Abstract
A genome-wide linkage scan has provided evidence for a chronic lymphocytic leukemia (CLL) susceptibility locus at 2q21 to which the chemokine receptor CXCR4 gene maps. Recent data provide some evidence for common variation in CXCR4 according to the polymorphic variant rs2228014 defining CLL risk. To examine the role of genetic variation in CXCR4 on CLL risk, we screened 188 familial CLL cases and 213 controls for germline mutations in the coding regions of CXCR4 and genotyped rs2228014 in 1058 CLL cases and 1807 controls. No association between rs2228014 and risk of CLL was seen (P = .83). One truncating (W195X) and 2 missense mutations with possible functional consequences (V139I and G335S) were identified among 186 familial cases and 0 in 213 controls sequenced. Our analysis provides no evidence that common variation in CXCR4 defined by rs228014 influences the risk of CLL, but that functional coding mutations in CXCR4 may contribute to familial CLL.
Introduction
Although the etiology of chronic lymphocytic leukemia (CLL) is largely unknown, the disease is characterized by a strong familial basis with an 8-fold increased risk observed in relatives of cases.1
Familial clustering of CLL and related B-cell lymphoproliferative disorders has provided the motivation for seeking to identify a susceptibility gene through genetic linkage. A recent linkage scan of 206 CLL families has provided evidence for a disease locus at 2q21, conferring a moderate-high risk of disease.2 The genetic basis of this linkage signal is, however, presently unknown. The gene encoding chemokine receptor 4 (CXCR4) maps to 2q21 and represents a potential candidate for the basis of the linkage signal. CXCR4 plays a key role in B lymphopoiesis by retaining immature B cells in the bone marrow. In addition, CXCR4 is important for trafficking and survival of CLL and other leukemia cells, through attachment to CXCL12-secreting stromal and nurse-like cells,3 and has extensively been studied as a coreceptor for HIV-1.4 Moreover, CXCR4 is up-regulated by interferon regulatory factor 4,5 and a recent genome-wide association study has shown that variation in interferon regulatory factor 4 influences the development of CLL.6 A recent association study of the single nucleotide polymorphism (SNP) rs2228014, which maps to exon 2 of CXCR4, provides some support for CXCR4 having a role in CLL risk, albeit marginally significant (P < .002, Padjusted = .09).7
To further examine the role of genetic variation in CXCR4 in CLL, we conducted a comprehensive analysis of the coding sequence and intron-exon boundaries in 188 familial CLL cases and 213 controls. In addition, we compared the genotype frequency of rs2228014 in 1058 cases and 1807 controls.
Methods
Mutational analysis of the coding and splice site regions of CXCR4 was conducted in 188 CLL patients (134 patients with family history of CLL only, 54 with family history of CLL and other B-cell lymphoproliferative disorders) ascertained through the International CLL Linkage Consortium. Mouthwash samples were available for 63 of the cases. The association study was based on 1058 CLL cases collected through International CLL Linkage Consortium and the Royal Marsden National Health Service Hospitals trust. The diagnosis of CLL was based on World Health Organization guidelines.8 Peripheral blood samples obtained from 1807 healthy persons, recruited through the National Study of Colorectal Cancer Genetics,9 were used as controls. Cases and controls were British residents and self-reported to be of European Ancestry. All biologic samples were obtained from patients and controls with informed consent, in accordance with the tenets of the Declaration of Helsinki, and approval from the Institute of Cancer Research ethical review board.
A search for mutations in the coding regions and splice sites of CXCR4 was performed by sequencing amplified polymerase chain reaction (PCR) fragments using BigDye Terminator chemistry implemented on an ABI 3730xl sequencer (Applied Biosystems). PCR primers were designed using Primer 3 software to facilitate the investigation of all intron-exon boundaries (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Sequence traces were aligned and compared with the gene consensus sequence using Mutation Surveyor (Version 3.0; SoftGenetics). Two in silico algorithms, PolyPhen and SIFT, were used to predict the putative impact of missense variants on protein function. Scores were classified as tolerated or deleterious according to proposed criteria.10-12
Genotyping of rs2228014 was conducted by competitive allele-specific PCR KASPar chemistry (KBiosciences Ltd). Primers (supplemental Table 1) were designed using PrimerPicker software (KBiosciences Ltd).
The relationship between categorical variables was determined by Fisher exact test. Deviation of the genotype frequencies in the controls from those expected under Hardy-Weinberg equilibrium was calculated using the χ2 test statistic. The risk of CLL associated with rs2228014 was calculated by deriving allelic, heterozygous, and homozygous odds ratios by unconditional logistic regression. All statistical manipulations were undertaken using STATA (Version 8.0; Stata Corporation).
