Abstract
Protein Z (PZ) binds to PZ-dependent inhibitor (ZPI) and accelerates the inhibition of the coagulation protease, activated factor X (FXa), in the presence of phospholipids and Ca2+. A 2.3Å resolution crystal structure of PZ complexed with ZPI shows that ZPI is a typical serine protease inhibitor and that PZ has a serine protease fold with distorted oxyanion hole and S1 pocket. The 2 molecules bind with fully complementary surfaces spanning over 2400Å2 and involving extensive ionic and hydrophobic interactions. ZPI has an unusual shutter region with a negatively charged residue buried within the hydrophobic core of the molecule. This unique Asp213 is critical in maintaining the balanced metastability required for optimal protease inhibition, especially when PZ is bound, with its replacement with Asn resulting in increased thermal stability, but decreased efficiency of protease inhibition. The structure of ZPI shows negatively and positively charged surfaces on top of the molecule, in keeping with mutagenesis studies in this work indicating exosite interactions with FXa when it docks on top of ZPI. As modeled in this study, the γ-carboxy-glutamic acid-containing domains of PZ and FXa enable them to bind to the same phospholipid surfaces on platelet and other membranes, with optimal proximity for the inhibition of FXa by the complexed ZPI.
Introduction
Blood coagulation is tightly controlled by members of the serine protease inhibitor (serpin) family of serine protease inhibitors.1-3 Activated factor X (FXa), the key enzyme in activating prothrombin, is mainly regulated by 2 serpins, antithrombin3,4 and protein Z (PZ)–dependent inhibitor (ZPI).5,6 However, both serpins are relatively inactive toward FXa in the absence of their corresponding cofactors, heparin and PZ. Heparin activates antithrombin and accelerates the interaction between antithrombin and FXa by approximately 300-fold.3 PZ is a vitamin K–dependent plasma protein and is homologous to blood coagulation factors VII, IX, X, and protein C.7,8 It has an N-terminal γ-carboxy-glutamic acid (Gla)–containing domain, which binds phospholipids, 2 epidermal growth factor–like (EGF) domains, and a serine protease domain that lacks catalytic activity. PZ binds ZPI with high affinity and accelerates the interaction between ZPI and FXa by more than 1000-fold in the presence of Ca2+ and phospholipids.9-11 Thus, it appears that antithrombin mainly targets FXa on the surface of the endothelium, where heparin-like glycosaminoglycans are anchored, whereas ZPI, located to phospholipid surfaces by its binding to PZ, mainly inhibits FXa on platelet and other membrane surfaces.12 Mice lacking ZPI or PZ developed enhanced thrombosis after arterial injury,13,14 and the deficiency of ZPI or PZ in humans has been associated with venous thrombosis and peripheral arterial disease.15-18 In plasma, PZ circulates as a complex with ZPI.19 It has been suggested that PZ binds ZPI through its C-terminal protease domain;11 however, the detailed interactions between PZ and ZPI are unknown, and no structure of either PZ or ZPI is available. In this study, we have prepared recombinant ZPI and a truncated PZ containing the EGF2 and SP domains, and solved the crystal structure of their complex.
Methods
Proteins
Recombinant full-length human ZPI (wild-type) was prepared from Escherichia coli using the SUMO fusion expression system, according to similar protocols for preparing recombinant corticosteroid binding globulin, as previously described.20 The concentration of ZPI was calculated from the absorbance at 280 nm using a molar absorption coefficient of 31 525 M−1 cm−1.10 The prepared ZPI was fully active in inhibiting FXa (see Table 2). Recombinant glycosylated PZ (∼ 50 kDa) containing residues 84-360 (starting with amino acid sequence LAKNECHP) of mature PZ and a C-terminal His-tag (referred to as PZ hereafter) was expressed in HEK293.EBNA cells21 and purified from the culture medium (Freestyle 293 medium; Invitrogen) by a HisTrap column and a subsequent S200 gel filtration column (GE Healthcare). After mixing ZPI with PZ, PZ/ZPI complex was purified by gel filtration. Human plasma PZ and human FXa were from Haematological Technologies. Rabbit brain phospholipids were purchased from Pel-Freez Biologicals. Mutagenesis of ZPI was performed using Quikchange kits (Stratagene), and ZPI mutants were prepared using the same procedure as for the wild-type.
Thermal stability
Thermal stability studies of ZPI variants by circular dichroism were performed in a JASCO J-810 spectropolarimeter. Samples of ZPI were prepared in 50 mM Tris-HCl and 0.1 M NaCl, pH 7.4, and thermal unfolding experiments were performed by monitoring the circular dichroism signal at 222 nm between 25°C and 98°C using a heating rate of 2°C/min at a concentration of 0.25 mg/mL. Thermal melting points (Tm) were calculated using an expression for a 2-state transition, as previously described.22
Stoichiometries and rates of inhibition
Stoichiometries of inhibition were determined by incubating increasing concentrations of ZPI or ZPI/PZ complex with a fixed concentration of FXa (2 μM). Reactions were incubated at 37°C for 15 minutes, and the residual amidolytic activity was determined by the addition of 0.2 mM S-2765 substrate in assay buffer (50 mM Tris-HCl, 100 mM NaCl, pH 7.4, with 0.1% polyethylene glycol 8000, 0.1 mg/mL bovine serum albumin). Linear regression analysis of the decrease in protease activity with an increasing concentration of ZPI yielded the estimates for the stoichiometry of inhibition as the intercept on the abscissa. The rates of inhibition of FXa by recombinant ZPI variants were determined at room temperature by a discontinuous assay procedure, as previously described.22 Briefly, under pseudo–first-order conditions, 10 μL of 2 or 5 μM ZPI variants was mixed with 10 μL of 10 nM FXa. The residual protease activity was determined at timed intervals by diluting the reaction mixture into 200 μL of the assay buffer containing 0.2 mM S-2765 substrate. The observed rate constant, kobs, for the reaction was obtained from the slope of a semilog plot of the residual protease activity against time, and the second-order rate constant, kapp, was calculated by dividing kobs by the initial ZPI concentration. The inhibition rates of ZPI on FXa in the presence of catalyzing concentrations of PZ and other cofactors (25 μM phospholipids and 5 mM Ca2+) were measured using a similar discontinuous procedure, as described.10 The kobs of the reactions with PZ were plotted against the PZ concentration (0 to ∼2 nM) with the linear regression fit yielding the second order rate constant of PZ-mediated ZPI inhibition of FXa.
Crystallization, data collection, and structure determination
Crystals were obtained by mixing 1 μL of PZ/ZPI complex (5 mg/mL) with 1 μL of precipitation solution (20%-30% polyethylene glycol 3000, 0.1 M citrate, pH 7) using the sitting drop method. Crystals grew to full size over 3 weeks. Crystals were quickly soaked (< 2 seconds) in cryosolution (the precipitation solution plus 15% ethylene glycol) and snap frozen in liquid nitrogen. Diffraction data of the PZ/ZPI complex were collected from a single crystal of PZ/ZPI at Diamond Synchrotron station I02 and processed with Mosflm.23 The structure was solved by molecular replacement with Phaser24 using antitrypsin (Protein Databank [PDB] 1QLP), from which the reactive loop was removed, and FXa (PDB 2UWP) as search models. One copy of the PZ/ZPI complex was found in the asymmetric unit. The structure was built in COOT25 and refined using Refmac26 and Phenix.refine27 (Table 1). The structure was validated by MolProbity28 and analyzed by EBI-PISA (European Bioinformatics Institute) for interface interactions.29 The first 38 residues of ZPI, which include many acidic amino acids, are disordered and were not built in the structure. The electron density map indicated that Asn185, 193, and 292 of PZ are glycosylated, but carbohydrates for 185 and 292 were not built due to weak density. PZ contains 7 pairs of disulfide bridges, of which 3 pairs are within EGF2, 2 pairs are within protease domain, and 2 pairs link EGF2 and the protease domain. In contrast, FXa with the same domains has 8 disulfide bonds (3 within EGF2 domain, 4 within the protease domain, and 1 linking the 2 domains). ZPI has no disulfides, but 2 free cysteines. To date, no crystals have been obtained from ZPI or PZ alone.
Data collection and refinement statistics
| . | PZ:ZPI complex (PDB 3F1S) . |
|---|---|
| Data collection | |
| Space group | P 21 21 21 |
| Cell dimensions | A = 68.56 Å, b = 96.51 Å, c = 117.08 Å, α = β = γ = 90° |
| Molecules in the asymmetric unit | One PZ and one ZPI |
| Resolution, Å | 44.63-2.3 (2.42-2.3) |
| I/σI | 15.4 (1.5) |
| Total number of measured reflections | 174 258 (10 388) |
| Total number of unique reflections | 32 582 (3462) |
| Completeness, % | 92.6 (69.4) |
| Multiplicity | 5.3 (3.0) |
| R merge, % | 6.7 (56.1) |
| R meas, % | 8.0 (71.1) |
| Refinement | |
| R work | 0.225 |
| R free | 0.270 |
| No. of atoms | |
| Protein | 5143 |
| Water | 82 |
| Citrate | 13 |
| Chloride | 1 |
| Average B factor, Å2 | 29.2 |
| RMS deviation | |
| Bond length, Å | 0.0058 |
| Bond angles, ° | 0.936 |
| Ramachandran plot (residues) | |
| Ramachandran favored, % | 97.32 |
| Ramachandran outliers | 0 |
| MolProbity score | 98th percentile* (n = 8909, 2.05-2.55 Å) |
| . | PZ:ZPI complex (PDB 3F1S) . |
|---|---|
| Data collection | |
| Space group | P 21 21 21 |
| Cell dimensions | A = 68.56 Å, b = 96.51 Å, c = 117.08 Å, α = β = γ = 90° |
| Molecules in the asymmetric unit | One PZ and one ZPI |
| Resolution, Å | 44.63-2.3 (2.42-2.3) |
| I/σI | 15.4 (1.5) |
| Total number of measured reflections | 174 258 (10 388) |
| Total number of unique reflections | 32 582 (3462) |
| Completeness, % | 92.6 (69.4) |
| Multiplicity | 5.3 (3.0) |
| R merge, % | 6.7 (56.1) |
| R meas, % | 8.0 (71.1) |
| Refinement | |
| R work | 0.225 |
| R free | 0.270 |
| No. of atoms | |
| Protein | 5143 |
| Water | 82 |
| Citrate | 13 |
| Chloride | 1 |
| Average B factor, Å2 | 29.2 |
| RMS deviation | |
| Bond length, Å | 0.0058 |
| Bond angles, ° | 0.936 |
| Ramachandran plot (residues) | |
| Ramachandran favored, % | 97.32 |
| Ramachandran outliers | 0 |
| MolProbity score | 98th percentile* (n = 8909, 2.05-2.55 Å) |
RMS indicates root mean square.
Among structures of comparable resolution, 100th percentile is the best, 0th percentile the worst.
Results
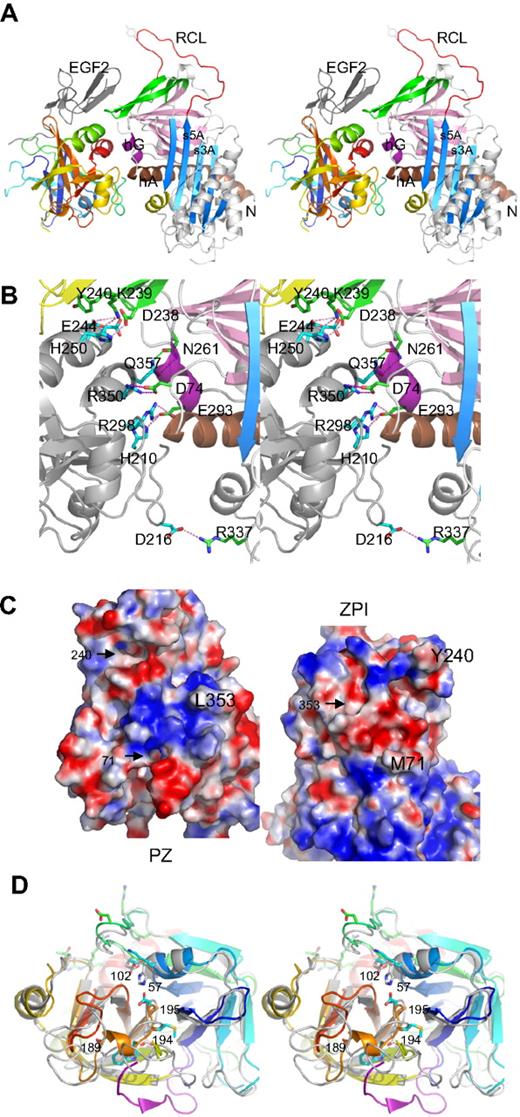
The structure of the complex shows (Figure 1A) that ZPI has a typical serpin fold, with 3 main β-sheets and a fully exposed reactive center loop (red). Both domains of PZ are clearly resolved, with the EGF2 domain closely packed with the serine protease domain, as observed in FXa and FVIIa.
Structure of PZ/ZPI complex. (A) Stereo view of PZ/ZPI complex. ZPI has a typical serpin fold with 3 main β-sheets: A (light blue), B (magenta), and C (green), and a fully exposed reactive loop (red). The protease domain of PZ is colored in rainbow from N (blue) to C terminus (red), and EGF2 domain is in dark gray. The P1 residue Y387 is shown in sticks. (B) Stereo view of the PZ/ZPI-binding interface. The PZ-binding site on ZPI is centered around hG (purple) and the C terminus of hA (brown). The salt bridges are shown in magenta dashes, and hydrogen bonds in green dashes. (C) The open-up view of the PZ/ZPI interface showing the complementary electrostatic surfaces of PZ (left) and ZPI (right) with positive charges in blue and negatives in red. In addition, the hydrophobic residues (Y240 and M71 of ZPI, and L353 of PZ) dock into corresponding surface cavities (black arrows). Other residues, such as L102, P103, M211, and K245 of PZ and L289, K284, and A290 of ZPI, also make significant hydrophobic interactions in the interface. (D) Stereo view of the overlaid structures of PZ (colored in rainbow) and FXa (PDB 2UWP, in gray). Ser195, Asp194, His57, and D189 of FXa are replaced (PZ numbering in brackets) by Asp313, Met312, Lys178, and Ala307 in PZ, respectively. The S1 pocket of PZ is filled by a 310 helix formed by residues 189-192 (307-310), a bulky Trp at 193 (311), and Q216 (334), and the activation peptide of FXa is in purple. In comparison with FXa, PZ lacks the disulfide bond of 191-220 around the S1 pocket and the disulfide bond22-27 linking the activation peptide of FXa, but has an extra disulfide bridge linking EGF2 to the protease domain (data not shown).
Structure of PZ/ZPI complex. (A) Stereo view of PZ/ZPI complex. ZPI has a typical serpin fold with 3 main β-sheets: A (light blue), B (magenta), and C (green), and a fully exposed reactive loop (red). The protease domain of PZ is colored in rainbow from N (blue) to C terminus (red), and EGF2 domain is in dark gray. The P1 residue Y387 is shown in sticks. (B) Stereo view of the PZ/ZPI-binding interface. The PZ-binding site on ZPI is centered around hG (purple) and the C terminus of hA (brown). The salt bridges are shown in magenta dashes, and hydrogen bonds in green dashes. (C) The open-up view of the PZ/ZPI interface showing the complementary electrostatic surfaces of PZ (left) and ZPI (right) with positive charges in blue and negatives in red. In addition, the hydrophobic residues (Y240 and M71 of ZPI, and L353 of PZ) dock into corresponding surface cavities (black arrows). Other residues, such as L102, P103, M211, and K245 of PZ and L289, K284, and A290 of ZPI, also make significant hydrophobic interactions in the interface. (D) Stereo view of the overlaid structures of PZ (colored in rainbow) and FXa (PDB 2UWP, in gray). Ser195, Asp194, His57, and D189 of FXa are replaced (PZ numbering in brackets) by Asp313, Met312, Lys178, and Ala307 in PZ, respectively. The S1 pocket of PZ is filled by a 310 helix formed by residues 189-192 (307-310), a bulky Trp at 193 (311), and Q216 (334), and the activation peptide of FXa is in purple. In comparison with FXa, PZ lacks the disulfide bond of 191-220 around the S1 pocket and the disulfide bond22-27 linking the activation peptide of FXa, but has an extra disulfide bridge linking EGF2 to the protease domain (data not shown).
The ZPI/PZ-binding interface
The binding site in PZ is centered around the anion-binding (heparin-binding) site of the serine protease near the C-terminal helix, and the binding site in ZPI is centered on helix G (hG, purple) and the C terminus of helixA (hA; Figure 1A-B; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The binding surfaces are fully complementary with the positively charged patches of one molecule matched by negatively charged patches from the other (Figure 1C). Ten residues (H250, D246, E244, R350, R298, and H210 of PZ and K239, D238, D74, and D293 of ZPI) form 3 clusters of salt bridges (Figure 1B; supplemental Figure 1) with those formed by R298, R350 of PZ being largely buried by surrounding hydrophobic interactions. The hydrophobic residues of Y240 and M71 of ZPI and L353 of PZ readily dock into cavities of the corresponding binding surfaces. Y240, located in the connecting loop between strands 3C and 4C (green) of ZPI, docks into the hydrophobic cavity formed by the EGF2 and the protease domains of PZ. The PZ and ZPI interface is further stabilized by 9 hydrogen bonds; notably, T297 of PZ forms both main-chain to main-chain and side-chain to main-chain hydrogen bonds to M71 in hA of ZPI (supplemental Figure 1).
The pseudo-protease domain of PZ
PZ has 33% sequence identity with FXa30 and has a typical serine protease fold (Figure 1A,D), with its inactivity resulting from the substitution of 2 key residues (S195 and H57, chymotrypsin numbering) of the catalytic cavity by D313 and K178 together with a completely different configuration around the S1 pocket and the oxyanion hole. PZ cannot form the typical interaction in the oxyanion hole of trypsin-like proteases, where I16 forms stabilizing interactions with D19431 because its activation peptide is 5 residues shorter than that of FXa, and PZ has a methionine (M312) at position 194 with its side chain pointing in the direction opposite to that of D194 of FXa. The S1 pocket of PZ is occupied by a glutamine (Q334) at position 216, a bulky tryptophan (W311) at 193 (G216 and G193 in FXa), and a half turn of 310 helix of residues 189-192 (residues 307-310 in PZ). Superimposition of PZ with thrombin shows that residues H210, R212, R350, L353, and Q357 in the binding interface of PZ correspond to H91, R93, R233, K236, and K240 of the anionic heparin-binding site (exocite II) of thrombin.32 Thus, PZ has adapted a cofactor-binding site of the serine protease for its own alternative cofactor function.
Structural features of ZPI
The relative positions of the reactive loop and β-sheet A of ZPI resemble those of antitrypsin and activated antithrombin (Figure 2A) with the reactive loop fully extended. ZPI, however, has a lengthened helix A (brown in Figure 1A), which is unusually bent, most likely due to the interactions of W46 with a hydrophobic pocket formed by P114 and surrounding leucines 112, 119, 120, and 123.
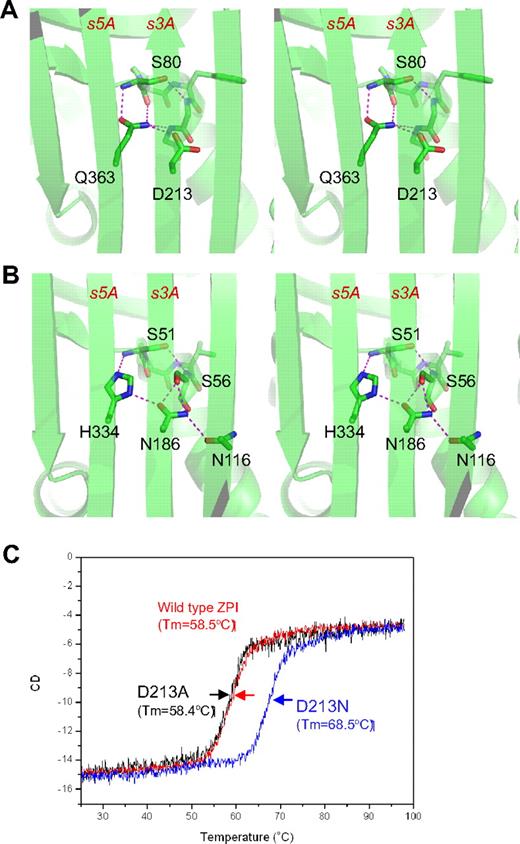
The unusual shutter region of ZPI. ZPI has a negatively charged D213 buried in the hydrophobic shutter region, where it forms only 1 hydrogen bond with neighboring Q363 (A). Antitrypsin has a different hydrogen-bonding network around the shutter region. This position is occupied by a conserved Asn186, which forms 4-hydrogen bonds with neighboring residues (B). The labeled residues are shown in sticks with carbon atoms in green, nitrogen in blue, and oxygen in red. Replacement of D213 of ZPI with Asn increased the Tm of ZPI by more than 10°C, as measured by circular dichroism (C), whereas D213A mutant has the same Tm as that of wild-type ZPI.
The unusual shutter region of ZPI. ZPI has a negatively charged D213 buried in the hydrophobic shutter region, where it forms only 1 hydrogen bond with neighboring Q363 (A). Antitrypsin has a different hydrogen-bonding network around the shutter region. This position is occupied by a conserved Asn186, which forms 4-hydrogen bonds with neighboring residues (B). The labeled residues are shown in sticks with carbon atoms in green, nitrogen in blue, and oxygen in red. Replacement of D213 of ZPI with Asn increased the Tm of ZPI by more than 10°C, as measured by circular dichroism (C), whereas D213A mutant has the same Tm as that of wild-type ZPI.
ZPI also has an unusual hydrogen bond network at the shutter region with the conserved Asn186 (antitrypsin numbering)22 of serpins being replaced by an aspartate (D213 in ZPI of all known species). This results in a changed H-bond pattern and the burial of the acidic residue D213 into a largely hydrophobic environment beneath β-sheet A (Figure 2A). D213 can form only 1 hydrogen bond in ZPI, whereas in other serpins, a conserved Asn occupies this position and forms 4 hydrogen bonds in antitrypsin (Figure 2B) and 3 hydrogen bonds in PAI-1. To check the significance of this unusual packing, we mutated D213 to an Asn and to an Ala and assessed the effects of each mutation on the thermal stability of ZPI and its inhibitory function. The ZPI-D213A mutant has a similar Tm of 58.4°C to the wild-type, indicating that the hydrogen bond formed by the side chain of Asp213 in wild-type ZPI could partly compensate for the free energy required for the burial of a negative charge in the shutter region. In contrast, the ZPI-D213N mutant has a Tm of 68.4°C, which is 10°C higher than that of wild-type ZPI (Figure 2C). In the absence of PZ, ZPI-D213A inhibits FXa with a similar inhibitory rate (kapp) and stoichiometry of inhibition (SI) as the wild-type, whereas the inhibition rate of ZPI-D213N decreased approximately 4-fold with SI increased 2-fold (Table 2). In the presence of PZ, there is a slight increase in SI (from 2.1 to 2.7) when wild-type ZPI interacts with FXa, which is consistent with the previous report10 ; however, there are dramatic increases in SIs of both ZPI-D213A (3.8-fold) and ZPI-D213N (3.5-fold). In other words, it takes only 2-3 molecules of wild-type ZPI to inhibit 1 molecule of FXa, whereas it will require 17 molecules of ZPI-D213N to inhibit 1 of FXa in the presence of PZ. Therefore, we conclude that D213, like Lys335 in antitrypsin,33 is designed to maintain the metastable conformation of ZPI and to allow the swift opening of β-sheet A that is necessary for protease inhibition, especially when PZ binds to ZPI. As expected, the metastability of ZPI will be tipped into instability by mutation to a bulkier charged residue in the same (shutter) region, as is observed with the natural mutation Q363R (supplemental Figure 2), which clinically predisposes to venous thrombosis.16 Similarly, a homologous mutation H338R in neuroserpin destabilizes the neuroserpin with consequent polymerization and subsequent neuron cell damage.34 The position and likely structural consequences of other known natural mutations/polymorphisms of PZ and ZPI associated with thrombosis are summarized in supplemental Figure 2.
Inhibitory activities of ZPI variants toward FXa and thermal stability of ZPI shutter region mutants
| . | Tm (°C) . | kapp (M−1s−1) . | SI . | kapp × SI(M−1s−1) . | SI (+PZ) . |
|---|---|---|---|---|---|
| ZPI | 58.5 ± 0.2 | 8.8 ± 2.1 × 103 | 2.1 ± 0.2 | 1.8 × 104 | 2.7 ± 0.2 |
| D213A | 58.4 ± 0.2 | 8.3 ± 2.4 × 103 | 1.9 ± 0.3 | 1.6 × 104 | 7.4 ± 0.2 |
| D213N | 68.5 ± 0.2 | 2.1 ± 0.7 × 103 | 4.6 ± 0.3 | 9.7 × 103 | 17.3 ± 0.2 |
| E231A | * | 3.7 ± 0.9 × 103 | 2.1 ± 0.3 | 7.8 × 103 | |
| D233A | 3.7 ± 1.4 × 103 | 1.9 ± 0.2 | 7.0 × 103 | ||
| E313A | 1.5 ± 0.3 × 103 | 4.3 ± 0.4 | 6.4 × 103 | ||
| K253A | 3.9 ± 0.5 × 103 | 1.7 ± 0.1 | 6.6 × 103 | ||
| K260A | 6.2 ± 1.6 × 103 | 1.7 ± 0.2 | 1.1 × 104 | ||
| K308/R310A | 3.6 ± 0.5 × 103 | 1.9 ± 0.4 | 6.8 × 103 |
| . | Tm (°C) . | kapp (M−1s−1) . | SI . | kapp × SI(M−1s−1) . | SI (+PZ) . |
|---|---|---|---|---|---|
| ZPI | 58.5 ± 0.2 | 8.8 ± 2.1 × 103 | 2.1 ± 0.2 | 1.8 × 104 | 2.7 ± 0.2 |
| D213A | 58.4 ± 0.2 | 8.3 ± 2.4 × 103 | 1.9 ± 0.3 | 1.6 × 104 | 7.4 ± 0.2 |
| D213N | 68.5 ± 0.2 | 2.1 ± 0.7 × 103 | 4.6 ± 0.3 | 9.7 × 103 | 17.3 ± 0.2 |
| E231A | * | 3.7 ± 0.9 × 103 | 2.1 ± 0.3 | 7.8 × 103 | |
| D233A | 3.7 ± 1.4 × 103 | 1.9 ± 0.2 | 7.0 × 103 | ||
| E313A | 1.5 ± 0.3 × 103 | 4.3 ± 0.4 | 6.4 × 103 | ||
| K253A | 3.9 ± 0.5 × 103 | 1.7 ± 0.1 | 6.6 × 103 | ||
| K260A | 6.2 ± 1.6 × 103 | 1.7 ± 0.2 | 1.1 × 104 | ||
| K308/R310A | 3.6 ± 0.5 × 103 | 1.9 ± 0.4 | 6.8 × 103 |
K308/R310A is a double mutant in which both K308 and R310 were replaced by Ala. In the presence of PZ, phospholipids, and Ca2+, these mutants also have similar ∼ 2- to 3-fold decreased inhibitory activity toward FXa compared with wild-type (data not shown). Tm of ZPI, second order inhibitory rate constant (kapp), and stoichiometry of ZPI-FXa reaction (SI) were measured, as described in “Methods.”
Missing values not determined.
Interactions between ZPI and proteases
The specificity of inhibition by serpins depends on the interaction of their reactive loop with the active site of target proteases, with an added specificity resulting from exosite interactions between the body of the serpin and various peptide loops of the protease. Thus, ZPI is also a potent inhibitor of activated factor XI (fXIa) in an interaction that does not require PZ or phospholipids.9 Positively charged residues in the autolysis loop (residues 143-154, chymotrypsin numbering) of both FXa and FXIa are known to play an important role in interacting with ZPI, especially R143 in FXa.35,36 Superposition of ZPI with the antithrombin-FXa Michaelis complex37 indicates that FXa could readily dock onto the top of the ZPI with an orientation similar to that in its complex with antithrombin. The negatively charged region on the top of ZPI, formed by residues E231, D233, and E313, will be well positioned to form exosite interactions with the autolysis loop of factor Xa (Figure 3A-B). Replacement of these residues in this study with Ala (Table 2) impairs the inhibition between ZPI and FXa with second order rate constants decreased by approximately 2- to 3-fold. Interestingly, the other half of the surface on the top of ZPI formed by K253, K260, K308, and R310 is positively charged (Figure 3B). Replacement of these residues with Ala also resulted in an approximately 2-fold decrease in inhibitory activity (Table 2). Thus, these residues are likely to be involved in the docking of FXa on ZPI through interactions with the acidic residues of the 36 loop (E36, E37, or E39) of FXa.
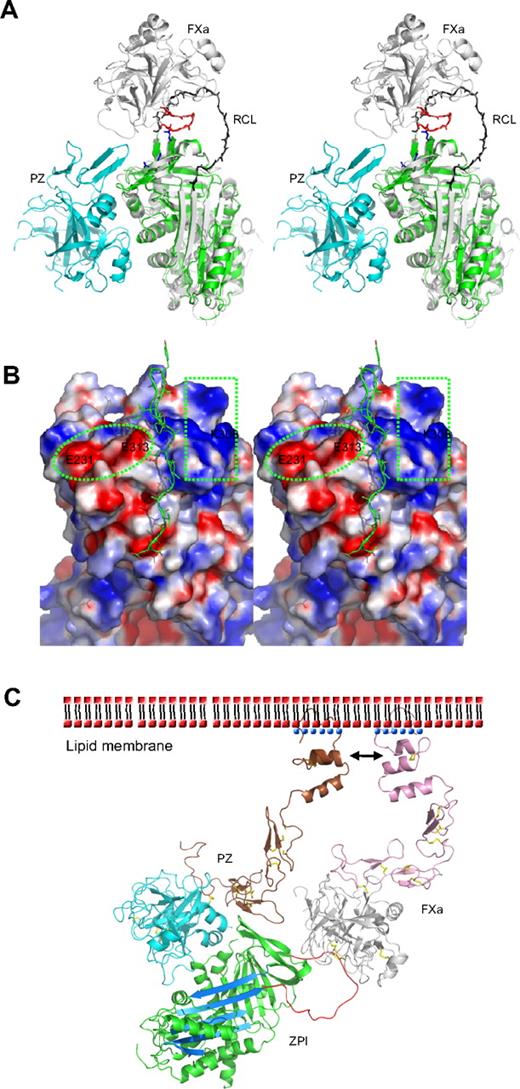
Protease inhibition mechanism of ZPI mediated by PZ. (A) PZ/ZPI complex overlaid with antithrombin/FXa (S195A) Michaelis complex (PDB 2GD4) using the serpin fold. For clarity, the reactive center loop of ZPI (green) was removed. The reactive center loop of antithrombin is shown in black. The 3 acidic residues (E313, E231, and D233) near the FXa docking site are shown in sticks (blue), and the autolysis loop (143-154) of FXa is in red. (B) The electrostatic surface on the top of of ZPI shows a negatively charged surface on the left (circled), formed by D233, E231, and E313, and a positive patch on the right (in rectangle), formed by residues K253, K260, H265, K308, and R310 of ZPI. The autolysis loop of FXa or FXIa most likely binds to the negative patch (left), whereas the 36 loop will interact with the positive charges. The reactive center loop is shown in lines. Although it has been indicated previously that substitution of glutamates of the 36 loop with glutamines has little effect on ZPI/FXa interaction, we speculate these mutations will only weaken, but not abolish these exosite interactions. Because the reactive loop of ZPI is 4 residues shorter than that of antithrombin, it is expected that FXa will dock onto ZPI in a slightly different orientation to that in antithrombin-FXa complex, which would allow potential interactions between the 36 loop of FXa and the positive surface on the top of ZPI. (C) Model of the ternary complex of PZ/ZPI/FXa on the phospholipid membrane surface. The relative positions of Gla, EGF1, EGF2, and protease domain of FXa were modeled using the coordinates of full-length FIXa (PDB 1PFX).41 Full-length PZ was modeled similarly, but the Gla domain (brown) was rotated approximately 180° along the flexible linker between EGF1 and Gla domains, which allows its interactions (indicated by arrows) with the Gla domain of FXa on the membrane surface, as suggested.11,15 ZPI is colored in green with reactive loop in red and A-β-sheet in light blue. The N-terminal domains (Gla, EGF1, and EGF2) are in brown in PZ and in pink in FXa. Ca2+ is shown as blue dots. The head (phosphate) groups of phospholipids are shown as red dots, and the hydrophobic tails as black dashes. The disulfide bonds are shown in yellow sticks. In the ternary complex model, the EGF2 domain of PZ is in close proximity to the protease domain of FXa, but it is unclear whether they can form further stabilizing interactions within the complex.
Protease inhibition mechanism of ZPI mediated by PZ. (A) PZ/ZPI complex overlaid with antithrombin/FXa (S195A) Michaelis complex (PDB 2GD4) using the serpin fold. For clarity, the reactive center loop of ZPI (green) was removed. The reactive center loop of antithrombin is shown in black. The 3 acidic residues (E313, E231, and D233) near the FXa docking site are shown in sticks (blue), and the autolysis loop (143-154) of FXa is in red. (B) The electrostatic surface on the top of of ZPI shows a negatively charged surface on the left (circled), formed by D233, E231, and E313, and a positive patch on the right (in rectangle), formed by residues K253, K260, H265, K308, and R310 of ZPI. The autolysis loop of FXa or FXIa most likely binds to the negative patch (left), whereas the 36 loop will interact with the positive charges. The reactive center loop is shown in lines. Although it has been indicated previously that substitution of glutamates of the 36 loop with glutamines has little effect on ZPI/FXa interaction, we speculate these mutations will only weaken, but not abolish these exosite interactions. Because the reactive loop of ZPI is 4 residues shorter than that of antithrombin, it is expected that FXa will dock onto ZPI in a slightly different orientation to that in antithrombin-FXa complex, which would allow potential interactions between the 36 loop of FXa and the positive surface on the top of ZPI. (C) Model of the ternary complex of PZ/ZPI/FXa on the phospholipid membrane surface. The relative positions of Gla, EGF1, EGF2, and protease domain of FXa were modeled using the coordinates of full-length FIXa (PDB 1PFX).41 Full-length PZ was modeled similarly, but the Gla domain (brown) was rotated approximately 180° along the flexible linker between EGF1 and Gla domains, which allows its interactions (indicated by arrows) with the Gla domain of FXa on the membrane surface, as suggested.11,15 ZPI is colored in green with reactive loop in red and A-β-sheet in light blue. The N-terminal domains (Gla, EGF1, and EGF2) are in brown in PZ and in pink in FXa. Ca2+ is shown as blue dots. The head (phosphate) groups of phospholipids are shown as red dots, and the hydrophobic tails as black dashes. The disulfide bonds are shown in yellow sticks. In the ternary complex model, the EGF2 domain of PZ is in close proximity to the protease domain of FXa, but it is unclear whether they can form further stabilizing interactions within the complex.
Is the PZ-mediated inhibition of FXa by ZPI an allosteric or template mechanism?
Heparin binds to antithrombin and accelerates the inhibition of FXa by an allosteric mechanism whereby heparin induces a conformational change in antithrombin with the full expulsion of its partially inserted reactive loop. But heparin also uses a template mechanism to accelerate the inhibition of thrombin, with the colocalization of thrombin and antithrombin on the same heparin chain. It has been previously proposed that PZ-mediated inhibition of FXa by ZPI is likely to involve a template mechanism, as the acceleration of inhibition of FXa by ZPI is critically dependent on the presence of both the Gla domain of PZ and the Gla domain of FXa, as well as phospholipids and Ca2+.5,11,15 Such Gla domain:phospholipid membrane interactions play a critical role in the assembly of many protein complexes of the blood coagulation system, with the Gla domain, stabilized by a linear array of Ca2+ ions, binding to phospholipid membranes through a solvent-exposed hydrophobic loop (ω-loop).38 The Gla domain of PZ has been shown to tightly bind to phospholipid vesicles39 and form a stabilizing interaction with FXa on the membrane surface.11,15 Our structural findings in this study support this template mechanism, as PZ is seen to bind to ZPI on the opposite side of the serpin molecule to the site where heparin binds to, and allosterically activates, antithrombin. Further evidence against a conformational activation of ZPI comes from our observation that the PZ fragment without the Gla domain, as seen in this study in the crystal structure, is unable to accelerate the inhibition of FXa by ZPI (data not shown). Therefore, as modeled in Figure 3C, the PZ promotion of the ZPI-FXa interaction will result from the anchoring of its Gla domain onto phospholipid surfaces, bringing the bound ZPI into close proximity to the Gla-anchored FXa with the formation of a ternary complex of PZ/ZPI/FXa.5,9,11
Discussion
The unique “suicidal” inhibitory mechanism of the serpins has the added advantage of allowing the modulation and localization of their inhibitory activity by cofactors.3,4 Heparin induces a conformational activation of antithrombin and localizes it onto the endothelium,3 and vitronectin anchors PAI-1 onto the surface of fibrin and stabilizes its active conformation.40 But, by comparison, the cofactor activation of ZPI appears to be a consequence of proximation rather than conformation, as activation is dependent on the possession by PZ of a Gla domain. PZ is a surrogate FX that has lost its catalytic activity, but gained a binding site for ZPI. Similarly, ZPI has uniquely adapted the serpin structure to maintain effective inhibitory activity even when its binding to PZ tightens its conformational flexibility. The linkage with PZ will inherently localize and anchor the bound ZPI in a proximity to FXa predictably optimal for inhibition, with the Gla domains of PZ and FXa binding to the same phospholipid surface (Figure 3C). Thus, the structure of PZ complexed with ZPI explains the apparent contradiction of the promotion of inhibition of activated FX by a FX-like molecule.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Professor Randy J. Read for expertise in crystallography. The coordinates have been deposited in the Protein Databank (www.rcsb.org, PDB 3F1S).
This work was supported by the British Heart Foundation and the Isaac Newton Trust.
Authorship
Contribution: Z.W. and Y.Y. performed the research; R.W.C. reviewed the data and wrote the paper; and A.Z. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aiwu Zhou, Department of Haematology, University of Cambridge, Cambridge Institute for Medical Research, Hills Rd, CB2 0XY, Cambridge, United Kingdom; e-mail: awz20@cam.ac.uk.
References
Author notes
Z.W. and Y.Y. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal