Abstract
Hairy cell leukemia (HCL) is generally responsive to single-agent cladribine, and only a minority of patients are refractory and with poor prognosis. HCLs generally express mutated (M) and, in a minority, unmutated (UM) IGHV. In a multicenter clinical trial in newly diagnosed HCL, we prospectively investigated clinical and molecular parameters predicting response and event-free survival after single-agent cladribine. Of 58 HCLs, 6 expressed UM-IGHV (UM-HCL) and 52 M-IGHV (M-HCL). Beneficial responses were obtained in 53 of 58 patients (91%), whereas treatment failures were observed in 5 of 58 patients (9%). Failures were associated significantly with UM-IGHV (5 of 5 failures vs 1 of 53 beneficial responses had UM-IGHV, P < .001), leukocytosis (3 of 5 vs 3 of 53, P = .006), and bulky spleen (4 of 5 vs 4 of 53, P < .001). The UM-HCL not benefiting from cladribine characteristically had bulky spleen (4 of 5, 80%), leukocytosis (3 of 5, 60%), and TP53 defects (2 of 5, 40%), and progressed rapidly after first treatment (median event-free survival, 7.5 months). Our data suggest that UM-HCLs identify the minor subgroup failing cladribine treatment and with more aggressive disease. High incidence of TP53 dysfunction indicates a potential mechanism of resistance to cladribine in the UM-HCL group. Overall, our data provide new molecular elements relevant for treatment concerns in HCL.
Introduction
Hairy cell leukemia (HCL) is a rare B-cell neoplasm generally responsive to purine analogs, including 2-chloro-desoxiadenosine (cladribine), and only a minority of patients are refractory.1,2 Cladribine is administered intravenously or subcutaneously for 5 to 7 days with equivalent efficacy in HCL.1,3 HCL patients who obtain either complete response (CR) or partial response (PR) are expected to have a normal life span and/or will have a higher chance of benefiting from second treatments if they relapse, and not to die of HCL.4,5 Conversely, the minority of patients who do not respond to cladribine often do not respond to subsequent treatments and have a poorer prognosis.1 The unresponsive group needs more accurate characterization for the definition of the effective treatment strategies, possibly at disease onset.6
Analysis of the tumor B-cell receptor (BCR) immunoglobulin (Ig) genes has provided elements essential to understand pathogenesis and behavior of B-cell tumors.7 In the most common B-cell neoplasm chronic lymphocytic leukemia (CLL), status of the tumor Ig heavy chain variable region genes (IGHV) has major impact on prognosis and is recommended for treatment stratification in clinical trials.8,9 Indeed, CLL with unmutated (UM) IGHV status associates with poor overall survival, BCR proliferation signals, and unfavorable genetic abnormalities that may cause resistance to purine analogs.8,10,11 Among these abnormalities, deletion or mutations of TP53, a gene located at 17p13 locus that is crucial in the mechanisms of cell proliferation and apoptosis, DNA repair, and sensitivity to purine analogs,12,13 emerge as a major prognostic parameter of poor outcome.14,15 Similar to CLL, UM-IGHV indicates poor prognosis also in other lymphomas.16,17
HCL has many unique features among B-cell tumors, and one of these is the expression of multiple isotype transcripts.18,19 In the majority of HCLs, the expressed tumor IGHV are mutated (M), whereas HCL cases with UM-IGHV represent a minor group.20-22 However, clinical significance of the mutational status of the tumor IGHV has not been investigated in HCL.
In HCLs, several studies have investigated clinical parameters that predict relapse, progression, or overall survival after purine analogs.5 However, there is no consensus on factors predicting response, and the molecular features that associate with clinical behavior of HCL are largely unknown.5,23
In a national multicenter clinical trial using single-agent cladribine in newly diagnosed HCL, we prospectively investigated (1) clinical and molecular parameters predicting response and (2) the immunogenetic and molecular profile of the refractory patients.
Methods
Patient eligibility and diagnosis
Patients were enrolled in the multicenter clinical trial Italian Cooperative Group for HCL2004 (protocol EudraCT code: Italian Cooperative Group for HCL2004, ethical approval obtained in September 2004 from the Università di Siena Institutional Review Board). All patients had a diagnosis of HCL, had not received previous therapies, and had given informed consent in accordance with the Declaration of Helsinki before entering the trial. Diagnosis of HCL was according to the World Health Organization 2001 Classification of Tumors of Hematopoietic and Lymphoid Tissues.24 Diagnosis was verified centrally by morphology (review panel, F.F., F.L.) and immunophenotype (review panel, D.R., E. Sozzi, M.T.) of the peripheral blood and bone marrow of all patients before treatment initiation. In the patients with leukocytosis (hairy cells [HCs] > 10 000/μL), with bulky spleen (> 10 cm below costal margin [bcm]), and/or who subsequently failed cladribine treatment, diagnosis was also centrally reviewed by immunohistochemistry of the paraffin-embedded bone marrow specimens (review panel, S.A.P., E. Sabattini, L.L.) according to the new World Health Organization 2008 Classification of Tumors of Hematopoietic and Lymphoid Tissues.19 ANXA1 was detected in leukemic cells of formalin-fixed paraffin sections of bone marrow by immune-alkaline phosphatase technique as described.25
Analysis of the tumor IGHV, IGKV, and IGLV transcript rearrangements
Peripheral blood and bone marrow aspirate were collected in ethylene diamine tetra-acetic acid at each participating center and centralized at the Hematology and Transplant Section, University of Siena before treatment initiation. Percentage of HCs and surface Ig kappa (IgK) or lambda (IgL) chain on the tumor cells was determined by triple staining of peripheral blood mononuclear cells or bone marrow mononuclear cells as described.26 The expressed tumor IGHVDJ, IGKVJ, and IGLVJ transcripts were amplified by reverse-transcript polymerase chain reaction, cloned, and sequenced, as described.22,26,27 The cloned sequences were aligned to the ImMunoGeneTics database to identify the tumor transcript and determine tumor gene usage and homology.28 Sequences with more than or equal to 98% homology to germline were defined as “UM.”8
Analysis of TP53 mutations
Mutation analysis of TP53 exons 2 through 10 was performed by polymerase chain reaction and DNA direct sequencing, as described.15 Mutations were confirmed on both strands on independent amplimers and validated by the IARC and UMD TP53 mutation databases (http://www-p53.iarc.fr/StructureAnalysis.html and http://p53.free.fr/Database/p53_recomendations.html).29,30 The distribution of mutations on the TP53 molecule was assessed using the IARC TP53 Mutation Database.30
Genome-wide DNA profiling
Genomic DNA extraction from HCs, integrity verification, and genome-wide DNA profiling with the GeneChip Human Mapping 250K-NspI arrays (Affymetrix) were performed as previously described.23 Briefly, data acquisition was performed using the Affymetrix GCOS 1.4 and GTYPE 4.1, and genotype calls were calculated using the BRLMM algorithm. The Affymetrix CNAT 4.01 was used to calculate copy numbers (CNs) starting from the BRLMM-CHP files. The CNAT hidden Markov model was used to estimate CN. CN gains and losses were defined in the presence of CN more than 2.15 and CN less than 1.85, respectively, corresponding to a P value of less than .001 after Bonferroni multiple test correction.
Treatment schedule, evaluation of response to treatment, and event-free survival
Treatment was indicated if hematopoietic insufficiency (hemoglobin < 11 g/dL, neutrophils < 1500/μL, or platelets < 100 000/μL), general symptoms (raised temperature of unknown etiology > 38°C, night sweats, > 10% loss of weight within 6 months), recurrent infections, or symptomatic splenomegaly were documented. Patients were assigned to receive 0.1 mg/kg per day cladribine for 5 or 7 consecutive days as a subcutaneous bolus injection.
Physical examination, blood cell, and HC counts were repeated weekly for the first month after treatment termination and every 4 to 6 months after final assessment of response. Response was assessed 2 and 6 months after treatment termination and included bone marrow biopsy. Evaluation of response was according to the 1987 Consensus Resolution Criteria plus inclusion of immunohistochemistry by DBA44 and/or TRAP staining for the determination of residual HCs in the bone marrow.31 Specifically, CR was defined when all the following criteria were fulfilled: (1) absence of HCs in the peripheral blood and bone marrow (< 1% HC by immunohistochemistry); (2) normalization of peripheral blood counts (hemoglobin, > 12 g/dL; white blood cells, 3000/μL; neutrophils, > 1500/μL; and platelets, > 100 000/μL); (3) regression to normal of all palpable organomegalies; and (4) absence of constitutional symptoms. PR required all of the following: (1) less than a CR, (2) circulating HCs less than 5% and reduction of more than 50% of HCs in the bone marrow, (3) normalization of peripheral blood counts, and (4) more than 50% reduction of abnormal adenopathy or hepatosplenomegaly. A minor response (mR) required (1) achieving less than a PR, (2) at least 50% reduction of HCs in the peripheral blood, and (3) improvement of one or more cell types in the peripheral blood. Achieving less than mR was classified as no response (NR). Responses that were considered clinically beneficial included CR and PR; mR and NR were rated as treatment failures. Event-free survival (EFS) was defined as the interval between the first treatment day to the first sign of disease progression, or treatment for relapse or progression, or death from HCL (whichever occurred first). Progression was defined as increase in one or more tumor parameters by at least 25%, or occurrence of new tumor manifestations or increase in the frequency and severity of symptoms of disease. Patients who did not fulfill the conditions for a progression were defined as stable.
Clinical correlations and statistical analysis
Response to treatment and EFS were chosen as indicators of clinical behavior and used to scan prognostic parameters of statistical significance. Statistical analyses were performed using the R statistical package (http://www.r-project.org/). Response categories (divided into the 2 groups, beneficial responses, or treatment failures) and any clinical and molecular categorical variable were compared by Fisher exact test or χ2 test, as appropriate. Survival (EFS) analysis was performed by the Kaplan-Meier method using the global log-rank test for significant (P < .05) associations.
Results
Characteristics of the patients
Of 62 patients recruited, 4 cases were excluded from the analysis because central diagnostic revision did not confirm diagnosis of HCL (2 cases) or had variant CD25−, CD27−, ANXA1− HCL (2 cases) according to the WHO 2008 classification.19 The leukemic population of the remaining 58 patients had a surface immunophenotype of classic HCL.
Clinical, laboratory, and molecular characteristics of the 58 investigated patients with classic HCL (46 males, 12 females; male/female ratio = 3.8:1) are described in Table 1. Median age was 51 years (range, 30-83 years). Splenomegaly was documented at physical examination in 34 of 58 patients (58%) and was bulky in 8 of 58 cases (14%). Lymph nodes larger than 1.5 cm were palpable in 6 of 58 patients (10%). Anemia was present in 33 of 58 patients (57%), thrombocytopenia in 44 of 58 (76%), and leukopenia in 38 of 58 (66%). Leukocytosis with HC more than 10 × 103/μL was observed in 6 of 58 patients (10%; range, 10.5-38.4 × 103 HC/μL; median, 12.9 × 103 HC/μL). Leukocytosis and bulky spleen were concomitant in 4 of 58 patients (7%). The most frequently used IGHV genes were IGHV3-30 (6 of 58, 10%), IGHV4-34 (5 of 58, 9%), IGHV3-21 (4 of 58, 7%), IGHV3-33 (4 of 58, 7%), and IGHV4-39 (4 of 58, 7%; supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Tumor IGHV genes were M in 52 of 58 patients (90%) and UM in 6 of 58 patients (10%). Mutations causing dysfunction of the TP53 gene were observed in 2 of 58 HCL (one insertion InsG14032 with Lys235 frameshift and one A13167G/Tyr163Cys missense mutation). Median time from diagnosis to treatment was 31 days (range, 4-376 days) and median follow-up after treatment was 24 months (range, 6-40 months).
Clinical and molecular characteristics of HCL patients before treatment
| Characteristic . | n/total (%) . | Median (range) . |
|---|---|---|
| Age, y | 51 (30-83) | |
| Males | 46/58 (79) | — |
| Females | 12/58 (21) | — |
| ECOG performance status | ||
| 0 | 45/58 (77) | — |
| 1 | 10/58 (17) | — |
| 2 | 2/58 (6) | — |
| Time from diagnosis to treatment, d | 31 (4-376) | |
| Follow-up, mo | 24 (6-40) | |
| Splenomegaly | 32/58 (55) | — |
| Splenomegaly, cm bcm | 2 (0-16) | |
| Bulky spleen (≥ 10 cm bcm) | 8/58 (14) | — |
| Lymph nodes > 1.5 cm | 6/58 (10) | |
| Hemoglobin | 11.4 (5.2-16) | |
| Less than 12 g/dL | 33/58 (57) | — |
| Less than 10 g/dL | 12/58 (21) | — |
| Less than 9 g/dL | 9/58 (16) | — |
| Less than 8 g/dL | 5/58 (9) | — |
| Platelets | 75 (30-237) | |
| Less than 100 × 103/μL | 44/58 (76) | — |
| Less than 50 × 103/μL | 9/58 (16) | — |
| WBCs | 3.1 (0.9-48) | |
| More than 10 × 103/μL | 6/58 (10)* | — |
| Less than 4 × 103/μL | 38/58 (66) | — |
| Neutrophils | 0.9 (0.104-4.8) | |
| Less than 1.5 × 103/μL | 50/58 (86) | — |
| Less than 0.5 × 103/μL | 15/58 (26) | — |
| Percentage of HCs in peripheral blood counts* | 11 (1-93) | |
| Percentage of HCs of all peripheral blood B cells* | 80 (30-99) | |
| B2M > × 1 ULN | 27/46 (59) | — |
| B2M> × 2 ULN | 3/46 (7) | — |
| LDH > × 1 ULN | 4/58 (7) | — |
| Hairy cell index, n = 55 | 0.42 (0.06-0.9) | |
| Molecular characteristics | ||
| IGHV homology ≥ 98% | 6/58 (10) | — |
| TP53 dysfunction† | 2/58 (2) | — |
| Characteristic . | n/total (%) . | Median (range) . |
|---|---|---|
| Age, y | 51 (30-83) | |
| Males | 46/58 (79) | — |
| Females | 12/58 (21) | — |
| ECOG performance status | ||
| 0 | 45/58 (77) | — |
| 1 | 10/58 (17) | — |
| 2 | 2/58 (6) | — |
| Time from diagnosis to treatment, d | 31 (4-376) | |
| Follow-up, mo | 24 (6-40) | |
| Splenomegaly | 32/58 (55) | — |
| Splenomegaly, cm bcm | 2 (0-16) | |
| Bulky spleen (≥ 10 cm bcm) | 8/58 (14) | — |
| Lymph nodes > 1.5 cm | 6/58 (10) | |
| Hemoglobin | 11.4 (5.2-16) | |
| Less than 12 g/dL | 33/58 (57) | — |
| Less than 10 g/dL | 12/58 (21) | — |
| Less than 9 g/dL | 9/58 (16) | — |
| Less than 8 g/dL | 5/58 (9) | — |
| Platelets | 75 (30-237) | |
| Less than 100 × 103/μL | 44/58 (76) | — |
| Less than 50 × 103/μL | 9/58 (16) | — |
| WBCs | 3.1 (0.9-48) | |
| More than 10 × 103/μL | 6/58 (10)* | — |
| Less than 4 × 103/μL | 38/58 (66) | — |
| Neutrophils | 0.9 (0.104-4.8) | |
| Less than 1.5 × 103/μL | 50/58 (86) | — |
| Less than 0.5 × 103/μL | 15/58 (26) | — |
| Percentage of HCs in peripheral blood counts* | 11 (1-93) | |
| Percentage of HCs of all peripheral blood B cells* | 80 (30-99) | |
| B2M > × 1 ULN | 27/46 (59) | — |
| B2M> × 2 ULN | 3/46 (7) | — |
| LDH > × 1 ULN | 4/58 (7) | — |
| Hairy cell index, n = 55 | 0.42 (0.06-0.9) | |
| Molecular characteristics | ||
| IGHV homology ≥ 98% | 6/58 (10) | — |
| TP53 dysfunction† | 2/58 (2) | — |
WBCs indicates white blood cells; HCs, hairy cells; B2M, β2-microglobulin; LDH, lactic dehydrogenase; —, not applicable; and ULN, upper limit of normal.
The percentage of HCs in peripheral blood was determined by morphologic examination of the peripheral blood smear; the percentage of HCs of all peripheral blood B cells was calculated in flow cytometry by double staining of PBMCs with anti-CD20 and anti-CD103 monoclonal antibodies. CD20+ B cells were gated and the percentage of CD20+CD103+ HC cells was determined.
Twelve of 60 cases were investigated for the whole genome DNA profile by using the 250K Affymetrix array: these included 3 of 5 NR + mR and 9 of 53 PR + CR. Of the 5 NR + mR HCLs, 2 had TP53 dysfunction by p53 mutation.
Response to treatment and EFS
Cladribine was administered at the proposed regimen with no modifications in all patients. Fifty-three of 58 patients (91%) had a beneficial response to treatment (42 CR, 72%; 11 PR, 19%). Five of 58 patients (9%) had no beneficial response (3 mR and 2 NR). Responses were independent of treatment schedule and dose given.32
Six of 58 patients (5 of 5 treatment failures and 1 of 53 beneficial responses) progressed and required new treatment with 91% EFS at 1 year, 88% at 2 years, and 81% at 3 years (supplemental Figure 1A). EFS of the 5 failures was 7 months and significantly different (P < .001) from the 53 beneficial responses (median EFS not reached, one progression after 28 months; supplemental Figure 1B).
Identification of clinical prognostic parameters of response to treatment and EFS
The clinical and molecular parameters investigated for response prediction are indicated in supplemental Table 2. Among the clinical characteristics, sex, age, performance status, lymph node enlargement (in 5 of 53 beneficial responses vs 1 of 5 failures), anemia, thrombocytopenia, and hairy cell index showed no significant correlation with treatment outcome. On the contrary, leukocytosis (3 of 53 beneficial responses vs 3 of 5 failures, P = .005) and bulky spleen (4 of 53 beneficial responses vs 4 of 5 failures, P < .001) showed a statistically highly significant correlation with response to cladribine. Most interestingly, among molecular characteristics, UM-IGHV (1 UM-HCL only within the 53 HCL patients who had beneficial responses, and 5 UM-HCL of the 5 HCL patients who failed treatment, P < .001) and tumor TP53 dysfunction resulting from missense or frameshift mutation (no HCL with TP53 dysfunction by mutations in the 53 beneficial responses, and 2 HCL with TP53 mutations in the 5 failures, P = .006) predicted response to single-agent cladribine.
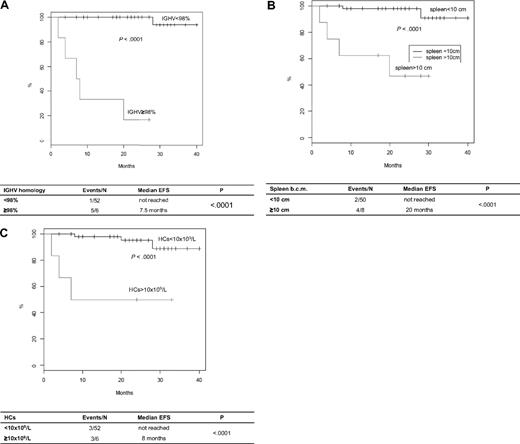
UM-IGHV, splenomegaly, and leukocytosis also predicted short EFS (Figure 1). Indeed, 5 of 6 (83%) UM-HCL versus 1 of 52 (2%) M-HCL (P < .001), 4 of 8 (50%) HCL with versus 2 of 50 (4%) without large splenomegaly (P < .001), and 3 of 6 (50%) HCL with versus 3 of 52 (6%) without leukocytosis (P < .001) progressed and/or required new treatment after a median time of 7.5 months versus not reached, 20 months versus not reached, and 14.5 months versus not reached, respectively.
EFS of HCL patients treated with cladribine according to prognostic parameters. (A) EFS in UM-HCL versus M-HCL. (B) EFS in HCL with large splenomegaly versus HCL without large splenomegaly. (C) EFS in HCL with leukocytosis versus HCL without leukocytosis.
EFS of HCL patients treated with cladribine according to prognostic parameters. (A) EFS in UM-HCL versus M-HCL. (B) EFS in HCL with large splenomegaly versus HCL without large splenomegaly. (C) EFS in HCL with leukocytosis versus HCL without leukocytosis.
Clinical and molecular profile of the UM-HCL failing cladribine treatment
Because of the very high power of UM-IGHV to predict response and short EFS, we specifically analyzed the clinical and molecular profile of the 5 UM-HCL that failed cladribine (Table 2; supplemental Figure 2).
Clinical and molecular profile of the HCL with unmutated IGHV not benefiting from cladribine
| Patient . | HCL11 . | HCL30 . | HCL49 . | HCL50 . | HCL119 . |
|---|---|---|---|---|---|
| WHO 2008 classification19 | Classic | Classic | Classic | Classic | Classic |
| Clinical parameters | |||||
| Spleen, cm bcm | 11 | 11 | 11 | 0 | 14 |
| Enlarged lymph nodes | − | + | − | − | − |
| Hemoglobin, g/dL | 15 | 11.1 | 5.2 | 10.9 | 13.4 |
| Platelets, ×109/L | 77 | 87 | 35 | 50 | 157 |
| WBCs, ×109/L | 3.5 | 10.9 | 36.4 | 1.2 | 19.4 |
| Neutrophils, ×109/L | 1.5 | 0.4 | 0.7 | 0.3 | 2.7 |
| Hairy cells, ×109/L | 0.5 | 10.5 | 28.8 | 0.3 | 10.7 |
| B2M, mg/dL | 1.0 | 1.1 | 2.2 | 1.9 | 2.9 |
| LDH, IU/L | 302 | 183 | 561 | 290 | 399 |
| Immunophenotype | |||||
| CD5 | − | − | − | − | − |
| CD11c | + | + | + | + | + |
| CD23 | − | − | − | − | − |
| CD25 | + | + | + | + | + |
| CD27 | − | − | − | − | − |
| CD38 | − | − | − | − | − |
