Abstract
Vascular endothelial growth factor receptor 1 (VEGFR1) is a marker for endothelial-specific gene expression. We previously reported that the human VEGFR1 promoter (between −748 and +284) contains information for expression in the intact endothelium of transgenic mice. The objective of this study was to dissect the cis-regulatory elements underlying VEGFR1 promoter activity in vitro and in vivo. In primary endothelial cells, binding sites for E74-like factor 1 (ELF-1; between −49 and −52), cyclic adenosine monophosphate response element binding (CREB; between −74 and −81), and early growth response factor 1/3 (EGR-1/3; between −16 to −25) were shown to play a positive role in gene transcription, whereas a putative E26 transformation-specificsequence (ETS) motif between −36 and −39 had a net negative effect on promoter activity. When targeted to the Hprt locus of mice, mutations of the ELF-1 binding site and the CRE element reduced promoter activity in the embryonic vasculature and resulted in a virtual loss of expression in adult endothelium. Postnatally, the EGR binding site mutant displayed significantly reduced promoter activity in a subset of vascular beds. In contrast, mutation of the −39 ETS site resulted in increased LacZ staining in multiple vascular beds. Together, these results provide new insights into the transcriptional regulatory mechanisms of VEGFR1.
Introduction
Vascular endothelial growth factor A (VEGF-A) binds to 2 receptors, VEGFR1 (also known as Flt-1) and VEGFR2 (also known as Flk-1 or KDR). Expression of both receptors is largely restricted to endothelial cells. The human VEGFR1 promoter has been previously cloned and characterized. A 1-kb region between −748 and +248 was shown to direct cell type–specific expression in cultured endothelial cells.1 Transient transfection assays revealed several positive-acting elements, including consensus sites for E26 transformation-specific sequence (ETS; between −49 and −52), cyclic adenosine monophosphate response element binding (CREB)/activating transcription factor (between −74 and −81), and early growth response factor (EGR; between −16 and −25).1 In contrast, a putative ETS consensus element between −36 and −39 was shown to repress promoter activity.1
Subsequently, we reported that a 1-kb fragment of the human VEGFR1 promoter (between −748 and +284) contains information for endothelial-restricted expression in transgenic mice.2 In the latter study, a single copy of the VEGFR1 promoter was targeted to the Hprt locus, thus providing a means to control for the effects of copy number and integration site on expression. In the current study, we used an identical Hprt targeting strategy to evaluate the activity of the VEGFR1 promoter containing a point mutation of 1 of the 2 ETS motifs, the CRE site, or the EGR binding element. Our data suggest that each of these DNA elements contributes in a unique way to the expression of VEGFR1 in vivo.
Methods
Plasmid constructions
Construction of VEGFR1 wild-type and mutant promoter constructs used in transient transfection assays and Hprt-locus targeting is detailed in supplemental Methods (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell culture
Human umbilical vein endothelial cells (HUVECs), human coronary artery endothelial cells (HCAECs), and human pulmonary artery endothelial cells (HPAECs) were purchased from Lonza and cultured in endothelial cell medium supplemented with EGM-2-MV bullet kit (Lonza). Murine endothelial cells were harvested from the hearts of Hprt-targeted mice as detailed in supplemental Methods.
Transient transfection assays
Transient transfection and cotransfections were carried out as described previously3 and in supplemental Methods.
ChIP
Chromatin immunoprecipitation (ChIP) assay was performed using a chromatin immunoprecipitation assay kit (Upstate) according to the manufacturer's instructions and as detailed in supplemental Methods.
Electrophoretic gel mobility shift assays
Nuclear extraction and electrophoretic gel mobility shift assays (EMSAs) were carried out as described previously4 and in supplemental Methods.
Transfection with siRNA
HUVECs were transfected with siRNAs against E74-like factor 1 (ELF-1) or ETS-1 (Dharmacon) as previously described.5 siRNA sequences are shown in supplemental Table 1.
Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (PCR) was carried out as previously described.5 Primer sequences are shown in supplemental Table 1.
Generation and analysis of Hprt-targeted transgenic mice
The generation and analysis of transgenic mice carrying promoter-reporter genes were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center. BK4 ES cells were transfected with the Hprt targeting vectors and injected into blastocysts as previously described2 (and supplemental Methods). Resulting chimeric males were bred with C57BL/6 females to obtain agouti offspring. The agouti female offspring were backcrossed with C57BL/6 males to generate hemizygous male F2 mice. Genotyping was performed by PCR analysis. Whole-mount and tissue section LacZ and CD31 staining of embryonic and adult tissues, β-galactosidase activity assays, and tumor xenografts were carried out as previously described6,7 (and supplemental Methods).
Statistical analyses
Data were expressed as mean plus or minus SE. The statistical significance of differences of the means was determined by 1-way analysis of variance and multiple comparisons by Tukey-Kramer multiple range test.
Results
Phylogenic footprinting of VEGFR1 promoter
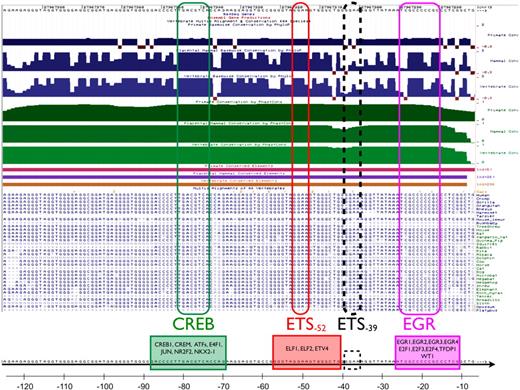
Previous studies have implicated a role for 2 ETS consensus elements, a CREB/ATF binding site and an EGR binding motif, in mediating basal or inducible expression of VEGFR1.1,8,9 Recognizing that functional regulatory regions are more highly conserved than neutrally evolving sequences, we used a comparative genomics approach to determine the extent to which these sites are evolutionarily conserved and to identify other potentially relevant cis-regulatory elements. Using University of California Santa Cruz (UCSC) Genome Browser10-12 and ECR Browser,13,14 we identified 3 regions of significant conservation between −1300 and +300: region 1 between −6 and −124, region 2 between −244 and −486, and region 3 between −659 and −1199 (supplemental Figures 1-2). Region 1 contains the EGR binding element (between −16 and −25), the ETS motif between −49 and −52 (referred to hereafter as −52 ETS), the −36 to −39 ETS site (referred to hereafter as −39 ETS), and the CRE element between −74 and −81 (Figure 1). These 4 sites demonstrate remarkable conservation across all vertebrae. A conserved transcription factor binding site search using MultiTF15,16 identified potential transcription factors with binding sites overlapping 3 of the 4 sites. The EGR site could potentially bind EGR1, EGR2, EGR3, EGR4, E2F1, E2F3, E2F4, TFDP1, or WT1. The −52 ETS could bind to several ETS family members, including ELF1/2 and ETV4. The −39 ETS site is not directly identified by MultiTF as a consensus binding site, likely due to the mismatch between the nucleotides flanking its core GGAA sequence (GGGAAA) and the consensus AGGAAG required by most ETS-type matrices in TRANSFAC.17 Finally, the CRE-type site could potentially bind CREB1, CREM, ATF1, ATF2, ATF3, ATF4, ATF5, ATF6, ATF7, JUN, NR2F2, and NKX2-1.