Results and discussion
Complete sequence data of the coding and splice site regions of CXCR4 were generated for 186 of the 188 patient samples submitted for mutational analysis. Mouthwash DNA genotypes were completely concordant with blood DNA genotypes in 100% of cases (data not shown). Together with the fact that CXCR4 do not map to any of the regions of the genome commonly associated with copy number variation in CLL13 mitigates against bias from differential genotyping as a consequence of allelic imbalance influencing study findings.
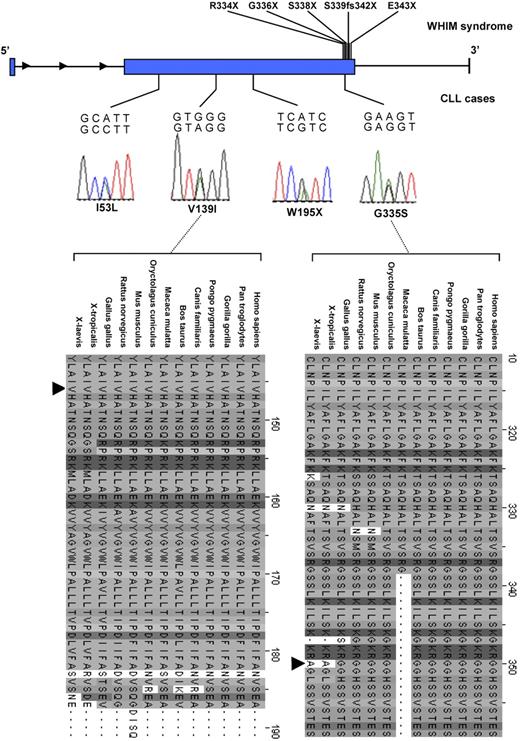
Mutational analysis of the coding regions of CXCR4 in the 186 CLL cases identified 8 sequence variants (Table 1). Two of these, rs2228014 (414C>T) and rs56400844 (I53L), have previously been documented by dbSNP as polymorphisms. The frequency of rs2228014 was identical in the 213 controls screened. V139I, W195X, and G335S were identified in single persons (Figure 1). As the 2 nonsynonymous mutations V139I and G335S effect sequence change in the expressed protein at amino acid positions, which are evolutionary highly conserved, it favors these being functional (Figure 1). The in silico algorithm SIFT, but not PolyPhen, is consistent with these mutations being deleterious. No missense mutations were identified among any of the 213 controls analyzed.
Sequence variants identified in CXCR4
| Nucleotide change* . | dbSNP MAF . | Protein change . | Frequency (n) . |
|---|---|---|---|
| Cases (n = 188) | |||
| rs56400844:157 A>C | No data† | I53L | 0.005 (1) |
| rs2228014: 414 C>T | 0.043 | Synonymous | 0.037 (14) |
| 415 G>A | — | V139I | 0.005 (1) |
| 584 A>G | — | W195X | 0.005 (1) |
| 1003 A>G | — | G335S | 0.005 (1) |
| 1295 C>G | — | 3′ UTR | 0.005 (1) |
| 1350 G>A | — | 3′ UTR | 0.005 (1) |
| 1430 C>T | — | 3′ UTR | 0.005 (1) |
| Controls (n = 213) | |||
| 153 T>A | — | Synonymous | 0.004 (1) |
| 294 C>T | — | Synonymous | 0.004 (1) |
| rs2228014: 414 C>T | 0.043 | Synonymous | 0.08 (19) |
| 783 C>T | — | Synonymous | 0.004 (1) |
| 1336 G>A | — | 3′ UTR | 0.009(2) |
| Nucleotide change* . | dbSNP MAF . | Protein change . | Frequency (n) . |
|---|---|---|---|
| Cases (n = 188) | |||
| rs56400844:157 A>C | No data† | I53L | 0.005 (1) |
| rs2228014: 414 C>T | 0.043 | Synonymous | 0.037 (14) |
| 415 G>A | — | V139I | 0.005 (1) |
| 584 A>G | — | W195X | 0.005 (1) |
| 1003 A>G | — | G335S | 0.005 (1) |
| 1295 C>G | — | 3′ UTR | 0.005 (1) |
| 1350 G>A | — | 3′ UTR | 0.005 (1) |
| 1430 C>T | — | 3′ UTR | 0.005 (1) |
| Controls (n = 213) | |||
| 153 T>A | — | Synonymous | 0.004 (1) |
| 294 C>T | — | Synonymous | 0.004 (1) |
| rs2228014: 414 C>T | 0.043 | Synonymous | 0.08 (19) |
| 783 C>T | — | Synonymous | 0.004 (1) |
| 1336 G>A | — | 3′ UTR | 0.009(2) |
MAF indicates minor allele frequency of SNP in whites (as reported in dbSNP); —, not applicable; and UTR, untranslated region.
Nucleotide position is taken from the first base of the start codon; dbSNP accession number is shown where available.
No frequency data available for whites.
Nonsynonymous mutations in CXCR4 in CLL and WHIM syndrome. Nonsynonymous mutations identified in CLL (chronic lymphocytic leukemia) patients are indicated at their relative positions as sequence traces with corresponding amino acid substitutions. Sites of truncating mutations associated with WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome are also shown.15,18 Cross-species sequence conservation is shown for the SNPs postulated to be functionally deleterious.
Nonsynonymous mutations in CXCR4 in CLL and WHIM syndrome. Nonsynonymous mutations identified in CLL (chronic lymphocytic leukemia) patients are indicated at their relative positions as sequence traces with corresponding amino acid substitutions. Sites of truncating mutations associated with WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome are also shown.15,18 Cross-species sequence conservation is shown for the SNPs postulated to be functionally deleterious.
Three 3′ untranslated region variants were identified in single cases and one in the controls. The truncating mutation was identified in a female patient diagnosed with CLL at age 79; family history included a sister diagnosed with non-Hodgkin lymphoma and nephew with CLL. This mutation is predicted to lead to loss of the terminal 157 amino acids of the expressed protein, a 7 transmembrane-spanning protein. Truncating mutations in the C-terminus cytoplasmic tail of CXCR4 cause the WHIM syndrome (warts, hypogammaglobulinemia, infections, and myelokathexis), characterized by B-cell dysfunction.14,16 Although we have no documentary evidence of WHIM in the patient harboring W195X, WHIM is a clinically and genetically heterogeneous disorder and hypogammaglobulinemia and verrucosis can be absent, making diagnosis problematic.17 Moreover, WHIM leukocytes can express wild-type CXCR4 receptor, suggesting involvement of additional genetic factors in phenotype development.18 All CXCR4 mutations causing WHIM documented to date result in loss of the terminal 9 to 18 amino acids of CXCR4. This is thought to cause gain of function and increased responsiveness to CXCL12.15 In contrast, the consequence of W195X is probably loss of function, which is more probable as a basis for a role in CLL development. Loss of CXRC4 expression has been shown to lead to more progressive CLL.19 Moreover, inactivation of CXCR4 in hematopoietic stem cells has been documented to cause excessive hematopoietic stem cell proliferation.20
rs2228014 genotypes were obtained for 1041 cases (98.5%) and 1763 (97.6%) of the controls. Genotypes obtained by KASPar genotyping were 100% concordant with those obtained by sequencing. No association was observed between rs2228014 and risk of developing CLL (supplemental Table 2). There are differences in minor allele frequencies observed in the Spanish and United Kingdom populations, 0.14 and 0.04, respectively. Hence, failure to replicate the findings of Enjuanes et al7 could be the result of differences in allelic architecture of the 2 populations.
Our analysis provides no support for the assertion that common variance in CXCR4 as defined by the SNP rs228014 influences the risk of CLL. In contrast, albeit not statistically significant, our findings provide evidence of an overrepresentation of functional coding mutations in CXCR4 (in keeping with the rare variant hypothesis, it may afford a more profound risk of CLL), and these may be a rare cause of the disease, possibly accounting for approximately 1.6% of familial disease (95% confidence interval, 0.3%-4.6%).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all patients, their clinicians, and other persons for their participation in this study.
This work was supported by Leukemia Research and the Arbib Foundation. D.C.-S. received a PhD studentship from the Institute of Cancer Research.
Authorship
Contribution: D.C.-S. designed and performed research, analyzed data, and wrote the paper; M.Q. performed research; M.J.S.D., E.M., and C.D. performed sample acquisition; D.C. obtained funding; R.S.H. designed research, obtained funding, and wrote the paper; and all authors contributed to the final paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, Section of Cancer Genetics, Institute of Cancer Research, 15 Cotswold Rd, Sutton, United Kingdom; e-mail: richard.houlston@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal