Abstract
Cyclic adenosine monophosphate response element binding (CREB)–binding protein (CBP) and p300 are multidomain transcriptional coactivators that help assemble large regulatory complexes at sites of active transcription. Nullizygosity of CBP or p300 results in pervasive defects in hematopoiesis. To systematically assess the structural domains of p300 required for normal hematopoiesis, we used recombinase-mediated cassette exchange to create an allelic series of coisogenic embryonic stem cells, each expressing a different mutant of p300 from the endogenous locus. We found that deletion of either the KIX or CH1 domain caused profound and pervasive defects in hematopoiesis, whereas the loss of most other domains had only lineage-restricted effects. When expressed from the p300 locus, an extra copy of CBP largely compensated for a lack of p300. Surprisingly, mutation of the p300 histone acetyltransferase (HAT) domain had minimal effects on hematopoiesis, and actually increased progenitor and stem cell numbers and proliferative potential. Our results suggest that, in distinct contrast to other organ systems, HAT activity does not provide a critical function for hematopoietic development and emphasizes the importance of enzyme-independent functions of p300.
Introduction
The paralogous proteins p300 and cyclic adenosine monophosphate response element binding (CREB)–binding protein (CBP) regulate transcriptional programs for a wide variety of developmental and physiologic processes. They function in part through endogenous histone acetyltransferase (HAT) activity, and in part through association with transcription factors, chromatin remodeling complexes such as SWI/SNF, pCAF, and p160 proteins, as well as general transcriptional machinery including TBP, TFIID, and RNA polymerase II.1 Eight distinct functional domains (N-terminal, CH1, KIX, Bromo, CH2, HAT, CH3, glutamine-rich)2,3 within p300 and CBP mediate interactions with hundreds of proteins and allow p300 and CBP to bridge together disparate transcription factors on the same promoter and serve as scaffolds to nucleate large regulatory complexes that activate transcription.2
Transcriptional programs that regulate cell fate decisions are integral to the establishment and maintenance of most organ systems, including the hematopoietic system.4 Clinical data suggest that the functions of p300 and CBP can be perturbed in disorders of the blood: leukemia-inducing proteins such as mutant Tal1 and translocation products such as NUP98-HoxA9 and MOZ-TIF2 recruit p300 and CBP to aberrantly activate transcription of proliferation-associated genes,5-7 whereas other translocations directly target CBP and p300 (eg, MLL-CBP, MOZ-CBP, MOZ-p300).8,9 Alternatively, AML1-ETO, the product of the t(8;21) translocation product, has been found to block p300/CBP activity and down-regulate transcription of differentiation-inducing genes.10 In addition, patients with Rubinstein-Taybi syndrome, a genetic disorder characterized by monoallelic loss of CBP or less commonly p300, have an increased incidence of cancers, including those of hematologic origin.11
Studies in mice have begun to shed light on the various ways in which p300 and CBP regulate normal and malignant hematopoiesis in vivo. Whereas nullizygosity of either gene leads to embryonic lethality,12,13 studies with heterozygous and chimeric mice demonstrated requirements for CBP and p300 in suppression of hematologic tumors, and normal stem cell self-renewal and differentiation.14,15 Analyses of lineage-specific knockout mice indicated clear roles for p300 and CBP in B- and T-cell development and in the prevention of T-cell lymphomas.16-18 Although point mutation and domain swapping studies have begun to elucidate the roles of specific p300 and CBP functional domains in hematopoiesis,19-22 except for the KIX domain, little is known of the role of specific p300 domains in vivo.19-22 In addition, the ability of an extra copy of CBP to replace p300 function (or vice versa) when under control of p300 locus regulatory elements is unknown, leaving the issue of functional redundancy between these 2 homologous proteins unresolved.
In this study, we used recombinase-mediated cassette exchange (RMCE)23 to circumvent the inefficiency of conventional gene targeting methods to rapidly generate an allelic series in which mutants of p300 were expressed from the endogenous p300 locus. We used this reverse genetic approach to systematically assess the biologic role of each p300 functional domain in hematopoiesis, and to determine the ability of CBP to compensate for a lack of p300.
Methods
Plasmids
Full-length mouse p300 cDNA (exons 1-31) was cloned from CBP−/− embryonic stem cells (ESCs)14 using reverse-transcription–polymerase chain reaction (RT-PCR), inserted into the Gateway system pDEST6.2-V5 vector (Invitrogen) and verified by sequencing. Deletions in p300 cDNA were created by standard restriction enzyme digestion and ligation methods. HAT domain point mutations were created with the Quick Change in vitro mutagenesis kit (Stratagene). pBS2 served as the backbone for both the master LoxP/Lox511 hygromycin targeting vector and the p300 donor vectors.
Creation of ESC lines
The master “recipient” ESC line was created by linearizing pKO-p300 with BspDI and electroporating it into previously described C57/BL6 p300−/− ESCs.12 Hygromycin (250 μg/mL) was used for selection and isolation of homologous recombination ESC clones. For all RMCE events, an expression vector for CRE recombinase and a p300 donor vector were electroporated into the master recipient ESC line. Selection was performed with 0.75 μg/mL puromycin. Nested PCR of genomic DNA from individual ESC clones determined proper recombination. Primers are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
ESC culture and immunoblotting
ESCs were grown in Dulbecco modified Eagle medium (Sigma) plus 20% ESC-grade fetal calf serum, penicillin/streptomycin, nonessential amino acids, l-glutamine, sodium pyruvate (all from Gibco), and leukemia inhibitory factor (Chemicon) in a 37°C, 10% CO2 incubator. Whole-cell lysates were made in phosphate-buffered saline plus 0.5% Triton X-100 containing protease inhibitors (Complete; Roche) and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting with anti-V5 (Invitrogen) and anti-p300 (clone RW128 [Millipore]; clone N15 [Santa Cruz Biotechnology]) antibodies. Histone extracts were made according to standard procedures and immunoblotting was performed with a pan-H3 (Abcam), anti-H3K56ac (Epitomics), anti-H3K4me3 (Abcam), or anti-H3K14ac (Millipore) antibody. Quantification of immunoblot bands was performed using the Photoshop histogram function (Adobe Systems). Absolute band intensity was calculated by multiplying mean by pixel values and relative band intensity was determined by normalizing absolute intensities to glyceraldehyde-3-phosphate dehydrogenase.
HAT assay
The 293T cells, grown in Dulbecco modified Eagle medium plus 10% fetal calf serum, were transiently transfected with the indicated pDEST6.2-p300-V5 vectors using Fugene 6 (Roche), according to the manufacturer's instructions. Forty-eight hours later, cells were lysed in 20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 150mM NaCl, 10mM ethyleneglycoltetraacetic acid, 40mM glycerophosphate, 1% NP40, 2.5mM MgCl2, 2mM sodium orthovanadate, 1mM dithiothreitol, with protease inhibitors, and immunoprecipitated with an anti-V5 antibody. Immunoprecipitates on beads were washed 6 times in NETN B (200mM NaCl, 1μM ethylenediaminetetraacetic acid, 20mM tris(hydroxymethyl)aminomethane [pH 8.0], 0.5% NP40, 10 μM ZnSO4) and once in 2× HAT buffer (100mM tris(hydroxymethyl)aminomethane-HCl, 20% glycerol, 2mM dithiothreitol, 2mM phenylmethylsulphonyl fluoride, 0.2mM ethylenediaminetetraacetic acid, 20mM butyric acid) before incubating in 1× HAT buffer plus 8 μg of acetyl coenzyme A (Roche) at 37°C for 1 hour. Acetylation of p300 itself or exogenous histone H4 protein (8 μg; data not shown) was examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting with an anti-acetyl lysine (Upstate Biotechnology) or anti-V5 antibody.
Mice
Chimeric mice were generated through the Dana-Farber Cancer Institute Transgenic Core Laboratory using standard methods.24 Briefly, C57/BL6 (Ly5.2+) ESCs were injected into FVB (Ly5.1+) blastocysts and implanted into pseudopregnant female mice. Two to 3 independent clones of each ESC line were used and resulted in a total of 2 to 4 litters and 5 to 16 chimeric mice for each ESC line. Coat color chimerism was visually assessed at 8 to 10 weeks of age by 4 independent observers. Hematopoietic chimerism was determined by allelic differences at the Ly5 locus (ie, Ly5.1 vs Ly5.2), using flow cytometry. Simple linear regression and best-fit line graphs were created with Microsoft Excel. Complete blood counts from peripheral blood (PB) were performed with a Beckman Coulter AcT 10 Coulter Counter and total white blood cell counts are shown in supplemental Figure 5. The Dana-Farber Cancer Institute Animal Care and Use Committee approved all animal protocols.
Flow cytometry and cell sorting
PB from 20-week-old mice was collected via a tail vein nick into heparinized capillary tubes (Fisher). Bone marrow (BM) was flushed from the femurs and tibias of chimeric mice, killed at age 24 to 30 weeks. Red blood cells were lysed with red blood cell lysis buffer (QIAGEN) and the remaining cells were stained with antibodies listed in supplemental Table 2. Flow cytometry was performed on a BD FACSAria, BD LSRII, or BD FACScan (PB only). Cell sorting was performed on a BD FACSAria or a Dako MoFlo high-speed sorter. Data analysis was performed with FlowJo (TreeStar) or BD CellQuest Pro software.
Bone marrow transplants
C57BL6 SJL PtPrcA Pep3b/Boy J (Ly5.1+) mice (The Jackson Laboratory), aged 8 to 10 weeks, were used as recipients and lethally irradiated with a split dose of 12.3 Gy (6.15 Gy each, separated by 5 hours) using a Cs137 source Gammacell-40 Exactor irradiator (Best Theratronics). Five hours after the second dose, a bolus of 3 × 105 donor bone marrow cells, from mice aged 10 to 30 weeks, was given via tail vein injection.
Stroma and LTC assays
Stroma cell layers, derived from chimeric mice, were established according to the StemCell Technologies protocol, “Isolation of Mesenchymal Progenitors from Mouse Compact Bone.”25 Immunofluorescence was performed according to standard procedures with an anti-p300 or anti-V5 tag primary antibody, fluorescein isothiocyanate–conjugated secondary antibody, and (4,6 diamidino-2-phenylindole) nuclear stain. Long-term culture (LTC) assays were performed according to a StemCell Technologies protocol in the Murine Long-Term Culture Initiating Cell Assay Manual.26
Serial replating assay
Assays were performed as previously described,8 with minor modifications. c-Kit+, Lineage−, Sca1+ (KLS) bone marrow cells were sorted from either wild-type or chimeric HATptmut mice and plated into duplicate 1-mL M3434 methylcellulose cultures for weekly colony and total cell number counting. Each subsequent week, 3000 cells were replated in duplicate cultures.
Teratoma formation
Ten NCr Nu/Nu mice (Charles River), aged 8 to 10 weeks, were used as recipients. ESCs (3 × 106) were injected subcutaneously into the flank of each recipient. Tumor measurements were taken every 3 to 4 days and harvested when their volume approached 800 mm2. Formalin-fixed samples were embedded in paraffin, sectioned, and mounted on slides. Hematoxylin/eosin staining and analysis were performed by the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core Facility. Standard immunohistochemistry was performed by the Dana-Farber/Harvard Cancer Center Specialized Histopathology Core Facility.
In vitro differentiation assays
Gelatin-adapted ESCs were either harvested for RNA isolation (day 0) using Trizol (Invitrogen) or replated in Iscove modified Dulbecco medium; 15% fetal bovine serum; penicillin/streptomycin (100 units, 0.1 mg/mL), 2 mM l-glutamine, 1× nonessential amino acids, 1mM sodium pyruvate (Gibco); 435nM monothioglycerol; 0.2 mg/mL iron-saturated transferrin, and 0.025 mg/mL ascorbic acid (Sigma) for embryoid body (EB) formation. EBs were established either in low-adherence 6-well plates (Costar; 300 000 cells/mL) or in 25-μL hanging drops (16 000 cells/mL) for 2 days before transferring to plates. EB RNA was isolated on days 2, 5, and 8. For hematopoietic differentiation, day-2 EBs were disaggregated and plated in M3434 methylcellulose (StemCell Technologies) for an additional 6 days and then harvested for RNA isolation. For neuronal differentiation, day-4 EBs were exposed to 5μM all-trans retinoic acid for 96 hours, disaggregated with 0.25% trypsin, and plated in neurobasal media (Invitrogen) containing N2 and B27 supplements (Invitrogen) on poly-d-lysine/laminin–coated plates (BD Falcon) for an additional 13 days and then harvested for RNA isolation. First-strand cDNA synthesis was performed with Superscript RT III (Invitrogen) and Quantitect Sybr green master mix (QIAGEN) was used for real-time PCR on a Stratagene Mx3000P machine. Primer sequences are listed in supplemental Table 4.
Statistical analysis
All of the indicated statistical tests were performed using Prism (GraphPad Software).
Results
Creation of a p300 allelic series in ESCs
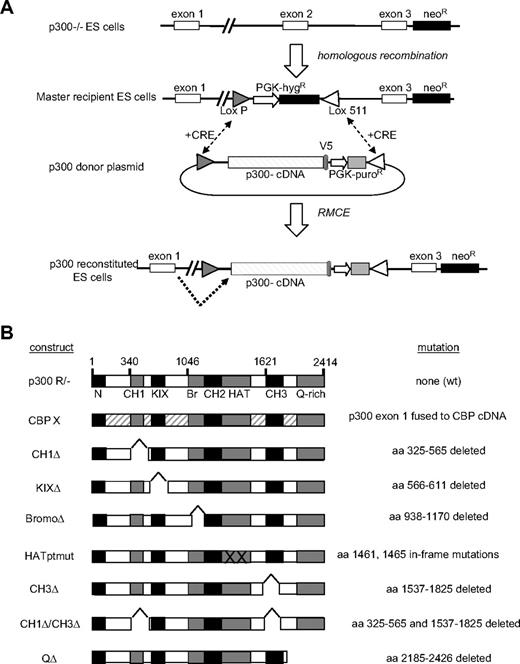
We created an allelic series for p300 using a hybrid approach combining RMCE27 with exon trapping.28 As a first step, p300 nullizygous ESCs12 were used to create a “master recipient” ESC line in which exon 2 of the p300 locus was replaced with a hygromycin-resistance (HygR) cassette flanked by heterologous (LoxP and Lox511)29 Lox sites (Figure 1A). To reconstitute expression of p300 and mutant alleles, CRE-mediated recombination was used to replace the HygR cassette with a donor plasmid cassette (ie, cassette exchange), resulting in targeted insertion of a p300 cDNA (exons 2-31) in place of exon 2 within the p300 locus (Figure 1A). Splicing information, encoded in the genomic sequence, was preserved during cassette exchange (supplemental Figure 1A), thus allowing the endogenous p300 promoter to drive expression of wild-type (wt) or mutant mRNAs by splicing of the native exon 1 to the cDNA encoding exons 2 to 31 (Figure 1A).
Creation of p300-reconstituted ESCs. (A) p300 locus is shown in parental p300−/−, master recipient, and reconstituted embryonic stem cells (ESCs). A neomycin cassette spanning exons 4 to 6 renders p300−/− and master recipient ESC lines nullizygous for p300. A loxP/lox511 flanked targeting cassette containing a hygromycin resistance (hygR) gene replaces exon 2 and provides exchange site for CRE-mediated recombination with p300 donor plasmids, resulting in the production of p300-reconstituted ESCs. Dotted arrow indicates splicing occurs from endogenous exon 1 into the p300 cDNA cassette. (B) Panel of p300 cDNA constructs used to create reconstituted ESC lines. The specific amino acid (aa) deletion, mutation, or CBP replacement design is indicated for each construct.
Creation of p300-reconstituted ESCs. (A) p300 locus is shown in parental p300−/−, master recipient, and reconstituted embryonic stem cells (ESCs). A neomycin cassette spanning exons 4 to 6 renders p300−/− and master recipient ESC lines nullizygous for p300. A loxP/lox511 flanked targeting cassette containing a hygromycin resistance (hygR) gene replaces exon 2 and provides exchange site for CRE-mediated recombination with p300 donor plasmids, resulting in the production of p300-reconstituted ESCs. Dotted arrow indicates splicing occurs from endogenous exon 1 into the p300 cDNA cassette. (B) Panel of p300 cDNA constructs used to create reconstituted ESC lines. The specific amino acid (aa) deletion, mutation, or CBP replacement design is indicated for each construct.
Two separate nested PCR reactions were used to confirm the loss of the HygR cassette and the insertion of the cDNA cassette at the p300 locus in individual ESC clones (supplemental Figure 1B). RMCE proved highly efficient, with overall 37% of isolated clones scoring positive for proper cassette integration (supplemental Figure 1C). In this manner, in addition to the nullizygous master recipient clone, 9 different “reconstituted” ESC lines were created, expressing either wt p300 (p300R/−), wt CBP (CBPX), or a specific mutant p300 protein (CH1Δ, KIXΔ, BromoΔ, HATptmut, CH3Δ, double CH1Δ/CH3Δ, or QΔ) from the endogenous p300 promoter (Figure 1B).
Immunoblotting was performed to verify expression of all V5-tagged mutant p300 proteins (supplemental Figure 2A-B) as well as HA-tagged CBP (supplemental Figure 2C). Although the level of p300 expressed in the reconstituted ESCs appeared to be slightly less than that of conventional p300+/− heterozygous ESCs (supplemental Figure 2D), substantial levels of V5-tagged p300 expression could be detected in bone marrow sorted from chimeric mice verifying in vivo expression in ESC-derived hematopoietic cells (supplemental Figure 2E). A HAT assay confirmed that the HATptmut p300 allele, using previously described point mutations,19,20 resulted in a loss of intrinsic acetyltransferase activity (supplemental Figure 3).
Analysis of hematopoietic system in p300-reconstituted chimeric mice
To circumvent the embryonic lethality resulting from nullizygosity of p300,14 we analyzed the hematopoietic contribution of p300-reconstituted cells in chimeric mice, similar to prior studies of Gata-2, SCL/Tal1, and Aml1.30-32 Ten ESC lines, comprising the entire p300 allelic series (Figure 1B), were injected into wt blastocysts, and hematopoiesis was analyzed in resulting chimeric mice. We used multicolor flow cytometry to examine 3 primitive and 3 mature hematopoietic populations for each chimeric line. Early progenitors were identified in bone marrow and included the c-Kit+ Lin−, Sca1+ (KLS) fraction, which is enriched for hematopoietic stem cells (HSCs),4 common myeloid progenitors (CMPs), defined immunophenotypically as c-Kit+, lineage−, Sca1−, CD34+, Fc gamma RII/IIIlo cells,33 and common lymphoid progenitors (CLPs) characterized as lineage− IL7Rα+, c-Kitlo, Sca1lo.34 More mature cell types were represented by myeloid (Gr1+) cells, B cells (B220+), and T cells (Ly1+) isolated from peripheral blood (PB).
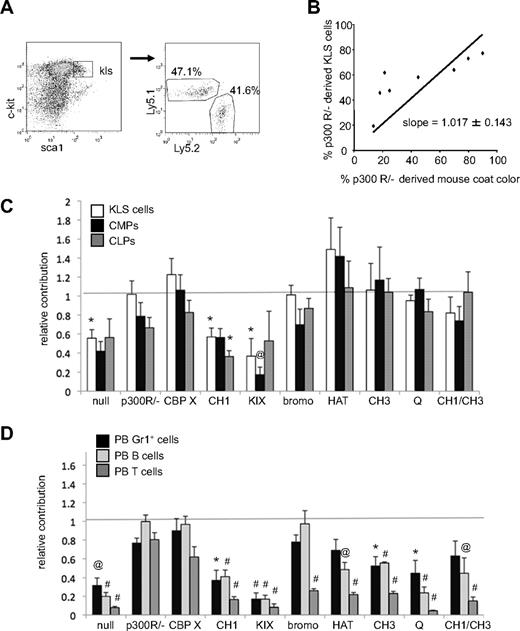
The percentage of each lineage derived from mutant p300 ESCs was determined by a congenic difference at the Ly5 locus. Specifically, wt blastocyst-derived cells expressed Ly5.1, whereas ESC-derived cells expressed Ly5.2 (Figure 2A and supplemental Figure 4). Previous studies have shown that for chimeric mice created from wt ESCs, hematopoietic contribution is equivalent to the level of coat color chimerism.14 Therefore, for each mutant ESC line, the percentage of Ly5.2+ cells was plotted against the percentage of coat color chimerism for linear regression analysis (shown for KLS cells in Figure 2B). In all cases analyzed, mice with no coat color chimerism were devoid of Ly5.2+ hematopoietic cells. A slope of 1 for the best-fit line through the origin was found for p300R/− ESC-derived KLS cells (Figure 2B), indicating that reconstitution of p300 expression using RMCE fully rescued hematopoiesis in vivo, resulting in equivalent contribution to both blood and coat color chimerism. Best-fit lines were calculated for each set of chimeric mice (supplemental Table 3) and a summary of their slopes plus or minus SE is shown as relative hematopoietic contribution in Figure 2C and D.
Contribution of p300-reconstituted cells to hematopoietic cell populations in chimeric mice. (A) Typical flow cytometry gating of lineage− cells for c-Kit and Sca1 expression identifies c-Kit+, Lineage−, Sca1+ (KLS) compartment. Gating of KLS cells determines the percentage that is Ly5.1+ (wt blastocyst-derived) versus Ly5.2+ (mutant p300 ESC-derived). (B) The contribution of p300-reconstituted (p300 R/−) ESC-derived cells to mouse coat color is compared with their contribution to the KLS compartment. Each dot represents 1 mouse. Simple linear regression with a zero y-intercept creates a best-fit line of the data (slope = 1.017 ± 0.143 SE). (C) Relative contribution of Ly5.2+ cells to KLS, CMP, and CLP compartments. Bars indicate the slope of each best fit line graph ± SE as depicted in panel B. (D) Relative contribution of Ly5.2+ cells to PB Gr1+ cells, B220+ (B cells), and Ly1+ (T cells) populations. Bars indicate the slope of each best fit line graph ± SE. Dashed line at 1 represents the expected level of hematopoietic contribution if no deficiencies exist (ie, when hematopoietic contribution is equivalent to coat color contribution). Unpaired t tests were used to compare hematopoietic contribution for each genotype with that of wt p300R/−. Samples that differed significantly from p300R/− populations are indicated with *P < .05; @P < .01, #P < .001. Number of mice used in each analysis is shown in supplemental Table 3.
Contribution of p300-reconstituted cells to hematopoietic cell populations in chimeric mice. (A) Typical flow cytometry gating of lineage− cells for c-Kit and Sca1 expression identifies c-Kit+, Lineage−, Sca1+ (KLS) compartment. Gating of KLS cells determines the percentage that is Ly5.1+ (wt blastocyst-derived) versus Ly5.2+ (mutant p300 ESC-derived). (B) The contribution of p300-reconstituted (p300 R/−) ESC-derived cells to mouse coat color is compared with their contribution to the KLS compartment. Each dot represents 1 mouse. Simple linear regression with a zero y-intercept creates a best-fit line of the data (slope = 1.017 ± 0.143 SE). (C) Relative contribution of Ly5.2+ cells to KLS, CMP, and CLP compartments. Bars indicate the slope of each best fit line graph ± SE as depicted in panel B. (D) Relative contribution of Ly5.2+ cells to PB Gr1+ cells, B220+ (B cells), and Ly1+ (T cells) populations. Bars indicate the slope of each best fit line graph ± SE. Dashed line at 1 represents the expected level of hematopoietic contribution if no deficiencies exist (ie, when hematopoietic contribution is equivalent to coat color contribution). Unpaired t tests were used to compare hematopoietic contribution for each genotype with that of wt p300R/−. Samples that differed significantly from p300R/− populations are indicated with *P < .05; @P < .01, #P < .001. Number of mice used in each analysis is shown in supplemental Table 3.
As shown in Figure 2C, p300-null cells were present in the KLS, CMP, and CLP compartments at 40% to 55% of expected levels. In contrast, both p300R/− and CBPX cells were represented in these early compartments at close to expected levels, demonstrating that RMCE reconstitution of p300 or ectopic CBP expression rescued hematopoiesis to levels observed in wt cells. CH1Δ and KIXΔ cells could not rescue the primitive cell defect and, like p300-null cells, had only limited contributions to KLS, CMP, and CLP populations. All other mutant p300 alleles rescued the p300-null defect, with HATptmut cells surprisingly contributing somewhat better than expected to KLS and CMP populations (Figure 2C).
Mature hematopoietic cell types were more drastically perturbed by the loss of p300 than primitive cell populations (Figure 2D), consistent with our prior studies demonstrating a critical requirement for p300 in hematopoietic differentiation.14 Similar to that for primitive populations (Figure 2C), reconstitution of wt p300 or wt CBP expressed from the p300 locus reversed these defects, whereas expression of CH1Δ and KIXΔ mutants did not. However, in contrast to the primitive cell populations, the production of mature myeloid and lymphoid cells could not be completely restored in any of the other p300 alleles, with the exception of B-cell recovery in the bromoΔ mutants. Collectively, these results demonstrate that expression of full-length p300 or CBP from the native p300 locus restored hematopoiesis in vivo, whereas specific deletion of either the CH1 or KIX domain was phenotypically similar to nullizygosity of p300. Given the central role of the HAT domain to p300 function, it was surprising that HAT activity was not required to establish or maintain hematopoietic progenitors (Figure 2C).
Competitive BM repopulation
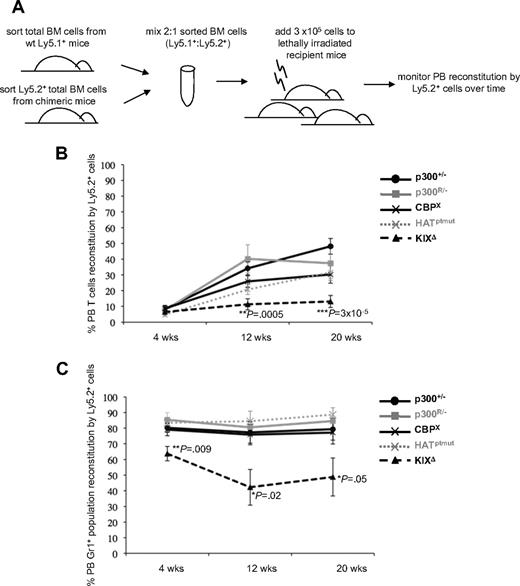
To further define whether p300 HAT activity was required for adult hematopoiesis, we next performed competitive BM repopulation assays (Figure 3A) to determine how well HAT-deficient p300 cells could repopulate the hematopoietic system in comparison with conventional p300+/− cells and other RMCE-reconstituted cells. Based on the chimeric mouse studies, we used p300R/− and CBPX cells as RMCE populations that are competent for hematopoiesis and KIXΔ cells as a RMCE population with pervasive defects in hematopoiesis. After transplantation, control cells (p300+/−, p300R/−, CBPX) were all able to reconstitute PB T cells (Figure 3B) and the myeloid (Gr1+) compartment (Figure 3C) to similar levels. As expected, KIXΔ BM cells showed a significantly reduced capacity to repopulate both lymphoid (Figure 3B) and myeloid (Figure 3C) compartments. HATptmut cells were able to contribute to both lymphoid (Figure 3B) and myeloid (Figure 3C) compartments at levels comparable with those observed for p300+/−, p300R/−, and CBPX controls. These results demonstrate that p300 HAT activity is dispensable for both establishment (Figure 2) and ablative reconstitution (Figure 3) of adult hematopoiesis.
Competitive bone marrow (BM) repopulation assays show KIXΔ cells have reduced reconstitution capacity. (A) Schematic of experimental design. (B) Repopulation of recipient PB T cells (Ly1+) by the indicated Ly5.2+ donor cells is shown at 4, 12, and 20 weeks after transplant. (C) Repopulation of PB myeloid cells (Gr1+) by the indicated Ly5.2+ donor cells is shown at 4, 12, and 20 weeks after transplant. Each line in graphs shows the average % of PB reconstitution for 7 to 11 recipient mice ± SD. Two to 4 sets of donors were used for transplantation, with each recipient receiving BM from 1 Ly5.1+ and 1 Ly5.2+ mouse. P values were calculated by Student t test using pairwise comparisons to the p300+/− values.
Competitive bone marrow (BM) repopulation assays show KIXΔ cells have reduced reconstitution capacity. (A) Schematic of experimental design. (B) Repopulation of recipient PB T cells (Ly1+) by the indicated Ly5.2+ donor cells is shown at 4, 12, and 20 weeks after transplant. (C) Repopulation of PB myeloid cells (Gr1+) by the indicated Ly5.2+ donor cells is shown at 4, 12, and 20 weeks after transplant. Each line in graphs shows the average % of PB reconstitution for 7 to 11 recipient mice ± SD. Two to 4 sets of donors were used for transplantation, with each recipient receiving BM from 1 Ly5.1+ and 1 Ly5.2+ mouse. P values were calculated by Student t test using pairwise comparisons to the p300+/− values.
Microenvironment and stem cell function
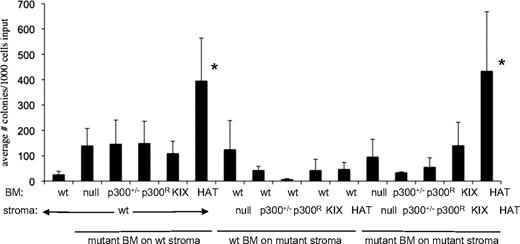
Hematopoiesis depends not only on hematopoietic cells, but also on interaction of those cells with the bone marrow microenvironment. For example, loss of the retinoblastoma protein results in abnormal hematopoiesis due to perturbations of the interaction of HSCs with the BM microenvironment.35 To determine whether p300-reconstituted bone marrow stromal cells were modulating hematopoiesis in chimeric mice (Figure 2) and mice that received a transplant (Figure 3), we used long-term culture (LTC) assays to evaluate the ability of wt and p300 mutant stroma to support lineage− Sca1+ cells ex vivo (supplemental Figure 6A). We derived 6 different primary stromal cell cultures (wt, p300+/−, p300R/−, HATptmut, KIX Δ, p300 null) from chimeric mice and used immunofluorescence to confirm the expression of p300 (wt or V5-tagged mutants) protein in each stroma type (supplemental Figure 6B). All stromal layers were verified to have a normal number of chromosomes (data not shown). We then tested the ability of each stromal layer to support lineage−, Sca1+ BM cells of wt and mutant origin. Clonogenic output from the LTC assays was significantly increased only in cultures of HATptmut BM, however, this occurred regardless of the genotype of the supporting stroma (Figure 4). These studies suggest that p300 mutant bone marrow stroma cells do not contribute to the hematopoietic deficits seen in chimeric mice (Figure 2). Moreover, these results further suggest that loss of HAT activity is not detrimental to hematopoietic function, and may in fact enhance growth in vivo (Figure 2B) and ex vivo (Figure 4).
Long-term culture (LTC) assay shows elevated colony production for HATptmut cells. Lineage−, Sca1+ wt or mutant p300 BM cells were sorted and plated on top of irradiated stromal layers with the indicated genotypes. Bars indicate the average number of colonies produced per 1000 input cells ± SD; n = 3 to 6 independent wells for each bar, using BM from 3 to 6 mice, and 2 independently established stroma layers per genotype. *P < .05 by 1-way ANOVA of all possible pairwise comparisons (Tukey-Kramer test) was used to determine significance.
Long-term culture (LTC) assay shows elevated colony production for HATptmut cells. Lineage−, Sca1+ wt or mutant p300 BM cells were sorted and plated on top of irradiated stromal layers with the indicated genotypes. Bars indicate the average number of colonies produced per 1000 input cells ± SD; n = 3 to 6 independent wells for each bar, using BM from 3 to 6 mice, and 2 independently established stroma layers per genotype. *P < .05 by 1-way ANOVA of all possible pairwise comparisons (Tukey-Kramer test) was used to determine significance.
Proliferative potential of KLS cells
Results of the LTC assay (Figure 4) suggest that mutation of the HAT domain affects early hematopoietic progenitor cell behavior in a cell-autonomous manner, and surprisingly may enhance proliferation. To specifically assess the effects of HAT mutation on KLS cell proliferation, we performed an in vitro serial replating assay with purified KLS cells.8 After 1 week in culture, the number of colonies produced by HATptmut KLS cells was equivalent to that of wt KLS cells (Figure 5A first set of bars), indicating that the initial number of colony-forming cells (CFCs) was the same. Likewise, the total number of cells produced during the first week was equivalent for HATptmut and wt cells (data not shown), indicating that that the population doubling time of HATptmut cells was not significantly shorter than in wt cells. Although the total number of colonies for both HATptmut and wt cells diminished with replating in subsequent weeks, there was a greater number of colonies in HATptmut cultures compared with wt through 5 weeks of serial replating (Figure 5A). On average, HATptmut KLS cells produced colonies for twice as long as wt cells (Figure 5B). Flow cytometric analysis of HATptmut cells after 6 weeks in culture showed most cells were lineage negative, CD34+, whereas 5% of these were also highly positive for both c-Kit and Sca1 expression (data not shown). HATptmut cells present for 6+ weeks in culture did not repopulate or cause malignant disease in sublethally irradiated recipient mice when assessed 7 weeks after transplant (data not shown). These results demonstrate that mutation of the HAT domain of p300 results in increased proliferative potential in hematopoietic stem or progenitor cells, although it does not result in frank transformation.
Serial replating assay shows that HATptmut KLS cells have greater proliferative potential than wt KLS cells. (A) The number of colonies produced by serially replating wt and HATptmut KLS cells directly in methylcellulose after 1 to 5 weeks in culture. Bars represent the average number of colonies on duplicate dishes for sorted KLS cells from 6 wt mice and 5 HATptmut chimeric mice. (B) The number of weeks that cultures generated colonies (≥ 1 colony per 1000 cells plated) is shown. Dots represent data from individual mice and the horizontal lines represent the average for each set of mice. P values were calculated using the Student t test.
Serial replating assay shows that HATptmut KLS cells have greater proliferative potential than wt KLS cells. (A) The number of colonies produced by serially replating wt and HATptmut KLS cells directly in methylcellulose after 1 to 5 weeks in culture. Bars represent the average number of colonies on duplicate dishes for sorted KLS cells from 6 wt mice and 5 HATptmut chimeric mice. (B) The number of weeks that cultures generated colonies (≥ 1 colony per 1000 cells plated) is shown. Dots represent data from individual mice and the horizontal lines represent the average for each set of mice. P values were calculated using the Student t test.
General proliferative and differentiation capacity of reconstituted p300 ESCs
The HAT domain of p300 has been previously shown to be critical for development of skeletal muscle, heart, lung, and small intestine.19,20 The unexpected finding that p300 HAT activity seemed dispensable for hematopoietic development led us to more broadly assess the ability of HAT-deficient cells to differentiate into other lineages. Because teratomas derived from ESC injections differentiate into all 3 germ layers, we examined the ability of KIXΔ, HATptmut, CBPX, p300+/−, p300R/−, and p300-null ESCs to form subcutaneous teratomas in nude mice. Histopathology and immunohistochemistry showed that KIXΔ and HATptmut ESCs preferentially differentiated into ectoderm (brain) tissue, whereas CBPX cells differentiated predominantly into mesoderm (skeletal muscle) tissue. p300+/−, p300R/−, and p300-null tumors were composed of a heterogeneous mix of all 3 (supplemental Figure 7A,C-D). Surprisingly, despite their poor performance in hematopoietic cell production, KIXΔ cells gave rise to the largest number and fastest growing teratomas (supplemental Figures 7A and B, respectively), suggesting that KIXΔ ESCs do not have a general impairment in proliferation, but perhaps an impediment for differentiation along certain lineages.
We then tested the ability of the various ESC lines to alter the expression of key regulatory genes under conditions that induce either hematopoietic or neuronal differentiation. The cytokines stem cell factor, erythropoietin, interleukin-3, and interleukin-6 have been found to induce expression of genes involved in hematopoietic differentiation. Expression of endothelial and early erythroid markers are among the first types of genes to be up-regulated in response to these cytokines.37,38 When exposed to these cytokines in a semisolid methylcellulose culture media, HATptmut, p300R/−, and p300-null cells all up-regulated expression of the endothelial marker Tie2 by 3- to 4-fold, whereas KIXΔ cells failed to respond (supplemental Figure 8A). However, all ESC lines were found to equally up-regulate expression of the early erythroid marker GATA1 (supplemental Figure 8B). We also subjected ESC lines to neuronal differentiation. RT-PCR analysis showed that although all ESC lines up-regulated expression of nestin in response to neuronal-inducing growth conditions, the induction in KIXΔ and HATptmut cells was 3.5 times higher than in p300R/− or p300-null cells (supplemental Figure 8C). Similarly, all 4 cell lines up-regulated expression of MAP2 in response to neuronal growth-inducing conditions, yet KIXΔ cells gave a 3-fold higher induction than any other cell type (supplemental Figure 8D).
Together, these studies demonstrate that the pervasive hematopoietic defects associated with mutation of the p300 KIX domain (Figures 2,Figure 3–4) are not due to an intrinsic defect in proliferation or a generalized block in differentiation. Furthermore, despite an apparent lack of requirement in hematopoiesis (Figures 2,Figure 3–4), mutation of the HAT domain does appear to impact differentiation of other tissue types (supplemental Figure 7).
Effects of HAT mutation on pluripotency and differentiation-related genes
We wondered whether the increased proliferation potential of p300 HAT-defective HSCs and progenitors (Figures 4–5) may be due to a block in transition from a pluripotent to a differentiated state. Alterations in epigenetic marks have been found to affect stem cell pluripotency and differentiation,39 and recent studies have shown that both p300 and CBP HAT activity regulate histone H3 lysine 56 (H3K56) acetylation.39,40 To determine whether loss of p300 HAT activity resulted in global perturbations in histone H3K56 acetylation, we isolated histones from wt and HATptmut ESCs and embryoid bodies. Global levels acetyl-H3K56, as well as those of 2 other epigenetic marks, were not grossly different in HATptmut ESCs compared with wt ESCs, and were similarly modulated in both cell types with differentiation into embryoid bodies (supplemental Figure 9).
To further assess the effects of p300 mutation on pluripotency and differentiation, we assessed a panel of pluripotency and differentiation-related factors.41,42 In response to the removal of LIF, HATptmut, p300R/−, as well as KIXΔ and p300-null embryonic cells all down-regulated expression of Oct4 and Nanog to similar degrees over the course of 5 days in liquid culture (first 3 time points in Figure 6A-B) and in cytokine-rich methylcellulose (MC) (last time point in Figure 6A-B). We then analyzed the expression of a broader panel of genes involved in either pluripotency (Dppa4, Tbx3, Gdf3, Pum2) or differentiation (the Wnt signaling molecules, Ctbp2, Spry4; integrins Itga9 and Itgb1; Nhlrc2). We noted that 2 of the pluripotency-related genes, Dppa4 and Gdf3, were consistently expressed at higher levels in HATptmut cells than in wt p300R/− cells when grown in cytokine-rich MC (Figure 6C). Interestingly, these differences were not present in the liquid cultures at day 2 or 8 (Figure 6D-E) in any cell type analyzed. These results suggest that mutation of the HAT domain may affect expression of certain pluripotency-associated genes under hematopoietic-inducing conditions.
Gene expression analysis of pluripotency and differentiation markers in differentiating ESCs. Normalized quantitative RT-PCR data from in vitro ESC differentiation assays. (A) Oct4 and (B) Nanog expression in ESCs, and in embryoid bodies (EBs) upon removal of LIF (day 2 [d2] liq, d5 liq). D8 MC time point is from EBs that were transferred to hematopoietic-inducing methylcellulose on d2 and grown for an additional 6 days. (C) Expanded gene expression analysis in d8 MC cultures of HATptmut and p300R/− cells. Gene expression levels in p300R/− cells were all set to 1 and expression in HATptmut cells is shown relative to this. (D) Dppa4 and (E) Gdf3 expression in d2 and d8 liquid EB cultures and after growth in cytokine-rich MC, d8 MC. Gene expression data were normalized to glyceraldehyde-3-phosphate dehydrogenase using the 2−ΔCt method.36 Bars indicate the average of triplicate samples; error bars indicate SD among triplicates. *Significant difference from p300R/− (P < .001), calculated by Student t test. Graphs are representative of 2 to 4 independent experiments.
Gene expression analysis of pluripotency and differentiation markers in differentiating ESCs. Normalized quantitative RT-PCR data from in vitro ESC differentiation assays. (A) Oct4 and (B) Nanog expression in ESCs, and in embryoid bodies (EBs) upon removal of LIF (day 2 [d2] liq, d5 liq). D8 MC time point is from EBs that were transferred to hematopoietic-inducing methylcellulose on d2 and grown for an additional 6 days. (C) Expanded gene expression analysis in d8 MC cultures of HATptmut and p300R/− cells. Gene expression levels in p300R/− cells were all set to 1 and expression in HATptmut cells is shown relative to this. (D) Dppa4 and (E) Gdf3 expression in d2 and d8 liquid EB cultures and after growth in cytokine-rich MC, d8 MC. Gene expression data were normalized to glyceraldehyde-3-phosphate dehydrogenase using the 2−ΔCt method.36 Bars indicate the average of triplicate samples; error bars indicate SD among triplicates. *Significant difference from p300R/− (P < .001), calculated by Student t test. Graphs are representative of 2 to 4 independent experiments.
Discussion
In the current study, we used a combination of RMCE and exon-trapping to create an allelic series of 10 coisogenic ESCs, each expressing a specific mutant p300 from an inserted cDNA cassette, and used them to create a corresponding series of mutant p300 chimeric mice. With an overall rate of 37% efficiency, RMCE provided a vast improvement over the notoriously inefficient rate of homologous recombination (< 1%). Importantly, this hybrid RMCE-exon trapping approach enabled transcription of each mutant p300 to be driven from the endogenous promoter, thus preserving natural expression patterns by comparison with random integration in transgenics created by pronuclear injection. Using both in vivo and in vitro assays, we used this allelic series to comprehensively define the structural requirements of p300 in hematopoiesis.
From these studies, we have determined that, of all the domains examined, loss of the KIX and CH1 domains had the most pervasive effects on hematopoiesis (Figure 7). Similar to the findings with a knock-in point mutation of KIX,22 we found an absolute requirement for the KIX domain in virtually all hematopoietic cell types. KIXΔ cells also showed a reduced capacity to reconstitute the hematopoietic system of lethally irradiated mice in competitive BM transplantation assays. The severely compromised ability of these cells to contribute to so many cell populations likely reflects the fact that the KIX domain provides critical binding sites for transcription factors involved in early hematopoietic differentiation, such as c-myb, CREB, and Aml1.43-46 Previous work by Kasper et al showed that the effects of mutating the KIX domain can be explained in part by the loss of c-myb activity.22 Our results demonstrate that disruption of KIX does not appear to result in a generalized proliferation or differentiation defect, although neuronal differentiation may also be altered in addition to hematopoiesis.
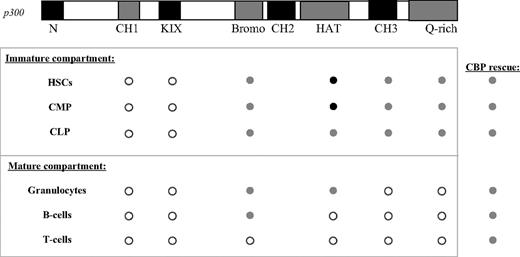
A summary of the p300 structure-function analysis in hematopoiesis. ○ indicates that loss of the domain is similar to the p300 nullizygous phenotype.  indicates that rescue of the nullizygous phenotype can be achieved in the absence of the domain. In these cases, rescue is similar to levels achieved with p300R/− cells. • indicates that, in the absence of the domain, rescue of the nullizygous phenotype surpasses that of p300R/− cells. The CBP column indicates rescue achieved by expressing an extra copy of CBP at the p300 locus is similar to that of p300R/− cells.
indicates that rescue of the nullizygous phenotype can be achieved in the absence of the domain. In these cases, rescue is similar to levels achieved with p300R/− cells. • indicates that, in the absence of the domain, rescue of the nullizygous phenotype surpasses that of p300R/− cells. The CBP column indicates rescue achieved by expressing an extra copy of CBP at the p300 locus is similar to that of p300R/− cells.
A summary of the p300 structure-function analysis in hematopoiesis. ○ indicates that loss of the domain is similar to the p300 nullizygous phenotype.  indicates that rescue of the nullizygous phenotype can be achieved in the absence of the domain. In these cases, rescue is similar to levels achieved with p300R/− cells. • indicates that, in the absence of the domain, rescue of the nullizygous phenotype surpasses that of p300R/− cells. The CBP column indicates rescue achieved by expressing an extra copy of CBP at the p300 locus is similar to that of p300R/− cells.
indicates that rescue of the nullizygous phenotype can be achieved in the absence of the domain. In these cases, rescue is similar to levels achieved with p300R/− cells. • indicates that, in the absence of the domain, rescue of the nullizygous phenotype surpasses that of p300R/− cells. The CBP column indicates rescue achieved by expressing an extra copy of CBP at the p300 locus is similar to that of p300R/− cells.
Deletion of the bromodomain had the least remarkable phenotype for p300 hematopoietic function, with major deficiencies appearing only in mature T cells (Figure 7). Interestingly, it was the only deletion to not affect B-cell development. Although not a major site of protein binding, the bromodomain is thought to assist p300 in its recognition of specific acetylated lysine residues in histones and transcription factors such as p53, MyoD, and STAT3.47-49
CBP and p300 are highly homologous, yet an increasing number of studies show that they have nonredundant functions as well. In the current study, expression of a CBP cDNA from the p300 locus (ie, resulting in no p300 and 3 copies of CBP) rescued nearly all of the hematopoietic defects associated with loss of p300 and the level of rescue was similar to that provided by p300-reconstituted cells (Figure 7). Although both CBP and p300 are expressed in the majority of cell types,50 differentiation of many hematopoietic lineages may be sensitive to disruptions in total gene dosage rather than to disruptions specifically of p300.
Surprisingly, our studies suggest that the intrinsic HAT activity of p300 is not required for establishment of the hematopoietic system. Previous in vivo studies have shown that elimination of p300 HAT enzymatic activity results in severe developmental defects in skeletal muscle, heart, lung, and small intestine.19,20 In contrast, loss of HAT activity in our studies resulted in increased numbers and proliferative potential of hematopoietic progenitors and stem cells. On a molecular level our studies suggest that HATptmut cells respond differently to hematopoietic-inducing cytokines. Given the fact that p300 serves as a coactivator for more than 100 transcription factors, and HAT activity plays a central role for transactivation, it is quite surprising that the loss of p300 acetyltransferase activity does not simply recapitulate the phenotype of loss of p300 in its entirety (nullizygosity). Overall, results from this study underscore the fact that p300 is a multifunctional transcriptional regulator and distinguishes the roles it has as a scaffolding protein from those it has as an acetyltransferase in hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank David Ziegler for assistance in assessing mouse coat color chimerism, Beatriz Ospina for technical assistance with teratomas, Lisa Moreau for cytogenetic assessment of chromosome numbers in primary stroma cells, and Andrei Krivtsov for advice on flow cytometry and serial replating assays.
Authorship
Contribution: E.A.K. designed, performed, and analyzed experiments and wrote the manuscript; X.X. and T.N.D. performed experiments; and M.E.L., V.I.R., and A.L.K. consulted in the design and analysis of experiments and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew L. Kung, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Children's Hospital Boston and Harvard Medical School, 44 Binney St, Mayer 649, Boston, MA 02115; e-mail: andrew_kung@dfci.harvard.edu.


![Figure 6. Gene expression analysis of pluripotency and differentiation markers in differentiating ESCs. Normalized quantitative RT-PCR data from in vitro ESC differentiation assays. (A) Oct4 and (B) Nanog expression in ESCs, and in embryoid bodies (EBs) upon removal of LIF (day 2 [d2] liq, d5 liq). D8 MC time point is from EBs that were transferred to hematopoietic-inducing methylcellulose on d2 and grown for an additional 6 days. (C) Expanded gene expression analysis in d8 MC cultures of HATptmut and p300R/− cells. Gene expression levels in p300R/− cells were all set to 1 and expression in HATptmut cells is shown relative to this. (D) Dppa4 and (E) Gdf3 expression in d2 and d8 liquid EB cultures and after growth in cytokine-rich MC, d8 MC. Gene expression data were normalized to glyceraldehyde-3-phosphate dehydrogenase using the 2−ΔCt method.36 Bars indicate the average of triplicate samples; error bars indicate SD among triplicates. *Significant difference from p300R/− (P < .001), calculated by Student t test. Graphs are representative of 2 to 4 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/23/10.1182_blood-2009-04-217794/4/m_zh89990945500006.jpeg?Expires=1764960313&Signature=ArzBu~hs1ffm97bvMYEDD7NKUaLs6A8QjQOnSb5bPipD0pelMTDUS-I6Bqb5XnLXowmoucR6PsxNHG71FkJ0BIRTkeq889phmM9KhF9OBqhP5I4c9Ojfsz3lD-xHKzohMXcE0r2GTc9KU~R9jMqwtexH7eoPssyYPNwEj1-ftCqGwAtTr~sRY9j4X5Bz2iK-BiNwmkfkXCpwivQ18BE9CCMS1~8xPqRlf037EGhSX3MJMOeSGh9r6S9wsCZAYITGGWNo0M3J3JpqNrYpHmAU86hOiiDGDCsSEofemaVbd7OybjbzW3KGoKwMQ6ME4A9vJEXX3Dtn~Dn~WJd6LD~JrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)