Sequence conservation of the immediate upstream VEGFR1 promoter. Top part shows evolutionary conservation of the promoter region between −7 and −125 (chr13:27967,272-27967,390) using the UCSC Gene Browser. Blue/green tracks on the top part of the figure illustrate conservation of the sequence in primates (top tracks), placental mammals (middle tracks), and vertebrae (bottom, light blue/light green tracks) using the PhyloP and PhastCons packages (Cornell University), respectively. Conserved elements identified by PhastCons are shown as magenta (primates), purple (placental mammals), and brown (vertebrae) bars. Sequence alignments show conservation of the CRE motif at −81, the −52 and −39 ETS motifs, and the EGR binding site at −25. Although highly conserved, the −39 ETS site was not identified as a binding site by MultiTF.
Sequence conservation of the immediate upstream VEGFR1 promoter. Top part shows evolutionary conservation of the promoter region between −7 and −125 (chr13:27967,272-27967,390) using the UCSC Gene Browser. Blue/green tracks on the top part of the figure illustrate conservation of the sequence in primates (top tracks), placental mammals (middle tracks), and vertebrae (bottom, light blue/light green tracks) using the PhyloP and PhastCons packages (Cornell University), respectively. Conserved elements identified by PhastCons are shown as magenta (primates), purple (placental mammals), and brown (vertebrae) bars. Sequence alignments show conservation of the CRE motif at −81, the −52 and −39 ETS motifs, and the EGR binding site at −25. Although highly conserved, the −39 ETS site was not identified as a binding site by MultiTF.
Consensus motifs for ETS, CREB/ATF, and EGR are involved in mediating VEGFR1 promoter activity in cultured human primary endothelial cells
To confirm a functional role for the −25 EGR binding element, the −52 and −39 ETS motifs, and the −81 CRE site in mediating VEGFR1 expression in vitro, we generated VEGFR1 promoter luciferase constructs containing point mutations of each these motifs (Figure 2A). The resulting constructs were transiently transfected into primary human endothelial cells, including HUVECs, HCAECs, and HPAECs. Mutation of the −52 ETS motif resulted in complete loss of expression, whereas mutation of the CRE and EGR elements resulted in a significant, though less dramatic, reduction in promoter activity (Figure 2B). In contrast, a mutation of the −39 ETS site yielded increased promoter activity (Figure 2B). Although the latter result was consistently observed in multiple experiments and endothelial cell types, it reached statistical significance in HCAEC alone. Thus, consensus elements for ETS (−49 to −52), CREB/ATF (−74 to −81), and EGR (−16 to −25) play a positive role in VEGFR1 expression, whereas a putative ETS motif between −36 and −39 has a net negative effect on promoter activity.
ETS, CRE, and EGR binding sites contribute to basal expression of the VEGFR1 promoter in primary human endothelial cells. (A) Wild-type (WT) and mutant cis-regulatory sequences used in transfections and Hprt-targeted mice. The wild-type DNA sequence is shown on top (50-bp per line) with each of the ETS, CRE, and EGR binding sites individually colored, and the 5′ untranslated region underlined. The mutated sequences are shown underneath, underlined in parentheses. (B) WT or mutant VEGFR1 promoters were coupled to luciferase in PGL3 and the resulting plasmids were transiently transfected into HUVECs, HCAECs, or HPAECs. The results show the means and SDs of luciferase light units (relative to untreated cells) obtained in triplicate from at least 3 independent experiments. + indicates 0.1; **P < .01; ***P < .001.
ETS, CRE, and EGR binding sites contribute to basal expression of the VEGFR1 promoter in primary human endothelial cells. (A) Wild-type (WT) and mutant cis-regulatory sequences used in transfections and Hprt-targeted mice. The wild-type DNA sequence is shown on top (50-bp per line) with each of the ETS, CRE, and EGR binding sites individually colored, and the 5′ untranslated region underlined. The mutated sequences are shown underneath, underlined in parentheses. (B) WT or mutant VEGFR1 promoters were coupled to luciferase in PGL3 and the resulting plasmids were transiently transfected into HUVECs, HCAECs, or HPAECs. The results show the means and SDs of luciferase light units (relative to untreated cells) obtained in triplicate from at least 3 independent experiments. + indicates 0.1; **P < .01; ***P < .001.
ELF-1 and SP1 interact with the −52 ETS motif to transactivate the VEGFR1 promoter in cultured human primary endothelial cells
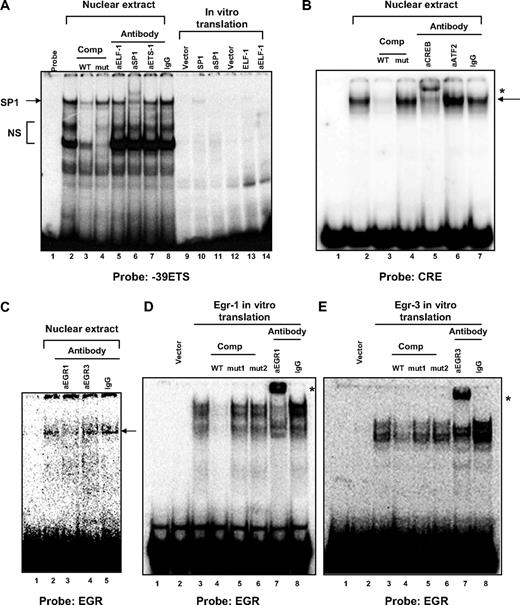
The transient transfection data suggested that the −52 ETS site plays a particularly important role in mediating VEGFR1 expression in endothelial cells. To identify the factor that binds to this site, we carried out EMSAs. Incubation of a radiolabeled probe spanning the −52 ETS site with nuclear extract from HUVECs resulted in several DNA-protein complexes (Figure 3A lane 2). Four of the complexes (Figure 3A labeled a-d) were inhibited by addition of cold wild-type ETS competitor, but not a mutant ETS competitor (Figure 3A lanes 3-6). Supershift assays were carried out with antibodies to ETS factors that have been previously implicated in endothelial cell gene regulation, including ETS-1, ETS-2, ELF-1, FLI-1, ERG, NERF, and PEA3 (Figure 3B shows ETS-1, ETS-2, and ELF-1). Of these antibodies, ELF-1 resulted in significant inhibition of a specific DNA-protein complex (Figure 3B lane 3). This complex migrated at the same distance as recombinant in vitro translated ELF-1 protein bound to DNA (Figure 3B compare lanes 12 and 2). Recombinant ELF-1–DNA complex was inhibited by anti–ELF-1 antibody (Figure 3B lane 13), indicating that the ELF-1 antibody does indeed inhibit rather than supershift the DNA-protein complex. Antibody to ETS-1 had a slight inhibitory effect on the specific DNA-protein complex (Figure 3B lane 5). However, given that ETS-1 is considerably smaller than ELF-1 (51 and 98 kDa, respectively), it is unlikely that the antibody is recognizing ETS-1 alone or bound together with ELF-1 to DNA (the DNA-protein complexes would migrate faster or slower than the ELF-1–DNA complex). As a positive control for ETS-1 binding and supershifting activity of the ETS-1 antibody, a radiolabeled probe spanning a consensus ETS-1 binding motif was incubated with recombinant ETS-1. As shown in Figure 3C, ETS-1 protein bound to the consensus ETS-1 binding, but not the −52 ETS probe. The resulting DNA-protein complex was supershifted by ETS-1 antibody. Thus, the −52 site binds ELF-1, but not ETS-1.
ELF-1 and SP1 bind to the −52 ETS motif in the human VEGFR1 promoter and induce promoter activity. (A) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-7) In competition assays, a 10-fold or 50-fold molar excess of unlabeled wild-type (lanes 3-4) or mutant (lanes 5-6) −52 ETS probe was added to the reaction mixture.  indicates specific DNA-protein complexes. (Based on EMSA in panel B, complexes a and c represent SP1 and ELF-1, respectively; the identity of complexes b and d is unknown). NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lanes 1, 8, 11) or presence of nuclear extract from HUVECs (lanes 2-7), recombinant SPI (lanes 9-10), or recombinant ELF-1 (lanes 12-13). In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lane 3, 13), SP1 (lane 4, 10), ETS-1 (lane 5), ETS-2 (lane 6), or control antibody (lane 7).
indicates specific DNA-protein complexes. (Based on EMSA in panel B, complexes a and c represent SP1 and ELF-1, respectively; the identity of complexes b and d is unknown). NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lanes 1, 8, 11) or presence of nuclear extract from HUVECs (lanes 2-7), recombinant SPI (lanes 9-10), or recombinant ELF-1 (lanes 12-13). In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lane 3, 13), SP1 (lane 4, 10), ETS-1 (lane 5), ETS-2 (lane 6), or control antibody (lane 7).  indicates SP1 and ELF-1 DNA-protein complexes. (C) EMSA was performed with 32P-labeled consensus ETS-1 binding probe (lanes 1-3) or −52 ETS (lanes 4-5) in the absence (lanes 1, 4) or presence of in vitro translated ETS-1 ± antibodies to ETS-1 (lane 3). (D) HUVECs were transfected with control siRNA or siRNA against ELF-1 and/or ETS-1. Real-time PCR was used to assay for mRNA expression of ELF-1, ETS-1, or VEGFR1. The results show the means and SDs of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. RQ indicates relative quantitation. *P < .05; **P < .01; ***P < .001, relative to control siRNA. (E) ChIP assay was performed using HUVECs. DNA was sheared and resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ELF-1, SP1, or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VEGFR1 proximal region, which included the −52 ETS site. (F) Cotransfection assay was carried out in HEK293 cells using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (G) Cotransfection assay was carried out in HUVECs using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (F-G) *P < .05; **P < .01; ***P < .001, relative to pCI + pcDNA.
indicates SP1 and ELF-1 DNA-protein complexes. (C) EMSA was performed with 32P-labeled consensus ETS-1 binding probe (lanes 1-3) or −52 ETS (lanes 4-5) in the absence (lanes 1, 4) or presence of in vitro translated ETS-1 ± antibodies to ETS-1 (lane 3). (D) HUVECs were transfected with control siRNA or siRNA against ELF-1 and/or ETS-1. Real-time PCR was used to assay for mRNA expression of ELF-1, ETS-1, or VEGFR1. The results show the means and SDs of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. RQ indicates relative quantitation. *P < .05; **P < .01; ***P < .001, relative to control siRNA. (E) ChIP assay was performed using HUVECs. DNA was sheared and resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ELF-1, SP1, or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VEGFR1 proximal region, which included the −52 ETS site. (F) Cotransfection assay was carried out in HEK293 cells using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (G) Cotransfection assay was carried out in HUVECs using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (F-G) *P < .05; **P < .01; ***P < .001, relative to pCI + pcDNA.
ELF-1 and SP1 bind to the −52 ETS motif in the human VEGFR1 promoter and induce promoter activity. (A) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-7) In competition assays, a 10-fold or 50-fold molar excess of unlabeled wild-type (lanes 3-4) or mutant (lanes 5-6) −52 ETS probe was added to the reaction mixture.  indicates specific DNA-protein complexes. (Based on EMSA in panel B, complexes a and c represent SP1 and ELF-1, respectively; the identity of complexes b and d is unknown). NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lanes 1, 8, 11) or presence of nuclear extract from HUVECs (lanes 2-7), recombinant SPI (lanes 9-10), or recombinant ELF-1 (lanes 12-13). In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lane 3, 13), SP1 (lane 4, 10), ETS-1 (lane 5), ETS-2 (lane 6), or control antibody (lane 7).
indicates specific DNA-protein complexes. (Based on EMSA in panel B, complexes a and c represent SP1 and ELF-1, respectively; the identity of complexes b and d is unknown). NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled −52 ETS probe in the absence (lanes 1, 8, 11) or presence of nuclear extract from HUVECs (lanes 2-7), recombinant SPI (lanes 9-10), or recombinant ELF-1 (lanes 12-13). In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lane 3, 13), SP1 (lane 4, 10), ETS-1 (lane 5), ETS-2 (lane 6), or control antibody (lane 7).  indicates SP1 and ELF-1 DNA-protein complexes. (C) EMSA was performed with 32P-labeled consensus ETS-1 binding probe (lanes 1-3) or −52 ETS (lanes 4-5) in the absence (lanes 1, 4) or presence of in vitro translated ETS-1 ± antibodies to ETS-1 (lane 3). (D) HUVECs were transfected with control siRNA or siRNA against ELF-1 and/or ETS-1. Real-time PCR was used to assay for mRNA expression of ELF-1, ETS-1, or VEGFR1. The results show the means and SDs of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. RQ indicates relative quantitation. *P < .05; **P < .01; ***P < .001, relative to control siRNA. (E) ChIP assay was performed using HUVECs. DNA was sheared and resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ELF-1, SP1, or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VEGFR1 proximal region, which included the −52 ETS site. (F) Cotransfection assay was carried out in HEK293 cells using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (G) Cotransfection assay was carried out in HUVECs using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (F-G) *P < .05; **P < .01; ***P < .001, relative to pCI + pcDNA.
indicates SP1 and ELF-1 DNA-protein complexes. (C) EMSA was performed with 32P-labeled consensus ETS-1 binding probe (lanes 1-3) or −52 ETS (lanes 4-5) in the absence (lanes 1, 4) or presence of in vitro translated ETS-1 ± antibodies to ETS-1 (lane 3). (D) HUVECs were transfected with control siRNA or siRNA against ELF-1 and/or ETS-1. Real-time PCR was used to assay for mRNA expression of ELF-1, ETS-1, or VEGFR1. The results show the means and SDs of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. RQ indicates relative quantitation. *P < .05; **P < .01; ***P < .001, relative to control siRNA. (E) ChIP assay was performed using HUVECs. DNA was sheared and resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ELF-1, SP1, or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VEGFR1 proximal region, which included the −52 ETS site. (F) Cotransfection assay was carried out in HEK293 cells using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (G) Cotransfection assay was carried out in HUVECs using 0.3 μg of pcDNA3-SP1 expression vector or empty vector (PCI-pcDNA), and either wild-type VEGFR1-luc (WT) or a similar construct containing a mutation of the −52 ETS site. Data represent mean ± SE of 6 replicates. Luciferase light units are expressed as fold induction over the empty expression vector. (F-G) *P < .05; **P < .01; ***P < .001, relative to pCI + pcDNA.
To provide further evidence for the selective involvement of ELF-1 in mediating VEGFR1 expression, HUVECs were transfected with siRNA against ELF-1 and/or ETS-1. ELF-1–siRNA resulted in 70% reduction in ELF-1 mRNA levels, and 30% reduction in VEGFR1 mRNA levels (Figure 3D). By contrast, siRNA-mediated knockdown of ETS-1 (80% reduction) had no effect on VEGFR1 expression. As a positive control, ETS-1 knockdown resulted in 70% reduction of its known target gene, monocyte chemoattractant protein 1 (not shown). ETS-1 knockdown did not lead to further reduction of VEGFR1 mRNA levels in ELF-1–deficient HUVECs, arguing against a functional interaction between these 2 factors (Figure 3D).
Unexpectedly, preincubation with anti-SP1 antibody resulted in a partial supershift of the slowest migrating DNA-protein complex (Figure 3B lane 4; the partial nature of the supershift may be explained by limiting amounts of antibody or the existence of a second DNA-protein complex that lacks SP1). Incubation of radiolabeled probe with in vitro translated SP1 resulted in a DNA-protein complex of similar size to that obtained with nuclear extracts (Figure 3B compare lanes 9 and 2). The latter complex was supershifted with anti-SP1 antibody (Figure 3B lane 10).
To investigate whether ELF-1 and SP1 bind to the VEGFR1 proximal region in endothelial cells, ChIP assay was performed. Formalin-fixed genomic DNA-protein complexes from HUVECs were sheared by sonication. Resulting small DNA-protein complexes were immunoprecipitated with antibodies to ELF-1 and SP1 (or control immunoglobulin G [IgG]), and the resulting products were used as template in a PCR reaction containing primers specific for the immediate upstream promoter of VEGFR1. Real-time PCR was used to calculate binding intensities. As shown in Figure 3E, ELF-1 and SP1, but not control IgG, bound to the proximal promoter region.
To determine whether ELF-1 and/or SP1 transactivate the VEGFR1 promoter, cotransfection assays were carried out in HEK293 cells. An advantage of using HEK293 cells is that they express low, nonsaturating levels of endothelial-associated ETS factors, including ELF-1. Overexpression of ELF-1 resulted in significant (3.4-fold) induction of VEGFR1 promoter activity (Figure 3F). Mutation of the −52 ETS site abrogated ELF-1–mediated transactivation of the promoter. Expression of SP1 also induced VEGFR1 activity (3.6-fold; Figure 3F). Interestingly, this effect was also abrogated by mutation of the −52 ETS site. Cotransfection of ELF-1 and SP1 resulted in an additive effect on promoter activity (6.0-fold). The cotransfection experiments were repeated in HUVECs. Although absolute levels of induction were lower compared with HEK293 cells, ELF-1 and SP1 each transactivated the wild-type, but not the −52 ETS mutant VEGFR1 promoter, and combined transfection with ELF-1 and SP1 had an additive effect (Figure 3G). Together with the EMSA results, these data suggest that ELF-1 and SP1 both bind to the −52 ETS motif and transactivate VEGFR1.
The −39 ETS motif is bound by SP1, but not ETS factor(s), in cultured human primary endothelial cells
Our transfection data suggested that the −39 ETS motif plays a negative role in VEGFR1 transcription. To determine whether this site binds nuclear protein, we performed EMSA using a radiolabeled probe spanning the −39 ETS site and nuclear extract from HUVECs. Several DNA-protein complexes were observed (Figure 4A lane 2). However, only 1 of these (the slowest migrating) was inhibited by cold wild-type ETS competitor, but not mutated ETS competitor (Figure 4A lanes 3-4). Supershift assays were carried out as described with the −52 ETS probe. Antibodies against the various ETS factors failed to supershift or inhibit the specific DNA-protein complex (Figure 4A lanes 5 and 7 show ELF-1 and ETS-1). In contrast, anti-SP1 antibody resulted in a supershifted complex (Figure 4A lane 6). In vitro translated SP1 protein resulted in a specific DNA-protein complex compatible with that observed with nuclear extracts (Figure 4A compare lanes 10 and 2). These findings, together with the observation that a mutation of the −39 ETS motif induces promoter activity, whereas a mutation of the −52 ETS motif (leaving the −39 ETS motif intact) abrogates SP1- and ETS-mediated transactivation of the VEGFR1 promoter, suggest that the −39 ETS site inhibits expression by binding and sequestering SP1. Consistent with this conclusion, binding of recombinant SP1 to the −52 ETS motif was inhibited to a similar degree by same-fold cold competitor containing either the −52 ETS or −39 ETS motif (supplemental Figure 3).
DNA-protein binding at the −39 ETS, CRE, and EGR binding motifs in the human VEGFR1 promoter. (A) EMSA was performed with 32P-labeled −39 ETS probe in the absence (lanes 1, 9, 12) or presence of nuclear extract from HUVECs (lanes 2-8), recombinant SPI (lanes 10-11), or recombinant ELF-1 (lanes 13-14). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 3) or mutant (lane 4) −39 ETS probe was added to the reaction mixture. In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lanes 5, 14), SP1 (lanes 6, 11), ETS-1 (lane 7), or control antibody (lane 8). The arrow indicates specific SP1-DNA complex. NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled consensus CRE probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-7). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 3) or mutant (lane 4) CRE probe was added to the reaction mixture. In supershift assays, nuclear extracts were incubated in the presence of antibodies to CREB (lane 5), ATF2 (lane 6), or control antibody (lane 7).  indicates specific CREB-DNA complex; *, supershifted complex. (C) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-5). In supershift assays, nuclear extracts were incubated in the presence of antibodies to EGR-1 (lane 3), EGR-3 (lane 4), or control antibody (lane 5).
indicates specific CREB-DNA complex; *, supershifted complex. (C) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-5). In supershift assays, nuclear extracts were incubated in the presence of antibodies to EGR-1 (lane 3), EGR-3 (lane 4), or control antibody (lane 5).  indicates DNA-protein complex. (D) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-1 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-1 (lane 7) or control antibody (lane 8). * indicates supershifted complex. (E) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-3 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-3 (lane 7) or control antibody (lane 8). * indicates supershifted complex.
indicates DNA-protein complex. (D) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-1 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-1 (lane 7) or control antibody (lane 8). * indicates supershifted complex. (E) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-3 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-3 (lane 7) or control antibody (lane 8). * indicates supershifted complex.
DNA-protein binding at the −39 ETS, CRE, and EGR binding motifs in the human VEGFR1 promoter. (A) EMSA was performed with 32P-labeled −39 ETS probe in the absence (lanes 1, 9, 12) or presence of nuclear extract from HUVECs (lanes 2-8), recombinant SPI (lanes 10-11), or recombinant ELF-1 (lanes 13-14). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 3) or mutant (lane 4) −39 ETS probe was added to the reaction mixture. In supershift assays, nuclear extracts or recombinant protein was incubated in the presence of antibodies (indicated by prefix “a”) to ELF-1 (lanes 5, 14), SP1 (lanes 6, 11), ETS-1 (lane 7), or control antibody (lane 8). The arrow indicates specific SP1-DNA complex. NS indicates nonspecific complex. (B) EMSA was performed with 32P-labeled consensus CRE probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-7). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 3) or mutant (lane 4) CRE probe was added to the reaction mixture. In supershift assays, nuclear extracts were incubated in the presence of antibodies to CREB (lane 5), ATF2 (lane 6), or control antibody (lane 7).  indicates specific CREB-DNA complex; *, supershifted complex. (C) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-5). In supershift assays, nuclear extracts were incubated in the presence of antibodies to EGR-1 (lane 3), EGR-3 (lane 4), or control antibody (lane 5).
indicates specific CREB-DNA complex; *, supershifted complex. (C) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lane 1) or presence of nuclear extract from HUVECs (lanes 2-5). In supershift assays, nuclear extracts were incubated in the presence of antibodies to EGR-1 (lane 3), EGR-3 (lane 4), or control antibody (lane 5).  indicates DNA-protein complex. (D) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-1 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-1 (lane 7) or control antibody (lane 8). * indicates supershifted complex. (E) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-3 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-3 (lane 7) or control antibody (lane 8). * indicates supershifted complex.
indicates DNA-protein complex. (D) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-1 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-1 (lane 7) or control antibody (lane 8). * indicates supershifted complex. (E) EMSA was performed with 32P-labeled EGR binding site probe in the absence (lanes 1-2) or presence of recombinant EGR-3 (lanes 3-8). In competition assays, a 50-fold molar excess of unlabeled wild-type (lane 4) or mutant (lanes 5-6) EGR probe was added to the reaction mixture. In supershift assays, recombinant protein was incubated in the presence of antibodies to EGR-3 (lane 7) or control antibody (lane 8). * indicates supershifted complex.
The CRE element is bound by CREB and the EGR binding element is bound by EGR-1 in cultured human primary endothelial cells
The transfection data pointed to a positive role for the CREB/ATF and EGR binding sites in mediating basal activity of the VEGFR1 promoter. To determine whether these sites bind to nuclear protein, we performed EMSA using radiolabeled probes spanning each motif. Incubation of the CRE probe with nuclear extract from endothelial cells resulted in a specific DNA-protein complex that was competed by self- but not mutant cold probe (Figure 4B lanes 3-4). The complex was supershifted by addition of anti-CREB antibody (Figure 4B lane 5), but not by antibody against ATF2 or IgG (Figure 4B lanes 6-7). Incubation of the probe containing the EGR binding site resulted in a DNA-protein complex that was partially inhibited by anti–EGR-1 antibody but not anti–EGR-3 antibody or control IgG (Figure 4C). The appearance of this band required a long exposure time (48 hours). To verify that the EGR binding site was capable of binding to EGR protein, the labeled probe was incubated with recombinant EGR-1 or EGR-3. Each factor formed specific complexes that were inhibited by cold wild-type but not mutant competitor and were supershifted by their respective antibody (Figure 4D-E). Thus, the EGR binding site has the capacity to bind both EGR-1 and EGR-3.
Consensus motifs for ETS, CREB/ATF, and EGR mediate VEGFR1 promoter activity in vivo
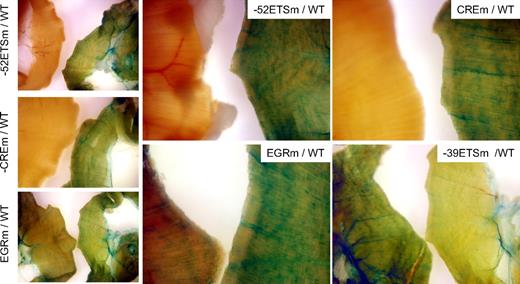
To determine the role of the various cis-regulatory motifs in directing expression of VEGFR1 in vivo, the wild-type or mutant VEGFR1 promoters were coupled to LacZ and the resulting constructs targeted to the Hprt locus of mice using homologous recombination. Hprt-targeted lines were bred to generate stable lines. Reporter gene activity was assayed in 6- to 8-week-old F2 males. To compare whole-mount LacZ staining of wild-type and mutant VEGFR1 mouse tissues, organs were harvested from age- and generation-matched male mice, and stained simultaneously with the same X-Gal–containing solution. LacZ staining of the diaphragm (a thin blood vessel–rich skeletal muscle that is particularly amenable to whole-mount staining) revealed vascular expression in wild-type VEGFR1 mice, but not in mice carrying a mutation of the −52 ETS or CRE sites (Figure 5). Compared with the wild-type promoter, the EGR mutant demonstrated lower expression in the diaphragm, whereas the −39 ETS mutant revealed higher activity (Figure 5). Whole-mount stains of the heart revealed LacZ expression in all lines, with highest expression in the −39 ETS mutant line followed by wild-type, EGR mutant, −52 ETS mutant, and CRE mutant lines (supplemental Figure 3). The pattern was similar in the lung, with the notable exception that reporter gene activity was comparable in EGR mutant and wild-type lines (supplemental Figure 4).
LacZ staining of diaphragm from Hprt-targeted mice carrying wild-type or mutant VEGFR1 promoters. Diaphragms were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying wild-type (WT) or mutant VEGFR1-lacZ transgenes and processed in parallel for whole-mount staining with X-Gal. In each panel, the diaphragm from WT VEGFR1 transgenic is on the right, and the diaphragm from the mutant mouse is on the left. The exposure time in bottom right panel was reduced compared with other panels. Whole-mount diaphragms were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
LacZ staining of diaphragm from Hprt-targeted mice carrying wild-type or mutant VEGFR1 promoters. Diaphragms were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying wild-type (WT) or mutant VEGFR1-lacZ transgenes and processed in parallel for whole-mount staining with X-Gal. In each panel, the diaphragm from WT VEGFR1 transgenic is on the right, and the diaphragm from the mutant mouse is on the left. The exposure time in bottom right panel was reduced compared with other panels. Whole-mount diaphragms were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
In tissue sections of Hprt-targeted adult mice carrying wild-type, −39 ETS mutant, or EGR mutant VEGFR1 promoters, LacZ staining colocalized with CD31 in the endothelial lining of vessels in all organs examined except the liver (Figure 6 shows heart, lung, kidney, and skeletal muscle). In addition, reporter gene expression was detectable in cardiomyocytes and occasional arterial smooth muscle cells. In the −52 ETS and CRE mutant mice, LacZ expression was detected in cardiomyocytes and vascular smooth muscle cells, but was undetectable in the endothelium of all organs examined (supplemental Figure 5 shows kidney, spleen, and lung). None of the Hprt-targeted transgenes was expressed in bone marrow or peripheral blood cells (not shown).
LacZ staining of tissue sections. Cryosections were collected from heart, lung, kidney, and skeletal muscle (Sk mu) harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying wild-type, −39 ETSm, or EGRm VEGFR1 transgenes and stained with X-Gal. Serial sections were processed for CD31 immunohistochemistry. Images were obtained using a 20× objective. Bar, 100μM. Lac-Z-stained sections were counterstained with eosin. Slides were analyzed under a Zeiss Axio Imager upright microscope, and photomicrographs were collected using a Zeiss Axiocam MRc camera and Axiovision 4.6.3 image acquisition software.
LacZ staining of tissue sections. Cryosections were collected from heart, lung, kidney, and skeletal muscle (Sk mu) harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying wild-type, −39 ETSm, or EGRm VEGFR1 transgenes and stained with X-Gal. Serial sections were processed for CD31 immunohistochemistry. Images were obtained using a 20× objective. Bar, 100μM. Lac-Z-stained sections were counterstained with eosin. Slides were analyzed under a Zeiss Axio Imager upright microscope, and photomicrographs were collected using a Zeiss Axiocam MRc camera and Axiovision 4.6.3 image acquisition software.
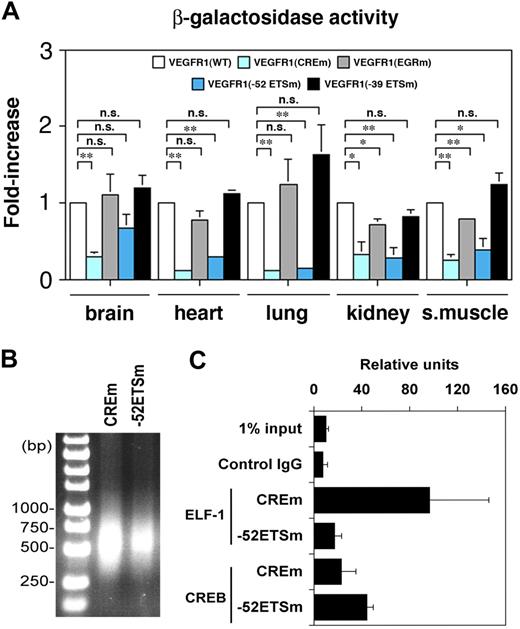
To quantitate differences in transgene expression, tissues were assayed for β-galactosidase activity. As shown in Figure 7A, β-galactosidase activity in the −52 ETS mutant line was significantly reduced in the heart, lung, kidney, and skeletal muscle. In the CRE mutant line, reporter gene activity was lower in the brain, heart, lung, kidney, and skeletal muscle. The EGR mutant demonstrated reduced β-galactosidase activity in the kidney and skeletal muscle, but not lung. In contrast to a clear increase in X-Gal reaction product in whole-mount organs of −39 ETS mutant mice, there were no significant differences in activity between wild-type and −39 ETS mutant mice. Given that the whole-mount staining of mutant and wild-type organs was always carried out in parallel in multiple independent animals and the results were documented side by side, we conclude that LacZ staining is a more sensitive assay than β-galactosidase activity measurements.
β-Galactosidase activity and CREB/ELF-1 binding assays. (A) β-Galactosidase activity of protein extracts from various organs of the Hprt-targeted mice. Data represent mean ± SE of 4 replicates. *P < .05; **P < .01; **P < .001. n.s., not significant. (B-C) ChIP assay was performed using mouse heart endothelial cells from Hprt-targeted mice carrying a mutation of the CRE site (CREm) or −52 ETS site (−52ETSm). The sheared DNA samples were analyzed by 1.7% agarose gel. Resulting DNA-protein complexes were immunoprecipitated in the absence or presence antibodies to ELF-1 or CREB. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for the human proximal promoter and LacZ cDNA (C).
β-Galactosidase activity and CREB/ELF-1 binding assays. (A) β-Galactosidase activity of protein extracts from various organs of the Hprt-targeted mice. Data represent mean ± SE of 4 replicates. *P < .05; **P < .01; **P < .001. n.s., not significant. (B-C) ChIP assay was performed using mouse heart endothelial cells from Hprt-targeted mice carrying a mutation of the CRE site (CREm) or −52 ETS site (−52ETSm). The sheared DNA samples were analyzed by 1.7% agarose gel. Resulting DNA-protein complexes were immunoprecipitated in the absence or presence antibodies to ELF-1 or CREB. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for the human proximal promoter and LacZ cDNA (C).
To determine whether the profound changes associated with mutation of −52 ETS motif and the CRE site manifested during development, we examined LacZ expression in utero. Embryonic day (E)–9.5 to E10.5 embryos carrying the wild-type VEGFR1 promoter demonstrated LacZ expression throughout the vasculature (supplemental Figure 6). A similar pattern was observed in mice carrying a mutation of the −39 ETS and EGR sites (data not shown). Mutation of the −52 ETS motif and the CRE site resulted in significantly reduced expression of LacZ (supplemental Figure 6). The majority of β-galactosidase activity was detected in the endothelium of yolk sac blood vessels and the endothelium of caudal intersomitic vessels.
We previously demonstrated that the Flt-1 promoter drives expression in the endothelium of tumor xenografts.2 To determine activity of the mutant promoters in tumor vasculature, B16-F1 melanoma and Lewis lung carcinoma cells were implanted subcutaneously into the flank of Hprt-targeted mice. When tumors reached approximately 2.5 cm3 in volume, the xenografts were harvested, sectioned, and stained for LacZ. The wild-type and all 4 mutant promoters displayed detectable activity within tumor endothelium (supplemental Figure 7). The presence of X-Gal reaction product in tumor vessels of the −52 ETS and CRE mutant lines is in striking contrast to the absence of detectable promoter activity in surrounding flank tissue including skin, subcutaneous fat, and abdominal wall muscle (not shown). These findings suggest that the −52 ETS and CRE mutant promoters become reactivated in the context of the tumor microenvironment and indicate that other cis-regulatory elements are sufficient for directing expression in this pathologic vascular bed.
ELF-1 and CREB bind to the VEGFR1 transgenic promoter
To investigate whether ELF-1 and CREB bind to the −52 ETS and CRE motifs in vivo, ChIP assay was performed using endothelial cells cultured from the hearts of −52 ETS and CRE mutant mice. Formalin-fixed genomic DNA-protein complexes from the murine endothelial cells were sheared by sonication (Figure 7B). DNA-protein complexes were immunoprecipitated with antibodies to ELF-1, CREB, or control IgG, and the resulting products were used as template in a PCR reaction containing primers specific for the VEGR1 transgene. One primer set comprised a forward primer that lies in the human VEGFR1 promoter and a reverse primer that is complementary to a sequence in the 5′ end of LacZ cDNA. Because the VEGFR1 gene contains a relatively long 5′ untranslated region (284 bp), we considered the possibility that the latter primers would not adequately cover the −52 ETS and CRE sites in sheared DNA. Thus, we confirmed the results using a second set of primers, both of which lie in the immediate upstream promoter region of human VEGFR1 and have low homology with the mouse VEGFR1 promoter (supplemental Table 1). In these assays, ELF-1 preferentially bound to the CREm transgenic promoter, whereas CREB demonstrated preferential binding to the −52 ETS transgenic promoter (Figure 7C shows the first primer set; similar results were obtained with the second primer set). These findings demonstrate that ELF-1 and CREB bind to the −52 ETS and CRE sites of the VEGFR1 promoter, respectively.
Discussion
Morishita et al first reported that a −748 to +248 region of the human VEGFR1 promoter directs lineage-specific expression in cultured endothelial cells.1 Deletion of 4 base pairs from a consensus CREB/ATF binding motif (ACGT out of TGACGTCA) resulted in an 85% reduction in promoter activity in bovine aortic endothelial cells.1 In a subsequent study, a mutation of the same CRE element (TGACGTCA to TCTGCTCA) in the context of a VEGFR1 promoter spanning the region between −90 and +8 resulted in 90% reduction in reporter gene expression in HUVECs.8 A mutation of the ETS motif between −49 and −52 (GGAA to CCAA) was also shown to impair reporter gene expression,8 whereas mutation of the ETS consensus site between −36 and −39 (GGAA to CCAA) resulted in increased promoter activity (≈1.85-fold).8 Our in vitro data confirm a positive role for the −52 ETS and CRE sites and a negative role for the −39 ETS motif in mediating VEGFR1 promoter activity in primary human endothelial cells.
Using EMSA and ChIP assays, we have shown that the −52 ETS motif is bound by the ETS factor, ELF-1, as well as SP1. A functional role for these DNA-protein interactions was evidenced by the ability of ELF-1 and SP1 to transactivate the wild-type VEGFR1 promoter, but not a promoter containing a mutation of the −52 ETS site. In EMSA, there was no evidence that ELF-1 and SP1 bind together to the −52 ETS oligonucleotide probe (SP1 represented the largest/slowest migrating complex). However, cotransfection with ELF-1 and SP1 had an additive effect on VEGFR1 promoter activity, suggesting that the 2 transcription factors interact functionally to promote expression. ELF-1 was originally described as a regulator of T-cell–specific genes.18 Subsequent studies revealed a role for ELF-1 in endothelial cell–specific gene regulation. For example, ELF-1 has been shown to bind to and/or transactivate the promoters for Tie-1, Tie-2, and endothelial nitric oxide synthase.19,20 The current study adds VEGFR1 to the list of ELF-1–responsive genes in endothelial cells.
Similar to −52 ETS, the negatively acting −39 ETS consensus site also bound SP1. However, we failed to detect binding of any ETS protein to this element. Indeed, the −39 ETS site was not directly identified by MultiTF as a consensus binding site. The finding that the −52 ETS mutant (with an intact −39 ETS site) was unresponsive to ELF-1 and/or SP1 expression in cotransfection assays suggests that SP1 binds to, but does not activate, the proximal ETS site. Thus, it is possible that the −39 ETS motif sequesters SP1, thereby inhibiting functional interactions between SP1 and the upstream −52 ETS motif.
Our results support the importance of the CREB/ATF binding element in mediating VEGFR1 expression. The data confirm earlier studies demonstrating a positive role for this site in transient transfection assays.1,8 In the latter studies, forskolin and dibutyryl-cyclic adenosine monophosphate failed to activate the VEGFR1 promoter, suggesting that the CRE motif functioned independently of cyclic adenosine monophosphate signaling.1,8 Wakiya et al showed that a specific DNA-protein complex formed on the CRE motif.8 Using nuclear extracts from primary human endothelial cells, we have shown that this complex consists of CREB, but not ATF2. The finding that CREB binds to an Hprt-targeted human VEGFR promoter containing an intact CRE site, but not an identical promoter with a mutation of CRE, provides further evidence for the role of CREB in mediating VEGFR1 expression in vivo.
It was previously demonstrated that an EGR binding site between −16 and −24 plays a role in mediating VEGFR1 expression in response to injury. In an in vitro wounding assay, both VEGFR and EGR-1 were expressed at the edge of the denuded area.9 A mutation of EGR binding site inhibited injury-induced activation of the promoter, but had no effect on basal activity.9 EMSA revealed inducible binding of EGR-1 to the consensus EGR binding element, and in cotransfection assays EGR-1 induced the expression of the VEGFR1 promoter.9
Previous studies of EGR proteins in vascular biology have focused primarily on EGR-1 (reviewed in Khachigian et al21 ). However, recent evidence points to an important role for EGR-3 in transducing signals in endothelial cells. For example, EGR-3 was one of the genes most strongly up-regulated by VEGF in DNA microarrays of HUVECs,22 and VEGF- and fibroblast growth factor 2 (FGF-2)–induced endothelial cell proliferation was attenuated by EGR-3 knockdown.23 We have demonstrated that EGR-3 has a far greater effect on VEGFR1 promoter activity compared with EGR-1 in cotransfection assays (17-fold vs 4-fold induction, respectively; T.M. and W.C.A., unpublished observations, March 2009). Moreover, siRNA-mediated knockdown of EGR-3 in VEGF-treated endothelial cells significantly inhibits VEGFR1 mRNA expression (T.M. and W.C.A., unpublished observations, March 2009). In the current study, we have shown that the EGR binding motif is capable of binding both recombinant EGR-1 and EGR-3. Given that both EGR-1 and EGR-3 are inducibly expressed in endothelial cells, it seems likely that both isoforms contribute to the expression of VEGFR1.
We previously reported that in Hprt-targeted mice the wild-type VEGFR1 promoter directed expression in blood vessels of all organs examined except the liver.2 Expression was observed primarily in endothelium. However, reporter gene activity was also detectable in some vascular smooth muscle cells as well as in cardiomyocytes. In the current study, we targeted the Hprt locus of mice with VEGFR1 promoter-reporter gene constructs containing mutations of the −39 ETS, −52 ETS, CRE, or EGR binding sites. We also generated a new line of mice targeted with wild-type VEGFR1-lacZ so that all analyses could be carried out in sex-, age-, and generation-matched mice.
Consistent with the transient transfection results, mutations of the −52 ETS, CRE, and EGR binding sites reduced reporter gene activity in vivo. Moreover, a mutation of the −39 ETS motif increased LacZ expression in the intact endothelium. These changes paralleled those observed in transient transfections. However, the in vivo results provided insights that were otherwise unapparent in vitro. For example, mutation of CREB/ATF binding site had an equal if not greater inhibitory effect on expression compared with mutation of −52 ETS, whereas the opposite was true in vitro. Despite the readily detectable activity of the VEGFR1 promoter and most of its variants in several types of cultured endothelial cells, expression was conspicuously absent in the liver of Hprt-targeted mice. Interestingly, mutations of the various cis-regulatory motifs resulted in time-dependent and/or vascular bed–specific changes in reporter gene activity. For example, the −52 ETS or CRE mutations were expressed in embryonic but not normal adult endothelium. The EGR binding site mutation resulted in a clear reduction in the endothelium of the diaphragm, but not the lung. Finally, the −52 ETS or CRE mutations are reactivated during tumor angiogenesis, suggesting that other regulatory elements are capable of promoting VEGFR1 expression in this pathologic setting. Indeed, phylogenetic footprinting of the VEGFR1 promoter reveals additional consensus sites that are highly conserved across species (please see supplemental Figures 1-2) and that may ultimately prove to play an important role in mediating spatial and temporal expression of VEGFR1. Together, our findings suggest that endothelial cells from different vascular beds use distinct regulatory mechanisms to control expression of VEGFR1.
To our knowledge, the current study is the first to systematically explore the functional relevance of promoter elements in the context of single-copy–single-locus gene targeting. By controlling for copy number and integration site, we can be confident that any differences in the pattern and/or levels of expression are reflective of differences in the promoter. It is formally possible that such differences are an artifact of fortuitous interactions between the targeted promoter and Hprt locus control elements. Indeed, the gold standard approach would be to mutate the cis-regulatory elements in the endogenous VEGFR1 locus. This limitation notwithstanding, the current study offers powerful proof of principle for the in vivo relevance of 4 regulatory elements in the VEGFR1 promoter.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Takeda Science Foundation in Japan (T.M.) and National Institutes of Health grant HL076540 (W.C.A. and P.O.).
National Institutes of Health
Authorship
Contribution: E.J., J.L., J.-i.S., L.Y., Y.O., V.N.-K., K.Y., L.J., D.B., K.C.S., D.L., E.R., and S.-C.S. designed and performed experiments and analyzed the data; P.O. and T.M. designed experiments and analyzed the data; and W.C.A. designed and performed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William C. Aird, Beth Israel Deaconess Medical Center, Molecular and Vascular Medicine; RN-227, 330 Brookline Ave, Boston, MA 02215; e-mail: waird@bidmc.harvard.edu.
References
Author notes
*E.J. and J.L. contributed equally to the work.