DNA methylation is a common mode of gene silencing in cancer, and MDS remains the poster child for a disease in which targeting the DNMT enzymes leads to clinical benefit.1-4 However, some important questions remain unanswered, 2 of which are directly addressed in this issue of Blood by Figueroa and colleagues5 : What pivotal gene sets are silenced by DNA methylation in MDS and de novo AML, a question that begs a reference to the patterns of normal marrow cells on a genome-wide scale, and what global changes in DNA methylation can be achieved when treating patients with DNMT inhibitors?
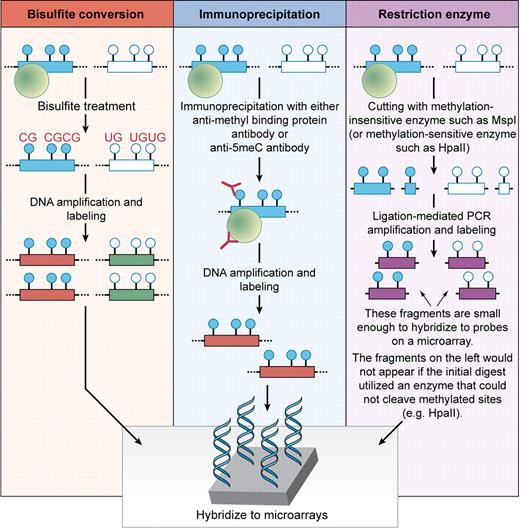
Genome-wide microarray technologies have been widely used for well over a decade and allow one to comprehensively assess gene expression. More recently, they have been applied to the study of DNA methylation.6 There are several ways to assess for DNA methylation changes using microarray-based platforms (see figure). The first uses hybridization of DNA that has been treated with sodium bisulfite, which converts unmethylated cystosines to uracils while methylated cytosines remain cytosines. A second approach uses antibodies directed against 5-methyl-cytosines or methyl-binding proteins to immunoprecipitate methylated DNA fragments. A third uses treatment of the DNA with methylation-specific restriction enzymes followed by hybridization to microarrays. In their report, Figueroa et al used an example of the latter technique called HELP (HpaII tiny fragment enrichment by ligation mediated PCR) to clarify the DNA methylation changes that exist in bone marrow samples ranging from normal to myelodysplasia (MDS) to de novo leukemia.7
Schema of commonly used approaches to assess global DNA methylation changes. The closed blue circles represent hypermethylated CpG dinucleotides in the promoter region of a given gene and the open circles represent unmethylated CpG dinucleotides for the same gene. The green circles represent methyl-binding proteins that bind specifically to methylated DNA sequences. In bisulfite conversion approaches, the DNA is chemically modified, which allows for generation of distinct sequences based on the original DNA methylation status of cytosines. In immunoprecipitation approaches, the DNA is immunoprecipitated with an antibody (shown in red) specific to methyl-binding proteins or 5-methyl-cytosines to enrich for segments with DNA methylation. In restriction enzyme approaches such as HELP used by Figueroa et al,5 the DNA is digested with enzymes that are either insensitive (cut methylated sequences) or sensitive (do not cut methylated sequences) to DNA methylation. In each assay, the DNA is labeled and amplified prior to hybridization to microarrays. Professional illustration by Kenneth X. Probst.
Schema of commonly used approaches to assess global DNA methylation changes. The closed blue circles represent hypermethylated CpG dinucleotides in the promoter region of a given gene and the open circles represent unmethylated CpG dinucleotides for the same gene. The green circles represent methyl-binding proteins that bind specifically to methylated DNA sequences. In bisulfite conversion approaches, the DNA is chemically modified, which allows for generation of distinct sequences based on the original DNA methylation status of cytosines. In immunoprecipitation approaches, the DNA is immunoprecipitated with an antibody (shown in red) specific to methyl-binding proteins or 5-methyl-cytosines to enrich for segments with DNA methylation. In restriction enzyme approaches such as HELP used by Figueroa et al,5 the DNA is digested with enzymes that are either insensitive (cut methylated sequences) or sensitive (do not cut methylated sequences) to DNA methylation. In each assay, the DNA is labeled and amplified prior to hybridization to microarrays. Professional illustration by Kenneth X. Probst.
The authors first focused on determining the DNA methylation differences between CD34− and CD34+ cells from bone marrow samples from both normal volunteers and those with MDS. Surprisingly, they found that there was a very high correlation between methylation patterns in CD34− and CD34+ cells within each group. This allowed the authors to examine DNA methylation changes in additional CD34− MDS clinical samples with greater confidence that any differences in methylation between these cells and those from normal or de novo acute myeloid leukemia (AML) samples would not be solely explained by lineage differences. They found that indeed there is a continuum of DNA methylation with the lowest amount of DNA methylation present in normal bone marrow cells, de novo AML cells in the middle, and MDS cells at the methylated end of the spectrum. Although it is unclear whether the DNA hypermethylation seen in MDS may account for the responsiveness of this disease to DNA methyltransferase (DNMT) inhibitors, it is intriguing to speculate that these changes may play a role.
To determine what changes are achievable with epigenetic therapy in humans, the authors next examined bone marrow biopsy specimens taken either before or after treatment with 5-azacytadine in combination with the histone deacetylase inhibitor entinostat; previously, the combination of these 2 classes of agents was shown to result in synergy of reexpression of DNA hypermethylated genes.8 They found that at early time points, significant changes in DNA methylation were achieved. The acquisition of samples at a very early time point was important because if they had waited too long, any differences seen might reflect an alteration in marrow composition (eg, reduced numbers of neoplastic cells). The numbers of patients in their series precluded an assessment of genes that might predict eventual hematologic response or clinical benefit, but their results do suggest the potential of this approach to identify predictors of eventual response or resistance in larger series. Also, data on the mRNA expression changes after treatment were not explored in their report, which is also crucial for understanding mechanisms of response/resistance.
It is clear that MDS is a distinct disease from de novo AML, but there is a good deal of heterogeneity in both categories of disease. Platforms and approaches such as those used by Figueroa et al enable one to peer through an informative looking glass to better clarify epigenetic differences between these 2 diseases based on DNA methylation changes. The same approach can clearly be applied serially in patient populations at risk of evolution to AML, studies that may reveal key epigenetic factors underlying clonal evolution of MDS to AML. When combined with analyses of gene expression and functional assessments of chromatin in normal bone marrow cells, MDS cells, and de novo leukemia cells, these methods should clarify whether there are critical epigenetic events that transform normal hematopoietic cells, induce genetic instability and clonal evolution, enhance survival of new clones, and induce resistance to therapy.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal