Abstract
In this issue of Blood, Gentles and colleagues used a computational model to investigate FL and transformed DLBCL, and identified the acquisition of an ESC-like signature as a key feature of transformation.1
Although follicular lymphoma (FL) is an indolent tumor, up to one-third of cases can progress to a more aggressive and usually fatal disease, with most cases transforming to diffuse large B-cell lymphoma (DLBCL). Transformation appears to be a complex process involving genomic, transcriptional, and epigenetic mechanisms that have lent themselves to whole-genome screening approaches. Unfortunately, analysis of gene expression to date has largely been restricted to the expression of single genes and cannot take into account the full complexity of the disease. Newer computational models analyze the biology of tumors by considering gene interaction networks and molecular pathways rather than separate genes. These approaches have already been applied to other epithelial tumors identifying signatures that drive cancer progression.2 In their paper, Gentles and colleagues performed a re-analysis of gene expression data from a study of FL-transformed DLBCL3 and generated a network that focused on the gene expression programs relevant to the transformation process. The power of models such as these is that they can capture modules—a group of genes regulated in concert by a shared regulation program—and thus the key events regulating transformation. It is reassuring to see that, within the module network, they also captured events already known to be important in the transformation process.
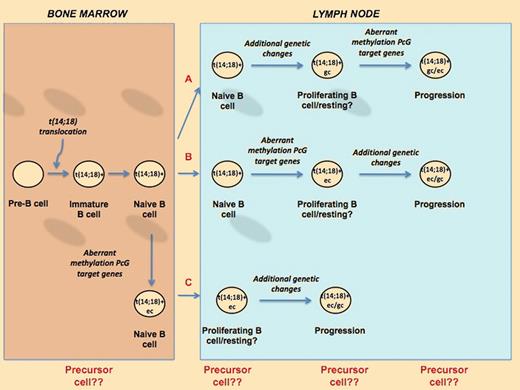
Schematic model of the possible patterns of evolution of FL. The cell of origin of FL and transformed DLBCL could be a cell that has already acquired the t(14;18) chromosomal aberration. After migrating from the bone marrow to the lymph node, this cell could acquire further genetic changes (gc) and subsequently the epigenetic aberrant methylation of the PcG genes (ec) before progressing to tumor (A). Alternatively, epigenetic changes of PcG target genes can occur in the BM (B) or in the lymph node (C) and subsequently evolve into FL or DLBCL after additional genomic events.
Schematic model of the possible patterns of evolution of FL. The cell of origin of FL and transformed DLBCL could be a cell that has already acquired the t(14;18) chromosomal aberration. After migrating from the bone marrow to the lymph node, this cell could acquire further genetic changes (gc) and subsequently the epigenetic aberrant methylation of the PcG genes (ec) before progressing to tumor (A). Alternatively, epigenetic changes of PcG target genes can occur in the BM (B) or in the lymph node (C) and subsequently evolve into FL or DLBCL after additional genomic events.
They first observed that multiple modules contained genes expressed in embryonic stem cells (ESCs). Further validation comes from the finding that the ESC1 module is a predictor of survival associated with propensity for transformation and that the ESC-like module was enriched for genes induced specifically by MYC overexpression in a transgenic mouse model of lymphoma. Finally, they derived a 3-module model. Two modules had an ESC-like related signature and one a stromal signature. These models were able to stratify the patients in 2 different cohorts that discriminated FL patients prone to transform to DLBCL from those with a better clinical course. The value of such models is that they allow hypothesis generation regarding pathways important in lymphomagenesis that may enable better use of available therapies and help to identify putative targets of novel therapies.
The ESC-like signatures are in support of a model of a lymphoma precursor cell, which may arise at an earlier stage of B-cell development than the germinal center. Of particular relevance to the process of FL transformation is whether the transformed lymphoma arises directly from FL, or might arise from additional genetic events in a lymphoma progenitor cell. Such a model is supported in findings by Carlotti et al4 and those of Ruminy et al5 wherein intraclonal diversity, as defined by the patterns of somatic hypermutation occurring in FL and transformed lymphoma, is more complex than previously described.
Among the core signatures identified in their analysis on modules closely associated with histologic transformation were enrichment for genes usually repressed in ESC through targeting by polycomb group (PcG) complexes. The role of PcG target genes in FL and transformation is further supported by the recent finding that these target genes are significantly overrepresented among aberrantly hypermethylated genes in FL6 and in aggressive lymphomas.7 The similarities in the patterns of methylation in sequential biopsies taken from FL and subsequent transformed lymphoma suggest that the widespread methylation represents an early event in lymphogenesis.6 Intriguingly, such a model is also supportive of a PcG-regulated process of lymphoma generating changes arising in an earlier precursor cell, whereby the methylation of these genes is an early event that “locks in” the stem cell–like phenotype. These findings suggest that alterations in methylation represent a common feature of the initiating events in lymphoma.
The genetic/epigenetic characteristics of the precursor cell, the sequential steps leading to its transformation into a lymphoma cell, the genes driving these processes, and the anatomical sites where the lymphoma precursor cells originate are still a matter of speculation. As outlined in the figure, future studies performed on purified populations of cells or single cells should show whether these B precursors have features of an immature cell or whether they have already acquired additional genetic lesions or an aberrant methylation of PcG target genes, clarifying their role in tumor progression and transformation.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■