mAbs): CD5-phycoerythrin (PE), CD20-fluorescin isothiocyanate (FITC), CD22-PE, CD38-PE, CD2-FITC, CD3-FITC, CD4-allophycocyanin (APC), CD8-PE, CD79b-FITC, CD56-FITC, CD16-PE (from Becton Dickinson Immunocytometry Systems); CD18-FITC, CD11a-FITC (Invitrogen); FMC7-FITC, anti-κ chain–PE, anti-λ chain–FITC, CD23-FITC (Dako); anti-CD19–phycoerythrin/cyanin 5·1 (Beckman Coulter). A FACSCalibur flow cytometer (Becton Dickinson) and CellQuest software (Becton Dickinson) were used.
Samples purification
Highly purified CD19+ cells (> 98%) were obtained from PB mononuclear cells by standard positive selection with the use of an anti-CD19 antibody conjugated with magnetic beads (Miltenyi Biotec) and were used for IgVH mutation and microarray study. For the latter, samples with at least 95% of CD5+/CD19+ clonal cells after purification were selected. The CD19− fraction was cryopreserved in the presence of DMSO (10%) in liquid nitrogen and subsequently used to evaluate T-cell cytokine production.
Polymerase chain reaction amplification of immunoglobulin rearrangements and sequence analysis
Genomic DNA was extracted from leukemic cells with the use of Wizard Genomic DNA Purification kit (Promega) according to the manufacturer's instructions. IgHV-D-J, IgKV-J, and IgLV-J gene rearrangements were amplified by polymerase chain reaction (PCR) from genomic DNA with the use of Taq polymerase with family-specific primes hybridizing to leader or framework (FR) 1 sequences and JH, Jκ, or Jλ, as previously described.12 PCR products were size-selected by electrophoresis in 2% agarose gel containing 10 mg/mL ethidium bromide (Sigma-Aldrich). The expected products were excised from the agarose gel and purified by MinElute Gel Extraction Kit (QIAGEN).
Sequencing was performed with an automatic ABI PRISM 3100 AVANT DNA genetic analyzer (Applied Biosystems). IgHV-D-J, IgKV-J, and IgLV-J nucleotide sequences were aligned to V-BASE sequence directory with the use of the Mac Vector software version 6.0.1 (Oxford Molecular Group), to IgBLAST database13 and to the international ImMunoGeneTics information system.14 Sequences were considered unmutated if deviation from the closest germline gene was less than 2%.
Analyses of HCDR3 and LCDR3 rearrangements
Heavy chain third complementarity-determining region (HCDR3) length was determined according to Kabat et al15 by counting the number of amino acids between position H94 at the 3′ end of FR3 (usually 2 amino acids downstream of the conserved cysteine) and position H102 at the beginning of FR4 (a conserved tryptophan in all JH segments).
The length of VL CDR3 was determined by counting the number of amino acids between position L89 (preceded by a conserved cysteine) at the end of FR3 and the position L97 at the beginning of FR4 (a conserved phenylalanine in all JL segments).
Microarray preparation and data analysis
RNA was extracted from 22 CD19-enriched CLL cases and 3 CD19-enriched lymphocytes from the PB of healthy donors. Of the 22 patients analyzed, 4 had experienced a spontaneous regression of CLL, whereas the remaining 18 CLL and 3 normal B-lymphocyte samples were used for comparison purposes. The CLL samples were from patients in different phases of their disease (stable versus progressive). In the spontaneous regression samples, the CD19+ fraction was entirely represented by the CLL clone, with less than 2% of residual normal B lymphocytes in the CD19+ gate.
Total RNA was extracted with the use of the TRIzol reagent (Invitrogen). To assess RNA quality, 2 μL RNA from each sample was analyzed by electrophoresis on agarose gel containing 10 mg/mL ethidium bromide (Sigma-Aldrich); for all samples, the 260/280 ratio was more than 1.8, as required for microarray analysis. For oligonucleotide array analysis, the HGU133A.2 arrays were used. The detailed protocol for sample preparation and microarray processing is available from Affymetrix (www.affymetrix.com). For statistical analysis, Affymetrix gene expression data were processed with dChip software (www.dchip.org), which uses an invariant set normalization method. The array with the median overall intensity was chosen as the baseline for normalization. Model-based expressions were computed for each array and probe set, using the PM-MM model.16 For unsupervised analysis, the following nonspecific filtering criteria were used: (1) gene expression levels were required to be higher than 300 in at least 10% of the samples; (2) the ratio of the standard deviation to the mean expression across all samples was required to be between 0.5 and 1000. To compare the 4 samples with spontaneous regression analyzed versus the remaining IgVH-mutated cases, a t test was used; only the genes with a P value less than .05 and a fold change greater than 1.5 were retained. Furthermore, to compare patients with a different clinical scenario, namely spontaneous regression, stable disease, and progressive disease, an ANOVA was applied with a P value less than .001. The microarray data have been deposited in Gene Expression Omnibus under the accession number GSE15777.
Intracellular T-cell cytokine expression analysis
Circulating lymphocytes from patients with CLL in spontaneous regression (n = 6), CLL with stable disease (n = 5; 2 unmutated IgVH and 3 mutated IgVH), and with progressive disease (n = 7; 5 unmutated IgVH and 2 mutated IgVH), and normal controls (n = 5) were evaluated for T-cell cytokine production. Briefly, T cells were washed, resuspended in complete medium with the addition of phorbol 12-myristate 13-acetate 25 ng/mL (Sigma-Aldrich) and ionomycin 1 μg/mL (Iono; Sigma-Aldrich) for cell activation, in the presence of 10 μg/mL Brefeldin A (Sigma-Aldrich) to inhibit intracellular transports and incubated for 4 hours at 37°C in 5% CO2 in air. Cells were then washed and stained with cell surface–conjugated mAbs. For cell-surface marker analysis, T cells were washed twice in Ca++/Mg++-free phosphate-buffered saline (Cambrex) and subsequently incubated for 20 minutes at 4°C with FITC- and PE-CD4, APC-CD8, PE-CD56, and peridinin chlorophyll protein (PerCP)–CD3 (all from Becton Dickinson). For intracellular staining, cells were permeabilized with the use of the Fix and Perm cell permeabilization kit (Invitrogen), according to the manufacturer's instructions, and stained with the anti–human IL-4, TNF-α, and IFN-γ FITC-conjugated mAbs (Becton Dickinson). Cells were analyzed with the FACSCalibur flow cytometer and the CellQuest software (Becton Dickinson). The percentage and absolute number of activated CD3+ cells expressing IFN-γ, TNF-α, and IL-4 were determined.
Statistical analysis
Differences between series of continuous and discrete variables were evaluated for significance by the Student t test and χ2/Fisher exact test, respectively.
Results
PB morphology and immunophenotype
Morphologically, lymphocytes were mostly small in size with a proportion of large granular lymphocytes (LGLs) that varied from 0% to 35% (median, 17%).
Immunophenotype documented a variable proportion of clonal B cells that ranged between 3% and 63% (median, 44%) of total lymphocytes; they were positive for CD5, CD23, CD19, and CD20 and negative for CD79b, FMC-7 (5 of 6), CD18, and CD11a (5 of 6), with κ light chain restriction in 6 cases and λ light chain restriction in 3. Therefore, CLL cells showed a typical phenotype, with a Matutes immunophenotypic score of 4 to 5 in all cases.
CD38 on B lymphocytes was less than 7% in all cases, ranging from 0% to 6% (median, 1.5%). ZAP-70 RNA expression levels were low, as defined by quantitative PCR on CD19-enriched lymphocytes (data not shown).
Residual normal B cells represented 0.3% to 5% of total lymphocytes and 0.4% to 57% of CD19+ lymphocytes. The proportion of T cells ranged from 26% to 72% and natural killer (NK) cells from 1% to 21%. The CD4/CD8 ratio was normal in all but one case, ranging from 1.2 to 3.25 (median, 1.8; Table 2). The PB immunophenotype was evaluated during the course of the disease in cases no. 1, no. 2, and no. 6, showing a slowly progressive reduction of the CLL clone over time.
Patients' biologic features at the time of the study
| Case . | Lymphocyte count, ×109/L . | CLL cells, % . | Normal B cells, % . | T cells, % . | NK cells, % . | CD4/CD8 ratio . | LGL (morphology), % . | CD38+ cells, % . |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.39 | 3 | 5 | 72 | 7 | 0.52 | 35 | 4 |
| 2 | 2.1 | 27 | 2.3 | 42 | 21 | 1.9 | 28 | 3 |
| 3 | 3.16 | 33 | 2.5 | 49 | 10 | 1.78 | 20 | 2 |
| 4 | 3.5 | 63 | 0.66 | 26 | 5 | 1.8 | 0 | 1 |
| 5 | 3.86 | 61 | 0.3 | 34 | 1 | 3.25 | 17 | 0 |
| 6 | 4.0 | 52 | 0.31 | 30 | 8 | 2.5 | 10 | 6 |
| 7 | 2.53 | NA | NA | NA | NA | NA | NA | 1 |
| 8 | 1.4 | 44 | 0.9 | 34 | NA | 1.2 | 0 | 1 |
| Case . | Lymphocyte count, ×109/L . | CLL cells, % . | Normal B cells, % . | T cells, % . | NK cells, % . | CD4/CD8 ratio . | LGL (morphology), % . | CD38+ cells, % . |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.39 | 3 | 5 | 72 | 7 | 0.52 | 35 | 4 |
| 2 | 2.1 | 27 | 2.3 | 42 | 21 | 1.9 | 28 | 3 |
| 3 | 3.16 | 33 | 2.5 | 49 | 10 | 1.78 | 20 | 2 |
| 4 | 3.5 | 63 | 0.66 | 26 | 5 | 1.8 | 0 | 1 |
| 5 | 3.86 | 61 | 0.3 | 34 | 1 | 3.25 | 17 | 0 |
| 6 | 4.0 | 52 | 0.31 | 30 | 8 | 2.5 | 10 | 6 |
| 7 | 2.53 | NA | NA | NA | NA | NA | NA | 1 |
| 8 | 1.4 | 44 | 0.9 | 34 | NA | 1.2 | 0 | 1 |
NA indicates not available.
Molecular analysis of Ig gene rearrangements
Productive IgHV-D-J was successfully amplified and sequenced in 7 patients. Using a cutoff value of at least 2% mutation rate, all 7 proved mutated with a mean mutation frequency of 7.2% plus or minus 2.8% (median, 7.4%; range, 2.7%-10.7%). Interestingly, all patients but one showed an involvement of the VH3 family. The individual VH genes encountered were VH3-30 in 2 (29.0%), VH3-07 in 1 (14.2%), VH3-15 in 1 (14.2%), VH3-33 in 1 (14.2%), VH3-72 in 1 (14.2%), and VH4-34 in 1 (14.2%). The mean sequence length of the HCDR3 was 14.4 plus or minus 2.7 codons (Table 3).
Patients' heavy and light Ig gene pattern at the time of the study
| Case . | IgVH . | IgVL . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VH . | Mutation, % . | D . | JH . | Motif . | HCDR3 . | VL . | Mutation, % . | JL . | Motif . | LCDR3 . | |
| AA sequence . | AA sequence . | ||||||||||
| 1 | 3-15 | 10.7 | 2-2 | 3b | GYSSTFDGAFDI | 12 | Vλ1-47 | 0.8 | λ7 | AAWDDSLSGR | 10 |
| 2 | Vκ3-15 | 1.9 | κ1 | QQPNNWPPTS | 10 | ||||||
| 3 | 3-30 | 10.2 | 3-10 | 4b | GWFEKVPQEYFDS | 13 | Vκ2-30 | 3.1 | κ2 | MQGTHWIHTF | 10 |
| 4 | 3-30 | 2.7 | 3-9 | 4b | TLGDEYDILTGYSKGYFDS | 19 | Vκ4-1 | 2.0 | κ4 | QQYYSTPLTF | 10 |
| 5 | 3-33 | 7.4 | 5-24 | 4b | EKLRGDGYNQALDY | 14 | Vκ4-1 | 3.5 | κ3 | QQYYSPPYFT | 10 |
| 6 | 3-07 | 7.7 | 5-24 | 5b | DLWADRGGPPSTRFDP | 16 | Vλ2-11 | 3.8 | λ2 | CSYAGSPTF | 9 |
| 7 | 3-72 | 4.6 | 3-10 | 5b | VLDGGIHWFDP | 11 | Vκ4-1 | 8.3 | κ2 | QQYYSPPYTF | 10 |
| 8 | 4-34 | 7.2 | 3-10 | 5b | ASVPGGSGSGPNCLDP | 16 | Vλ1-51 | 4.2 | λ3 | GTWDSSLSGRVF | 12 |
| Case . | IgVH . | IgVL . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VH . | Mutation, % . | D . | JH . | Motif . | HCDR3 . | VL . | Mutation, % . | JL . | Motif . | LCDR3 . | |
| AA sequence . | AA sequence . | ||||||||||
| 1 | 3-15 | 10.7 | 2-2 | 3b | GYSSTFDGAFDI | 12 | Vλ1-47 | 0.8 | λ7 | AAWDDSLSGR | 10 |
| 2 | Vκ3-15 | 1.9 | κ1 | QQPNNWPPTS | 10 | ||||||
| 3 | 3-30 | 10.2 | 3-10 | 4b | GWFEKVPQEYFDS | 13 | Vκ2-30 | 3.1 | κ2 | MQGTHWIHTF | 10 |
| 4 | 3-30 | 2.7 | 3-9 | 4b | TLGDEYDILTGYSKGYFDS | 19 | Vκ4-1 | 2.0 | κ4 | QQYYSTPLTF | 10 |
| 5 | 3-33 | 7.4 | 5-24 | 4b | EKLRGDGYNQALDY | 14 | Vκ4-1 | 3.5 | κ3 | QQYYSPPYFT | 10 |
| 6 | 3-07 | 7.7 | 5-24 | 5b | DLWADRGGPPSTRFDP | 16 | Vλ2-11 | 3.8 | λ2 | CSYAGSPTF | 9 |
| 7 | 3-72 | 4.6 | 3-10 | 5b | VLDGGIHWFDP | 11 | Vκ4-1 | 8.3 | κ2 | QQYYSPPYTF | 10 |
| 8 | 4-34 | 7.2 | 3-10 | 5b | ASVPGGSGSGPNCLDP | 16 | Vλ1-51 | 4.2 | λ3 | GTWDSSLSGRVF | 12 |
Productive IgLV-J rearrangements were sequenced in 8 cases. The light chain was κ in 5 of 8 patients and λ in 3 of 8 patients. The Vκ genes expressed were Vκ4-1 in 3 (60%), Vκ2-30 in 1 (20%), and Vκ3-15 in 1 (20%). The Vλ genes displayed were Vλ1-47, Vλ1-51, and Vλ2-11. CLL cases displayed VL gene mutations in 6 of 8 patients, with an average mutation frequency, among mutated cases, of 4.1% plus or minus 2.2% (median, 3.6%; range, 2.0%-8.3%). The mean sequence length of the light complementarity-determining region 3 (LCDR3) was 10.7 plus or minus 0.7 codons (Table 3).
Microarray analysis
Unsupervised analysis of all 22 CLL samples, including 4 with spontaneous regression, showed a tight clustering of the latter (Figure 2), suggesting that these cases have a common genetic signature, distinct from the remaining cases, that is already evident when using unbiased filtering criteria. To identify specifically expressed genes in the spontaneous regression cases, different approaches were used. First, a t test was used to compare these cases (n = 4) with the remaining IgVH-mutated cases (n = 12); we chose to exclude IgVH-unmutated patients, as well as those cases whose mutational status was unknown, because they could provide misleading results. This analysis selected 204 genes differentially expressed in these cases; again, the spontaneous regression patients displayed a very homogeneous pattern of expression. Second, an ANOVA was performed that subdivided the patients according to their clinical scenario (spontaneous regression, n = 4; stable disease, n = 6; progressive disease, n = 6). Also in this analysis, cases experiencing a spontaneous regression displayed a very unique pattern; of interest, patients with a stable disease shared a certain degree of commonality with these cases (Figure 3). Among the genes that are exclusively overexpressed in spontaneous regression patients, a set of transcripts could be further classified in (1) ribosomal proteins (RPS8, RPL39, MRPL4), in line with previous findings17 ; (2) signal transduction (AKT2, INPP5D, ADRBK2, SYK, ARHGAP2); (3) regulation of transcription (TRIM66, ARID1A, LHX3); and (4) lipidic metabolism (LYPLA1, HADH). Finally, among the individual genes we identified UNC5B, involved in the apoptotic machinery, and, to some surprise, B2M and CCNI.
Unsupervised clustering of all CLL cases. The 4 spontaneous regression cases are labeled in orange. Relative levels of gene expression are depicted with a color scale: red indicates the highest levels of expression; blue, the lowest levels of expression.
Unsupervised clustering of all CLL cases. The 4 spontaneous regression cases are labeled in orange. Relative levels of gene expression are depicted with a color scale: red indicates the highest levels of expression; blue, the lowest levels of expression.
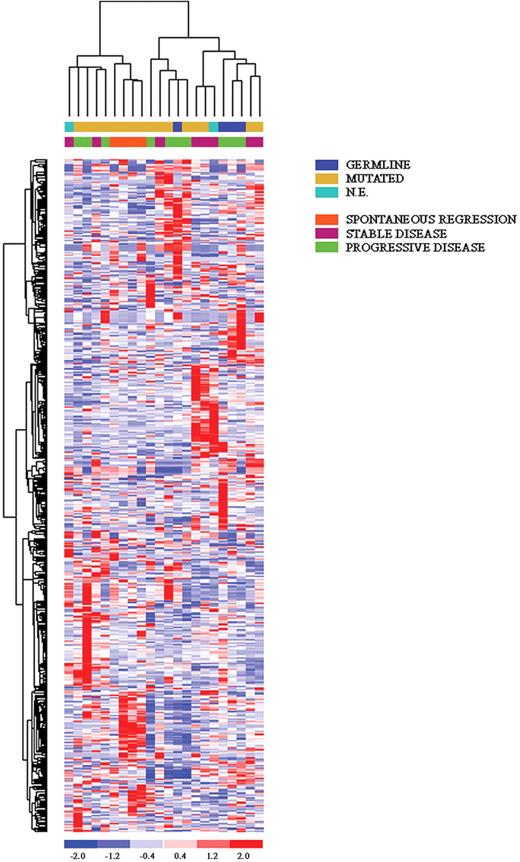
ANOVA analysis comparing patients with CLL in spontaneous regression, stable, or progressive disease. The first group displayed a very unique pattern, sharing a certain degree of commonality with the group of stable CLL.
ANOVA analysis comparing patients with CLL in spontaneous regression, stable, or progressive disease. The first group displayed a very unique pattern, sharing a certain degree of commonality with the group of stable CLL.
Among the genes consistently up-regulated in both spontaneous regression and stable disease patients it was possible to identify WWP1, known to regulate p53 stability; BPTF, which regulates apoptosis; RBP2, which participates to cell-cycle control through its direct interaction with pRb; and DICER. Of interest, the BCR signaling pathway is overrepresented, being, in general, more highly expressed in cases with spontaneous regression and stable disease; similarly, genes involved in the regulation of transcription are up-regulated in these 2 subsets of patients.
Among the down-regulated genes in both spontaneous regression and stable disease cases, it is worth mentioning DUSP1, a phosphatase specific of the MAPK pathway, which plays a role in apoptosis induction; GADD34, known to be involved in melanoma progression; at variance, NOL1 and SERTAD2, which are specifically expressed during the S phase, together with POLR2I and CREG1, which are involved in regulation of transcription, were specifically down-regulated in spontaneous regression cases. A detailed gene list is provided in Table 4.
Differentially expressed genes between spontaneous regression, stable, and progressive patients, as identified by ANOVA
| Probe set ID . | Gene symbol . | P . | Gene function . | Highly expressed in . |
|---|---|---|---|---|
| 210042_s_at | CTSZ | < .000 | Proteolysis | Spontaneous regression |
| 218105_s_at | MRPL4 | < .000 | Translation | Spontaneous regression |
| 200033_at | DDX5 | < .000 | RNA splicing | Spontaneous regression |
| 213748_at | TRIM66 | < .000 | Negative regulation of transcription | Spontaneous regression |
| 212152_x_at | ARID1A | < .001 | Chromatin modification | Spontaneous regression |
| 202525_at | PRSS8 | < .001 | Proteolysis | Spontaneous regression |
| 221670_s_at | LHX3 | < .001 | Regulation of transcription, DNA-dependent | Spontaneous regression |
| 216138_at | Unknown | < .001 | Unknown | Spontaneous regression |
| 203007_x_at | LYPLA1 | < .001 | Lipid metabolic process | Spontaneous regression |
| 212358_at | CLIP3 | < .001 | Unknown | Spontaneous regression |
| 208656_s_at | CCNI | < .001 | Cell cycle | Spontaneous regression |
| 203808_at | AKT2 | < .001 | Protein serine/threonine kinase activity | Spontaneous regression |
| 206989_s_at | SFRS2IP | < .001 | RNA splicing | Spontaneous regression |
| 207521_s_at | ATP2A3 | < .001 | Calcium ion transport | Spontaneous regression |
| 214250_at | NUMA1 | < .001 | Nuclear organization and biogenesis | Spontaneous regression |
| 203332_s_at | INPP5D | < .001 | Signal transduction | Spontaneous regression |
| 201891_s_at | B2M | < .001 | Immune response | Spontaneous regression |
| 218077_s_at | ZDHHC3 | < .001 | Unknown | Spontaneous regression |
| 204183_s_at | ADRBK2 | < .001 | Protein amino acid phosphorylation | Spontaneous regression |
| 204184_s_at | ADRBK2 | < .001 | Protein amino acid phosphorylation | Spontaneous regression |
| 207540_s_at | SYK | < .001 | B-cell receptor signaling pathway | Spontaneous regression |
| 211982_x_at | XPO6 | < .001 | Intracellular protein transport | Spontaneous regression |
| 200858_s_at | RPS8 | < .001 | Structural constituent of ribosome | Spontaneous regression |
| 207961_x_at | MYH11 | < .001 | Calmodulin binding | Spontaneous regression |
| 221318_at | NEUROD4 | < .001 | Nervous system development | Spontaneous regression |
| 208695_s_at | RPL39 | < .001 | Structural constituent of ribosome | Spontaneous regression |
| 208397_x_at | KCNJ5 | < .001 | Potassium ion transport | Spontaneous regression |
| 210260_s_at | TNFAIP8 | < .001 | Apoptosis | Spontaneous regression |
| 221030_s_at | ARHGAP24 | < .001 | Cellular morphology, angiogenesis | Spontaneous regression |
| 218720_x_at | SEZ6L2 | < .001 | Unknown | Spontaneous regression |
| 218949_s_at | QRSL1 | < .001 | Unknown | Spontaneous regression |
| 209269_s_at | SYK | < .001 | B-cell receptor signaling pathway | Spontaneous regression |
| 215883_at | LOC401210 | < .001 | Unknown | Spontaneous regression |
| 204265_s_at | GPSM3 | < .001 | Unknown | Spontaneous regression |
| 210288_at | KLRG1 | < .001 | Cellular defense response | Spontaneous regression |
| 221884_at | EVI1 | < .001 | DNA binding | Spontaneous regression |
| 215288_at | TRPC2 | < .001 | Unknown | Spontaneous regression |
| 211569_s_at | HADH | < .001 | Lipid metabolic process | Spontaneous regression |
| 215467_x_at | DHX9 | < .001 | ATP-dependant RNA helicase activity | Spontaneous regression |
| 212205_at | H2AFV | < .001 | Nucleosome assembly | Spontaneous regression |
| 211416_x_at | GGTLA4 | < .001 | Unknown | Spontaneous regression |
| 218666_s_at | STX17 | < .001 | Unknown | Spontaneous regression |
| 217683_at | HBE1 | < .001 | Heme binding | Spontaneous regression |
| 213042_s_at | ATP2A3 | < .001 | Calcium ion transport | Spontaneous regression |
| 212239_at | PIK3R1 | < .001 | Phosphatidylinositol binding | Spontaneous regression |
| 200972_at | TSPAN3 | < .01 | Unknown | Spontaneous regression |
| 41856_at | UNC5B | < .01 | Apoptosis | Spontaneous regression |
| 202757_at | COBRA1 | < .000 | Negative regulation of transcription | Spontaneous regression and stable disease |
| 206370_at | PIK3CG | < .000 | Negative regulation of apoptosis | Spontaneous regression and stable disease |
| 210556_at | NFATC3 | < .000 | Regulation of transcription from RNA II polymerase promoter | Spontaneous regression and stable disease |
| 207186_s_at | BPTF | < .000 | Chromatin remodeling | Spontaneous regression and stable disease |
| 209527_at | EXOSC2 | < .000 | rRNA processing | Spontaneous regression and stable disease |
| 202173_s_at | VEZF1 | < .001 | Angiogenesis | Spontaneous regression and stable disease |
| 209579_s_at | MBD4 | < .001 | DNA binding | Spontaneous regression and stable disease |
| 219980_at | C4orf29 | < .001 | Unknown | Spontaneous regression and stable disease |
| 212638_s_at | WWP1 | < .001 | Protein modification process | Spontaneous regression and stable disease |
| 219512_at | DSN1 | < .001 | Cell cycle | Spontaneous regression and stable disease |
| 217717_s_at | YWHAB | < .001 | Monooxygenase activity | Spontaneous regression and stable disease |
| 209682_at | CBLB | < .001 | Immune response (negative regulator | Spontaneous regression and stable disease |
| 201684_s_at | TOX4 | < .001 | Unknown | Spontaneous regression and stable disease |
| 212395_s_at | KIAA0090 | < .001 | Unknown | Spontaneous regression and stable disease |
| 202171_at | VEZF1 | < .001 | Angiogenesis | Spontaneous regression and stable disease |
| 206061_s_at | DICER1 | < .001 | miR processing | Spontaneous regression and stable disease |
| 215750_at | KIAA1659 | < .001 | Unknown | Spontaneous regression and stable disease |
| 218518_at | C5orf5 | < .001 | Unknown | Spontaneous regression and stable disease |
| 215075_s_at | GRB2 | < .001 | Epidermal growth factor receptor signaling pathway | Spontaneous regression and stable disease |
| 215698_at | RBP2 | < .001 | Regulation of transcription, DNA dependent | Spontaneous regression and stable disease |
| 219498_s_at | BCL11A | < .001 | Regulation of transcription, DNA dependent | Spontaneous regression and stable disease |
| 212073_at | CSNK2A1 | < .001 | Protein amino acid phosphorylation | Spontaneous regression and stable disease |
| 218607_s_at | SDAD1 | < .001 | Unknown | Spontaneous regression and stable disease |
| 214757_at | Unknown | < .001 | Unknown | Spontaneous regression and stable disease |
| 213229_at | DICER1 | < .001 | miR processing | Spontaneous regression and stable disease |
| 204028_s_at | RABGAP1 | < .001 | Regulation of GTPase activity | Spontaneous regression and stable disease |
| 216022_at | WNK1 | < .001 | Protein serine/threonine kinase activity | Spontaneous regression and stable disease |
| 201514_s_at | G3BP1 | < .001 | Helicase activity | Spontaneous regression and stable disease |
| 218930_s_at | TMEM106B | < .001 | Unknown | Spontaneous regression and stable disease |
| 221006_s_at | SNX27 | < .001 | Protein transport | Spontaneous regression and stable disease |
| 214808_at | Unknown | < .001 | Unknown | Spontaneous regression and stable disease |
| 201960_s_at | MYCBP2 | < .001 | Regulation of transcription, DNA-dependent | Spontaneous regression and stable disease |
| 217888_s_at | ARFGAP1 | < .001 | Vesicle-mediated transport | Spontaneous regression and stable disease |
| 202760_s_at | AKAP2/PALM2-AKAP2 | < .001 | Unknown | Spontaneous regression and stable disease |
| 219547_at | COX15 | < .001 | Mitochondrial electron transport, cytochrome C to oxygen | Spontaneous regression and stable disease |
| 202980_s_at | SIAH1 | < .000 | Apoptosis | Stable disease |
| 217322_x_at | Unknown | < .001 | Unknown | Stable disease |
| 202252_at | 7 | < .001 | Vesicle-mediated transport | Stable disease |
| 220974_x_at | SFXN3 | < .001 | Unknown | Stable disease |
| 200977_s_at | TAX1BP1 | < .001 | Apoptosis | Stable disease |
| 222029_x_at | PFDN6 | < .001 | Unknown | Stable disease |
| 205812_s_at | TMED9 | < .001 | Unknown | Stable disease |
| 202981_x_at | SIAH1 | < .001 | Apoptosis | Stable disease |
| 208971_at | UROD | < .001 | Heme biosynthetic process | Stable disease |
| 204103_at | CCL4 | < .001 | Chemotaxis | Stable disease |
| 212463_at | CD59 | < .001 | Immune response | Stable disease |
| 206352_s_at | PEX10 | < .001 | Peroxisome organization and biogenesis | Stable disease |
| 207024_at | CHRND | < .001 | Ion transport | Stable disease |
| 220305_at | MGC3260 | < .001 | Unknown | Stable disease |
| 221970_s_at | NOL11 | < .000 | Unknown | Stable and progressive disease |
| 214427_at | NOL1 | < .001 | Regulation of proliferation | Stable and progressive disease |
| 201200_at | CREG1 | < .001 | Regulation of cell growth | Stable and progressive disease |
| 208451_s_at | C4B | < .001 | Complement activation | Stable and progressive disease |
| 202433_at | SLC35B1 | < .001 | Carbohydrate transport | Stable and progressive disease |
| 202657_s_at | SERTAD2 | < .001 | Regulation of transcription, DNA-dependent | Stable and progressive disease |
| 214603_at | MAGEA2B | < .001 | Unknown | Stable and progressive disease |
| 215517_at | PYGO1 | < .001 | Unknown | Stable and progressive disease |
| 217600_at | SCUBE3 | < .001 | Calcium ion binding | Stable and progressive disease |
| 220538_at | ADM2 | < .001 | Hormone activity | Stable and progressive disease |
| 209129_at | TRIP6 | < .001 | Focal adhesion formation | Stable and progressive disease |
| 214777_at | Unknown | < .001 | Unknown | Stable and progressive disease |
| 212955_s_at | POLR2I | < .001 | Regulation of transcription | Stable and progressive disease |
| 204847_at | ZBTB11 | < .001 | Unknown | Stable and progressive disease |
| 201124_at | ITGB5 | < .001 | Cell adhesion | Stable and progressive disease |
| 206241_at | KPNA5 | < .000 | Protein transporter activity | Progressive disease |
| 218013_x_at | DCTN4 | < .000 | Unknown | Progressive disease |
| 208980_s_at | UBC | < .001 | unknown | Progressive disease |
| 208803_s_at | SRP72 | < .001 | Signal transduction | Progressive disease |
| 202014_at | GADD34 | < .001 | Cell-cycle arrest | Progressive disease |
| 37028_at | GADD34 | < .001 | Cell-cycle arrest | Progressive disease |
| 219898_at | GPR85 | < .001 | Unknown | Progressive disease |
| 212862_at | CDS2 | < .001 | Phospholipid biosynthetic process | Progressive disease |
| 208274_at | OCLM | < .001 | Visual perception | Progressive disease |
| 211999_at | H3F3B | < .001 | Nucleosome assembly | Progressive disease |
| 212225_at | EIF1 | < .001 | Regulation of translation | Progressive disease |
| 208931_s_at | ILF3 | < .001 | Transcription | Progressive disease |
| 200790_at | ODC1 | < .001 | Polyamine biosynthetic process | Progressive disease |
| 201104_x_at | NBPF11 | < .001 | Unknown | Progressive disease |
| 201044_x_at | DUSP1 | < .001 | MAP kinase tyrosine/serine/threoninephosphatase activity | Progressive disease |
| 203682_s_at | IVD | < .001 | Metabolic process | Progressive disease |
| 201041_s_at | DUSP1 | < .001 | MAP kinase tyrosine/serine/threoninephosphatase activity | Progressive disease |
| 203248_at | ZNF24 | .001 | Transcription factor activity | Progressive disease |
| Probe set ID . | Gene symbol . | P . | Gene function . | Highly expressed in . |
|---|---|---|---|---|
| 210042_s_at | CTSZ | < .000 | Proteolysis | Spontaneous regression |
| 218105_s_at | MRPL4 | < .000 | Translation | Spontaneous regression |
| 200033_at | DDX5 | < .000 | RNA splicing | Spontaneous regression |
| 213748_at | TRIM66 | < .000 | Negative regulation of transcription | Spontaneous regression |
| 212152_x_at | ARID1A | < .001 | Chromatin modification | Spontaneous regression |
| 202525_at | PRSS8 | < .001 | Proteolysis | Spontaneous regression |
| 221670_s_at | LHX3 | < .001 | Regulation of transcription, DNA-dependent | Spontaneous regression |
| 216138_at | Unknown | < .001 | Unknown | Spontaneous regression |
| 203007_x_at | LYPLA1 | < .001 | Lipid metabolic process | Spontaneous regression |
| 212358_at | CLIP3 | < .001 | Unknown | Spontaneous regression |
| 208656_s_at | CCNI | < .001 | Cell cycle | Spontaneous regression |
| 203808_at | AKT2 | < .001 | Protein serine/threonine kinase activity | Spontaneous regression |
| 206989_s_at | SFRS2IP | < .001 | RNA splicing | Spontaneous regression |
| 207521_s_at | ATP2A3 | < .001 | Calcium ion transport | Spontaneous regression |
| 214250_at | NUMA1 | < .001 | Nuclear organization and biogenesis | Spontaneous regression |
| 203332_s_at | INPP5D | < .001 | Signal transduction | Spontaneous regression |
| 201891_s_at | B2M | < .001 | Immune response | Spontaneous regression |
| 218077_s_at | ZDHHC3 | < .001 | Unknown | Spontaneous regression |
| 204183_s_at | ADRBK2 | < .001 | Protein amino acid phosphorylation | Spontaneous regression |
| 204184_s_at | ADRBK2 | < .001 | Protein amino acid phosphorylation | Spontaneous regression |
| 207540_s_at | SYK | < .001 | B-cell receptor signaling pathway | Spontaneous regression |
| 211982_x_at | XPO6 | < .001 | Intracellular protein transport | Spontaneous regression |
| 200858_s_at | RPS8 | < .001 | Structural constituent of ribosome | Spontaneous regression |
| 207961_x_at | MYH11 | < .001 | Calmodulin binding | Spontaneous regression |
| 221318_at | NEUROD4 | < .001 | Nervous system development | Spontaneous regression |
| 208695_s_at | RPL39 | < .001 | Structural constituent of ribosome | Spontaneous regression |
| 208397_x_at | KCNJ5 | < .001 | Potassium ion transport | Spontaneous regression |
| 210260_s_at | TNFAIP8 | < .001 | Apoptosis | Spontaneous regression |
| 221030_s_at | ARHGAP24 | < .001 | Cellular morphology, angiogenesis | Spontaneous regression |
| 218720_x_at | SEZ6L2 | < .001 | Unknown | Spontaneous regression |
| 218949_s_at | QRSL1 | < .001 | Unknown | Spontaneous regression |
| 209269_s_at | SYK | < .001 | B-cell receptor signaling pathway | Spontaneous regression |
| 215883_at | LOC401210 | < .001 | Unknown | Spontaneous regression |
| 204265_s_at | GPSM3 | < .001 | Unknown | Spontaneous regression |
| 210288_at | KLRG1 | < .001 | Cellular defense response | Spontaneous regression |
| 221884_at | EVI1 | < .001 | DNA binding | Spontaneous regression |
| 215288_at | TRPC2 | < .001 | Unknown | Spontaneous regression |
| 211569_s_at | HADH | < .001 | Lipid metabolic process | Spontaneous regression |
| 215467_x_at | DHX9 | < .001 | ATP-dependant RNA helicase activity | Spontaneous regression |
| 212205_at | H2AFV | < .001 | Nucleosome assembly | Spontaneous regression |
| 211416_x_at | GGTLA4 | < .001 | Unknown | Spontaneous regression |
| 218666_s_at | STX17 | < .001 | Unknown | Spontaneous regression |
| 217683_at | HBE1 | < .001 | Heme binding | Spontaneous regression |
| 213042_s_at | ATP2A3 | < .001 | Calcium ion transport | Spontaneous regression |
| 212239_at | PIK3R1 | < .001 | Phosphatidylinositol binding | Spontaneous regression |
| 200972_at | TSPAN3 | < .01 | Unknown | Spontaneous regression |
| 41856_at | UNC5B | < .01 | Apoptosis | Spontaneous regression |
| 202757_at | COBRA1 | < .000 | Negative regulation of transcription | Spontaneous regression and stable disease |
| 206370_at | PIK3CG | < .000 | Negative regulation of apoptosis | Spontaneous regression and stable disease |
| 210556_at | NFATC3 | < .000 | Regulation of transcription from RNA II polymerase promoter | Spontaneous regression and stable disease |
| 207186_s_at | BPTF | < .000 | Chromatin remodeling | Spontaneous regression and stable disease |
| 209527_at | EXOSC2 | < .000 | rRNA processing | Spontaneous regression and stable disease |
| 202173_s_at | VEZF1 | < .001 | Angiogenesis | Spontaneous regression and stable disease |
| 209579_s_at | MBD4 | < .001 | DNA binding | Spontaneous regression and stable disease |
| 219980_at | C4orf29 | < .001 | Unknown | Spontaneous regression and stable disease |
| 212638_s_at | WWP1 | < .001 | Protein modification process | Spontaneous regression and stable disease |
| 219512_at | DSN1 | < .001 | Cell cycle | Spontaneous regression and stable disease |
| 217717_s_at | YWHAB | < .001 | Monooxygenase activity | Spontaneous regression and stable disease |
| 209682_at | CBLB | < .001 | Immune response (negative regulator | Spontaneous regression and stable disease |
| 201684_s_at | TOX4 | < .001 | Unknown | Spontaneous regression and stable disease |
| 212395_s_at | KIAA0090 | < .001 | Unknown | Spontaneous regression and stable disease |
| 202171_at | VEZF1 | < .001 | Angiogenesis | Spontaneous regression and stable disease |
| 206061_s_at | DICER1 | < .001 | miR processing | Spontaneous regression and stable disease |
| 215750_at | KIAA1659 | < .001 | Unknown | Spontaneous regression and stable disease |
| 218518_at | C5orf5 | < .001 | Unknown | Spontaneous regression and stable disease |
| 215075_s_at | GRB2 | < .001 | Epidermal growth factor receptor signaling pathway | Spontaneous regression and stable disease |
| 215698_at | RBP2 | < .001 | Regulation of transcription, DNA dependent | Spontaneous regression and stable disease |
| 219498_s_at | BCL11A | < .001 | Regulation of transcription, DNA dependent | Spontaneous regression and stable disease |
| 212073_at | CSNK2A1 | < .001 | Protein amino acid phosphorylation | Spontaneous regression and stable disease |
| 218607_s_at | SDAD1 | < .001 | Unknown | Spontaneous regression and stable disease |
| 214757_at | Unknown | < .001 | Unknown | Spontaneous regression and stable disease |
| 213229_at | DICER1 | < .001 | miR processing | Spontaneous regression and stable disease |
| 204028_s_at | RABGAP1 | < .001 | Regulation of GTPase activity | Spontaneous regression and stable disease |
| 216022_at | WNK1 | < .001 | Protein serine/threonine kinase activity | Spontaneous regression and stable disease |
| 201514_s_at | G3BP1 | < .001 | Helicase activity | Spontaneous regression and stable disease |
| 218930_s_at | TMEM106B | < .001 | Unknown | Spontaneous regression and stable disease |
| 221006_s_at | SNX27 | < .001 | Protein transport | Spontaneous regression and stable disease |
| 214808_at | Unknown | < .001 | Unknown | Spontaneous regression and stable disease |
| 201960_s_at | MYCBP2 | < .001 | Regulation of transcription, DNA-dependent | Spontaneous regression and stable disease |
| 217888_s_at | ARFGAP1 | < .001 | Vesicle-mediated transport | Spontaneous regression and stable disease |
| 202760_s_at | AKAP2/PALM2-AKAP2 | < .001 | Unknown | Spontaneous regression and stable disease |
| 219547_at | COX15 | < .001 | Mitochondrial electron transport, cytochrome C to oxygen | Spontaneous regression and stable disease |
| 202980_s_at | SIAH1 | < .000 | Apoptosis | Stable disease |
| 217322_x_at | Unknown | < .001 | Unknown | Stable disease |
| 202252_at | 7 | < .001 | Vesicle-mediated transport | Stable disease |
| 220974_x_at | SFXN3 | < .001 | Unknown | Stable disease |
| 200977_s_at | TAX1BP1 | < .001 | Apoptosis | Stable disease |
| 222029_x_at | PFDN6 | < .001 | Unknown | Stable disease |
| 205812_s_at | TMED9 | < .001 | Unknown | Stable disease |
| 202981_x_at | SIAH1 | < .001 | Apoptosis | Stable disease |
| 208971_at | UROD | < .001 | Heme biosynthetic process | Stable disease |
| 204103_at | CCL4 | < .001 | Chemotaxis | Stable disease |
| 212463_at | CD59 | < .001 | Immune response | Stable disease |
| 206352_s_at | PEX10 | < .001 | Peroxisome organization and biogenesis | Stable disease |
| 207024_at | CHRND | < .001 | Ion transport | Stable disease |
| 220305_at | MGC3260 | < .001 | Unknown | Stable disease |
| 221970_s_at | NOL11 | < .000 | Unknown | Stable and progressive disease |
| 214427_at | NOL1 | < .001 | Regulation of proliferation | Stable and progressive disease |
| 201200_at | CREG1 | < .001 | Regulation of cell growth | Stable and progressive disease |
| 208451_s_at | C4B | < .001 | Complement activation | Stable and progressive disease |
| 202433_at | SLC35B1 | < .001 | Carbohydrate transport | Stable and progressive disease |
| 202657_s_at | SERTAD2 | < .001 | Regulation of transcription, DNA-dependent | Stable and progressive disease |
| 214603_at | MAGEA2B | < .001 | Unknown | Stable and progressive disease |
| 215517_at | PYGO1 | < .001 | Unknown | Stable and progressive disease |
| 217600_at | SCUBE3 | < .001 | Calcium ion binding | Stable and progressive disease |
| 220538_at | ADM2 | < .001 | Hormone activity | Stable and progressive disease |
| 209129_at | TRIP6 | < .001 | Focal adhesion formation | Stable and progressive disease |
| 214777_at | Unknown | < .001 | Unknown | Stable and progressive disease |
| 212955_s_at | POLR2I | < .001 | Regulation of transcription | Stable and progressive disease |
| 204847_at | ZBTB11 | < .001 | Unknown | Stable and progressive disease |
| 201124_at | ITGB5 | < .001 | Cell adhesion | Stable and progressive disease |
| 206241_at | KPNA5 | < .000 | Protein transporter activity | Progressive disease |
| 218013_x_at | DCTN4 | < .000 | Unknown | Progressive disease |
| 208980_s_at | UBC | < .001 | unknown | Progressive disease |
| 208803_s_at | SRP72 | < .001 | Signal transduction | Progressive disease |
| 202014_at | GADD34 | < .001 | Cell-cycle arrest | Progressive disease |
| 37028_at | GADD34 | < .001 | Cell-cycle arrest | Progressive disease |
| 219898_at | GPR85 | < .001 | Unknown | Progressive disease |
| 212862_at | CDS2 | < .001 | Phospholipid biosynthetic process | Progressive disease |
| 208274_at | OCLM | < .001 | Visual perception | Progressive disease |
| 211999_at | H3F3B | < .001 | Nucleosome assembly | Progressive disease |
| 212225_at | EIF1 | < .001 | Regulation of translation | Progressive disease |
| 208931_s_at | ILF3 | < .001 | Transcription | Progressive disease |
| 200790_at | ODC1 | < .001 | Polyamine biosynthetic process | Progressive disease |
| 201104_x_at | NBPF11 | < .001 | Unknown | Progressive disease |
| 201044_x_at | DUSP1 | < .001 | MAP kinase tyrosine/serine/threoninephosphatase activity | Progressive disease |
| 203682_s_at | IVD | < .001 | Metabolic process | Progressive disease |
| 201041_s_at | DUSP1 | < .001 | MAP kinase tyrosine/serine/threoninephosphatase activity | Progressive disease |
| 203248_at | ZNF24 | .001 | Transcription factor activity | Progressive disease |
Only the probe sets with an Affymetrix expression value greater than 100 in at least 1 clinical subgroup are reported. P values E−06 or E−05 are reported as < .000.
Furthermore, we compared the gene profile of this CLL cohort to that of circulating B lymphocytes from healthy donors. In the unsupervised analysis, CLL in spontaneous regression clustered with normal B lymphocytes (data not shown). To further explore this similarity, we analyzed the expression of the genes selected by ANOVA also on normal B lymphocytes; as already observed by unsupervised analysis, this approach highlighted a closer similarity between CLL in spontaneous regression and lymphocytes from healthy donors (Supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Intracellular T-cell cytokine expression
No significant differences were observed between CLL in spontaneous regression and normal persons in the absolute number of activated T lymphocytes expressing cytoplasmic IFN-γ, TNF-α, and IL-4 (Table 5). Furthermore, we compared the absolute number of T cells expressing IFN-γ, TNF-α, and IL-4 between patients in different phases of the disease, as shown in Table 5. CD3+ cells from progressive CLL expressed more IFN-γ than did CLL in spontaneous regression, stable CLL, and control donors, but the difference was not statistically significant except for control donors (P = .013). In contrast, a statistically significant increase in the number of TNF-α–expressing T cells was found in progressive CLL compared with CLL in spontaneous regression (P = .001), stable CLL (P = .011), and healthy persons (P < .001). Finally, the number of CD3+ cells that expressed IL-4 was similar in all groups studied.
Intracellular T-cell cytokine production
| Disease status . | Absolute number of cytokine-producing CD3+ cells/μL . | ||
|---|---|---|---|
| IFN-γ . | TNF-α . | IL-4 . | |
| Spontaneous regression (n = 6) | 387 ± 190 | 553 ± 171 | 25 ± 26 |
| Progressive disease (n = 7) | 646 ± 262 | 1099 ± 196 | 13 ± 30 |
| Stable disease (n = 5) | 401 ± 173 | 605 ± 265 | 21 ± 30 |
| Controls (n = 5) | 209 ± 59 | 454 ± 122 | 14 ± 21 |
| Disease status . | Absolute number of cytokine-producing CD3+ cells/μL . | ||
|---|---|---|---|
| IFN-γ . | TNF-α . | IL-4 . | |
| Spontaneous regression (n = 6) | 387 ± 190 | 553 ± 171 | 25 ± 26 |
| Progressive disease (n = 7) | 646 ± 262 | 1099 ± 196 | 13 ± 30 |
| Stable disease (n = 5) | 401 ± 173 | 605 ± 265 | 21 ± 30 |
| Controls (n = 5) | 209 ± 59 | 454 ± 122 | 14 ± 21 |
Discussion
In CLL, the occurrence of spontaneous regression of the disease is an exceptional event. Recognized since the 1970s,18,19 this phenomenon has been rarely described in the literature. In the largest series of spontaneous regressions,10 as well as in smaller series from other groups, no exhaustive data on the biologic features of the disease are reported.
In a series of 10 patients in stage A0, AI, AII, and B at diagnosis,10 a decrease in lymphocyte counts and palpable lymph nodes and spleen was observed in the absence of treatment after a median follow-up of 9 years (range, 4-21 years). Six of the 10 cases showed a clinical remission defined as complete absence of anemia or thrombocytopenia, lymphadenopathy, splenomegaly, or constitutional symptoms and a lymphocyte count less than 4 × 109/L. No BM assessment was performed, and there was evidence of persistence of the CLL clone in most cases, with 1% to 50% of circulating CLL cells. In the other 3 recent cases of spontaneous CLL regression, a clonal remission was documented by immunophenotype in both PB and BM in 2 cases,9,11 and molecularly in the PB with evidence of residual disease in the marrow in the third case.8
No concomitant clinical conditions that may lead to a spontaneous regression of CLL have been clearly identified, although antihypertensive therapy with the calcium antagonist verapamil has been associated to some of these cases.20 More recently, it has been reported that intake of green tea products may induce regression of low-grade B-cell malignancies, including CLL.21 A spontaneous remission of CLL can also precede the diagnosis of a primary second malignancy by months or years.22,23
We add to the literature with a series of 9 cases managed over the years at a single institution, in which the occurrence of a spontaneous regression of CLL occurred after a median follow-up of 11 years and persisted thereafter. In fact, after a follow-up of 16 years, all patients remain in persistent regression. Because we currently manage approximately 800 patients with CLL, this phenomenon can be estimated to occur in approximately 1% of cases. No evident clinical conditions, including second malignancies, autoimmune diseases, drug intake, or viral infections (including hepatitis B or C), could account for the regressions recorded in our series.
The CLL clone was still immunophenotypically evident in the PB, ranging from 3% to 60% (median, 44%) of circulating lymphocytes. The expression of CD38 and of ZAP-70 on CLL cells was, as expected, always negative. The IgVH genes were mutated in all cases, whereas the IgVL genes were mutated in 75% of them. Interestingly, there was an evident restriction in the IgVH family usage, with the VH3 family represented in all cases but one. Two (30%) of 7 cases showed the usage of the VH3-30 gene. The VH3 family is one of the most represented in both mutated and unmutated CLL,24,25 and the VH3-30 gene comprises approximately 7% of all CLL cases.24,25 However, comparing a series of stable (n = 78) and progressive (n = 74) CLL from our institute, VH3-30 was significantly overrepresented in patients with stable (n = 10; all mutated) rather than with progressive disease (n = 1; unmutated; P < .001; A.G. and R.F., unpublished data, December 2008); thus, VH3-30 might be suggested as a marker of very indolent disease. The Ig light chain genes represented among CLL in spontaneous regression were Vκ4-1, Vκ2-30, Vκ3-15, Vλ1-51, Vλ2-11, and Vλ1-47. Vκ4-1 and Vκ2-30 are frequently represented in unselected CLL,25 being the third and fourth most common κ chain genes, respectively. Vλ1-51 and Vλ2-11 genes also recur, each representing approximately 5% of all λ light chain genes, although reported as mostly unmutated. We previously reported on a series of 25 patients with highly stable and indolent CLL who never required treatment over a prolonged follow-up period.6 In that series, the IgVH and IgVL genes were mostly mutated, and a biased IgVH usage was highlighted, with a preferential usage of VH3 (40%) and VH4 (20%) families, and VH3-72 (24%) and VH2-05 (12%) genes. Among IgVL genes, Vκ1-5 (36%) and Vκ4-1 (13%) were the most represented. An association between Vκ4-1 and VH3-72 was recorded. Also in our series of spontaneous regressing cases, the usage of the Vκ4-1 gene occurred. However, we did not find recurrent associations between heavy and light chain genes nor characteristic HCDR3/LCDR3 amino acidic motifs.
Specific features in the gene machinery of the CLL clone in regression were identified. Microarray analysis showed a distinctive pattern of expression for such cases both by unsupervised and supervised approaches. The most intriguing and somehow unexpected finding comes from the overrepresentation of several BCR-related genes, suggesting that the BCR signaling may play an important role in the spontaneous regression scenario. These findings are supported by the recent study reporting that in IgVH-mutated CLL IgM stimulation induces an apoptotic signal.26 Another hypothesis could be that in these patients the BCR signaling cascade is simply intact, indicating that the B cell is properly functioning as an antigen-presenting cell. This possibility is further supported by 2 evidences: first, B2M, the β chain of the HLA class I complex, is highly expressed in these cases, in line with the fact that, in solid tumors, its lack induces a mechanism of tumor escape from cytotoxic lymphocyte recognition; second, the analysis of T-cell response in these patients shows a closer similarity of their profile of cytokine expression with that of T cells from healthy donors. Furthermore, in agreement with a previous report by Durig et al,17 in the spontaneous regression cases we documented an up-regulation of B2M and CCNI, as well as an up-regulation of ribosomal genes, thus confirming the effect of the translation-associated genes, in particular ribosomal genes, on the clinical behavior. Finally, given that microRNAs (miRs) appear to play a key role in CLL initiation or progression or both, the overexpression of DICER in spontaneous regression and stable disease cases is intriguing, because this gene is a regulator of miR processing. These tiny RNA molecules repress the expression of several target genes; down-regulation of DICER in progressive patients suggests that this mechanism of gene repression is impaired.
Comparison between CLL cases and B lymphocytes from healthy donors has highlighted a higher degree of similarity of the latter with cases in spontaneous regression rather than with the remaining patients with CLL. Of interest, B2M and CCNI were overexpressed also in normal B lymphocytes, as well as DICER, thus corroborating the hypothesis that B2M down-regulation is associated with disease progression and that down-regulation of DICER may be involved in tumor progression, as reported in patients with lung cancer.27 Among the genes that were discordant between CLL in spontaneous regression and normal B lymphocytes, SYK, a tyrosine kinase involved in BCR signaling and known to play a role in CLL pathogenesis, is more highly expressed in regression cases, suggesting that this molecule may contribute to the persistence of the malignant clone.
Focusing on the accessory cell compartment, an increase in LGLs was observed in CLL in spontaneous regression (17%; range, 0%-35%) compared with stable (5%; range, 1%-14%) or progressive CLL (1.5%; range, 1%-13%). The CD4/CD8 ratio in spontaneous regression cases was mostly normal, similar to what was observed in patients with a long-lasting indolent disease.5 These data further point to a primary role played by the host immune system in controlling the disease. Studies on the T-cell cytokine production in CLL have not consistently supported the hypothesis of an immune-mediated control of B-cell proliferation. Here, we could show that the amounts of TNF-α and IFN-γ expressed by activated T cells from CLL in spontaneous regression are lower than those measured in progressive CLL, being similar to the levels found in patients with stable CLL or in healthy persons. This is in line with what was reported in the literature,28-31 leading to the hypothesis that an increase in cytokine synthesis might be, among other factors, responsible for malignant cell accumulation in the BM microenvironment and for disease progression, preventing CLL cells from entering apoptosis.
In conclusion, our findings provide evidence that clinical regressions can occur in patients with early-stage CLL over a prolonged follow-up period. The use of mutated VH3-30 and Vκ4-1 might identify patients with a very indolent course of the disease and a possible spontaneous regression over time. The evidence of a peculiar gene profile suggests that BCR signaling may play a primary role in this scenario as the most significant feature of the leukemic clone in regression. Although no clear explanation of the regulatory mechanisms leading to a down-regulation of CLL proliferation up to spontaneous clinical remission is available, the role of antigenic stimulation or deprivation, via a functional BCR machinery, is possible.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan; Ministero dell'Università e della Ricerca Scientifica; Progetto PRIN (Programmi di ricerca di Rilevante Interesse Nazionale), Rome; Ministero della Salute; Progetto “Oncologia,” Rome; Compagnia di San Paolo, Turin; Italian Association against Leukemia, Section of Rome (RomAIL); and the Mediterranean Institute of Hematology.
Authorship
Contribution: I.D.G. designed the research, analyzed the data, and wrote the manuscript; S.C. performed microarray experiments and contributed to the revision of the manuscript; S.T. performed microarray experiments; M.S.D.P. performed immunophenotype; R.M. and N.P. performed intracellular T-cell cytokine production analysis; F.M. performed peripheral blood morphology; S.S. and M.M. performed molecular analysis of Ig gene rearrangements; F.R.M. analyzed patients and contributed to the revision of the manuscript; A.G. analyzed the data and contributed to the revision of the manuscript; and R.F. reviewed the design of the study, analyzed and discussed the results, and critically revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robin Foà, Division of Hematology, Department of Cellular Biotechnologies and Hematology, Sapienza University, Via Benevento 6, 00161 Rome, Italy; e-mail: rfoa@bce.uniroma1.it.