| CD103 | + | + | + | + | + |
| FMC7 | + | + | + | + | + |
| Surface Ig isotypes | MA | MDGA | MDGA | MDGA | MG |
| Bone marrow histology | |||||
| CD20/ANXA1 | + | + | + | + | + |
| DBA44 | + | + | + | + | + |
| CD11c | + | + | + | + | + |
| bcl1 | + | + | + | + | + |
| Molecular profile | |||||
| IGHV | 3-09 | 3-23 | 3-33 | 3-33 | 4-34 |
| Homology, percentage | 98.4 | 98.3 | 98.7 | 99.1 | 99.65 |
| IGHD | 6-13 | 3-10 | 1-26 | 1-26 | 3-03 |
| IGHJ | 5 | 6 | 3 | 4 | 5 |
| IGK/LV | IGKV6-21 | IGLV2-5 | IGLV1-47 | IGLV1-47 | IGKV3-15 |
| Homology, percentage | 99,24 | 91,22 | 98,33 | 98,23 | 99,64 |
| IGK/LJ | KJ1 | LJ1 | LJ3 | LJ3 | KJ4 |
| LCDR3 subset* | − | − | 1L | 1L | − |
| TP53 | Mut | WT | Mut | WT | WT |
| Clinical course | |||||
| Response to first line | mR | NR | NR | mR | mR |
| EFS, mo | 20 | 4 | 2 | 8 | 7 |
| Second-line treatment | Rituximab | Rituximab | IFN-α | DCF | IFN-α |
| Response to second line | NR | CR | NR | PR | − |
| Other lines | IFN-α | − | − | − | − |
| Last follow-up, mo | 38 | 17 | 3 | 33 | 13 |
| Status at last follow-up | IFN-α | Stable | Dead | Stable | IFN-α |
| Patient . | HCL11 . | HCL30 . | HCL49 . | HCL50 . | HCL119 . |
|---|---|---|---|---|---|
| WHO 2008 classification19 | Classic | Classic | Classic | Classic | Classic |
| Clinical parameters | |||||
| Spleen, cm bcm | 11 | 11 | 11 | 0 | 14 |
| Enlarged lymph nodes | − | + | − | − | − |
| Hemoglobin, g/dL | 15 | 11.1 | 5.2 | 10.9 | 13.4 |
| Platelets, ×109/L | 77 | 87 | 35 | 50 | 157 |
| WBCs, ×109/L | 3.5 | 10.9 | 36.4 | 1.2 | 19.4 |
| Neutrophils, ×109/L | 1.5 | 0.4 | 0.7 | 0.3 | 2.7 |
| Hairy cells, ×109/L | 0.5 | 10.5 | 28.8 | 0.3 | 10.7 |
| B2M, mg/dL | 1.0 | 1.1 | 2.2 | 1.9 | 2.9 |
| LDH, IU/L | 302 | 183 | 561 | 290 | 399 |
| Immunophenotype | |||||
| CD5 | − | − | − | − | − |
| CD11c | + | + | + | + | + |
| CD23 | − | − | − | − | − |
| CD25 | + | + | + | + | + |
| CD27 | − | − | − | − | − |
| CD38 | − | − | − | − | − |
| CD103 | + | + | + | + | + |
| FMC7 | + | + | + | + | + |
| Surface Ig isotypes | MA | MDGA | MDGA | MDGA | MG |
| Bone marrow histology | |||||
| CD20/ANXA1 | + | + | + | + | + |
| DBA44 | + | + | + | + | + |
| CD11c | + | + | + | + | + |
| bcl1 | + | + | + | + | + |
| Molecular profile | |||||
| IGHV | 3-09 | 3-23 | 3-33 | 3-33 | 4-34 |
| Homology, percentage | 98.4 | 98.3 | 98.7 | 99.1 | 99.65 |
| IGHD | 6-13 | 3-10 | 1-26 | 1-26 | 3-03 |
| IGHJ | 5 | 6 | 3 | 4 | 5 |
| IGK/LV | IGKV6-21 | IGLV2-5 | IGLV1-47 | IGLV1-47 | IGKV3-15 |
| Homology, percentage | 99,24 | 91,22 | 98,33 | 98,23 | 99,64 |
| IGK/LJ | KJ1 | LJ1 | LJ3 | LJ3 | KJ4 |
| LCDR3 subset* | − | − | 1L | 1L | − |
| TP53 | Mut | WT | Mut | WT | WT |
| Clinical course | |||||
| Response to first line | mR | NR | NR | mR | mR |
| EFS, mo | 20 | 4 | 2 | 8 | 7 |
| Second-line treatment | Rituximab | Rituximab | IFN-α | DCF | IFN-α |
| Response to second line | NR | CR | NR | PR | − |
| Other lines | IFN-α | − | − | − | − |
| Last follow-up, mo | 38 | 17 | 3 | 33 | 13 |
| Status at last follow-up | IFN-α | Stable | Dead | Stable | IFN-α |
WBCs indicates white blood cells; B2M, β2-microglobulin; LDH, lactic dehydrogenase; −, negative; +, positive; Mut, mutation leading to p53 dysfunction; WT, wild type; EFS, event-free survival; CR, complete response; PR, partial response; mR, minor response; cm, centimeter; bcm, below costal margin; and NR, no response.
Subsets were identified according to Forconi et al.22 The remaining one UM-HCL patient (case HCL74), who obtained a PR and has not progressed after a follow-up of 27 months from end of cladribine, had no leukocytosis (hemoglobin, 9.9 g/dL; platelets, 169 × 109/L; WBCs, 1.04 × 109/L), no bulky spleen (0 cm bcm), classic peripheral blood immunophenotype, and bone marrow immunohistochemistry, and no TP53 mutations.
Clinically, UM-IGHV status was frequently associated with bulky spleen (4 of 5, 80% UM-HCL had bulky spleen) and leukocytosis (3 of 5, 60% UM-HCL). Enlarged lymph nodes were absent in all patients but one, consistent with classic features of HCL. Immunohistochemical central revision of the diagnostic bone marrow biopsies confirmed the diagnosis of classic HCL according to the WHO 2008 classification in all UM-HCL cases, which showed a typical morphology and stained positive for DBA44 and ANXA1 antibodies (Table 2; supplemental Figure 2). Consistently, all cases expressed multiple surface Ig isotypes, which is a feature unique to tumor B cells of HCL19,26 and CD25.
Immunogenetically, IGHV3 family was used in 4 of 5 cases and IGHV4-34 in 1 of 5, lambda light chain was used in 3 of 5 UM-HCL with 2 of 3 using IGLV1-47/J3. Interestingly, the IGL1-47/J3 HCL used IGHV3-33, as for the previously described HCL subset 1L (Table 2).22 All UM-IGHV tumor genes paired with UM-IGLV or UM-IGKV genes.
DNA for high-density genome-wide DNA profiling was available from 3 of 5 refractory UM-HCLs and 9 of 52 responsive M-HCLs (supplemental Table 3). A stable profile was documented in all the UM-HCL and M-HCL cases investigated. However, combined TP53 mutational analysis and whole genomic DNA profile demonstrated TP53 dysfunction in at least 2 of 5 (40%) UM-HCL failing cladribine and not in the 52 responsive M-HCLs.
Discussion
The goals of the present study were (1) to identify clinical and molecular parameters predicting response to single-agent cladribine, and subsequently (2) to characterize the clinical and molecular features of the novel HCL subset with UM-IGHV. We documented that (1) leukocytosis and splenomegaly represented the clinical indicators of poor response and/or rapid progression after first cladribine treatment; (2) UM-IGHV status was an almost absolute discriminator of response failure to cladribine and rapid progression after treatment; and (3) the novel UM-HCL subset identified a group associated with leukocytosis, splenomegaly, and frequent TP53 dysfunction.
In this study, response to cladribine was similar to previous studies using intravenous or subcutaneous cladribine in HCL.1,33 In the Group C protocol of the National Cancer Institute with 928 patients by Cheson et al,33 performance status more than 3 and age older than 68 years emerged as the only clinical prognosticators of response. Other single-institution studies identified lymphadenopathy,34,35 hemoglobin,34,36,37 splenomegaly,37,38 leukopenia,34 or leukocytosis34,36 as possible factors predicting response, although no consensus has been achieved yet.5 Our analysis of patients from multiple centers points to leukocytosis and bulky spleen as significant predictors for failure and progression after treatment. If compared with the large multicenter study at NCI, the relatively smaller number of patients in our analysis (n = 12-68 years) and lack of patients with performance status more than 2 may justify the discrepancies.33 However, the general lack of consensual clinical prognostic parameters among different studies and the evidence that 40% patients with either leukocytosis or large splenomegaly can achieve beneficial responses to cladribine alone prompted us to look for molecular parameters associated with poor outcome.
Biologic parameters have rarely been investigated for the prognosis of HCL.39 The frequency of the minor group with UM-IGHV genes was defined in our extended series of 90 HCLs.22 Usage and status of tumor IGHV in the present analysis were consistent with our and other studies.21,22,40 The novel finding, that UM-IGHV status mirrored treatment failure and rapid progression after treatment, was remarkable and much more specific than bulky spleen and leukocytosis that were also observed in HCL responsive to cladribine. In CLL, UM-IGHV status associates with poor overall survival, BCR proliferation signals, and unfavorable genetic abnormalities.8,10,11 In addition, multivariate analyses in CLL indicate that UM-IGHV status is one of only 2 factors that predict refractoriness to purine analogs, the other factor being TP53 dysfunction by mutation and/or deletion.14,15 Consistent with the data in CLL, we found that our UM-HCLs resistant to cladribine frequently harbored genetic damages leading to TP53 dysfunction, which is a cause of resistance to purine analogs.12,13
HCs accumulate in the bone marrow, in the blood, and in the spleen by definition.19 The observation that our UM-HCLs associated with high tumor burden with leukocytosis and large splenomegaly at presentation suggested that the UM-HCL cases might have accumulated tumor cells more rapidly than M-HCL cases, in a similar manner to UM-CLL that are frequently diagnosed at advanced disease stages.41 Our cases did not accumulate in the lymph nodes, a peculiarity that is consistent with the diagnosis of classic HCL.19 We actively reviewed morphology, immunophenotype, and bone marrow histology in all cases either with leukocytosis or with splenomegaly or refractory to cladribine to rule out diagnostic doubts. All our UM-HCLs, including the 2 cases with TP53 mutations leading to p53 protein dysfunction, classified as classic HCL, that indeed can harbor TP53 defects.42,43
There is impelling evidence that new treatment modalities, including rituximab alone or in combination with purine analogs, can be highly effective in producing complete molecular remissions or in reverting refractoriness to single-agent purine analogs in some patients.37,44-47 A combination of rituximab with cladribine can probably eradicate minimal residual disease independently of IGHV mutational status.45 We had confirmed that the combined schedule could completely eradicate disease in a UM-HCL patient refractory to single-agent purine analogs.47 In addition, a recent update of the Royal Marsden Hospital database indicated rituximab as a tool to improve outcome in HCL even if given in relapsed and/or purine analog refractory patients.48 Based on these grounds, in the present study, we had the chance to administer rituximab after cladribine in 2 UM-HCLs rather than purine analogs again. The case (HCL30) that did not have TP53 defects was rituximab-sensitive, whereas the case (HCL11) with a TP53 frameshift mutation was rituximab-refractory (Table 2). Refractoriness to second-line treatment was also documented in the other TP53-defective UM-HCL patient (HCL49) who received interferon-α because of concurrent fungal infection during progressive cytopenia.49 Despite the irrelevant number, it would be interesting to verify whether treatment strategies should be based on selected molecular discriminators.
Overall, the new observation that HCLs with UM-IGHV genes are resistant to single-agent cladribine may provide a tool to identify the minor group of refractory HCL patients. It will need to be assessed in a larger group of UM-HCLs whether these patients should be treated differently. The evidence that a proportion of UM-HCL has defects of TP53 gene provides a mechanism of resistance in this group of patients.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Silvia Tavera (Cuneo), Enrico Morello (Bolzano), Piero Galieni (A. Piceno), Alessandra Caremani (Ematologia), and Sara Barulli (Pesaro) for patient referral and contribution to the clinical management of the patients evaluated for this study.
The work was supported by EHA Clinical Research Grant 2004, Hairy Cell Leukemia Research Foundation, Piano di Ateneo per la Ricerca 2005 (University of Siena), Cofinanziamento Ministero dell'Università e della Ricerca per Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (MUR-PRIN) 2006, Associazione Italiana per la Ricerca sul Cancro, and Associazione Italiana contro le Leucemie, Mielomi e Linfoma (Siena-AIL Section), Siena, Italy.
Authorship
Contribution: F.F. designed and performed research, collected, analyzed, and interpreted data, performed statistical analysis, and wrote the manuscript; E. Sozzi processed diagnostic samples, performed research, and analyzed data; E.C. collected and analyzed data; M.T. and D.R. performed research and analyzed data; F.Z., T.I., C.S., L.R., F.G., R.C. A.B., A.G., A.Z., A.P., and M.G. identified patients and shipped diagnostic material for central revision and performed research; S.A.P., E. Sabattini, and L.L. performed central histology revision; F.B. and A.R. performed genome-wide DNA profiling; and F.L. designed research and supervised research and manuscript writing.
The study was run on behalf of the Italian Cooperative Group for HCL. A complete list of the Italian Cooperative Group for HCL participants appears online on the Blood website.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Forconi, Sezione di Ematologia e Trapianti, Dipartimento di Medicina Clinica e Scienze Immunologiche, Università di Siena, AOUS, Viale Bracci, 53100 Siena, Italy; e-mail: forconif@unisi.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal