Abstract
The constitutive activation of nuclear factor-κB (NF-κB) has been implicated in tumorigenesis of lymphoid malignancies. We have previously shown that chromosome 6q was frequently deleted in ocular marginal zone B-cell lymphoma and identified TNFAIP3/A20, a negative regulator of NF-κB pathways, as the primary target for 6q deletion. In the study reported here, we extended the analysis to other subsets of non-Hodgkin lymphomas and found that A20 is frequently deleted in mantle cell lymphoma and diffuse large B-cell lymphoma. Importantly, A20 promoter methylation or gene mutation is also frequently detected in these lymphomas, raising the possibility that inactivation of A20 may be involved in lymphomagenesis. To address this question, we conducted overexpression experiments in lymphoma cell lines with A20 deletion and down-regulated expression of A20 with an siRNA technique in Epstein-Barr virus–infected lymphoblastoid cell lines. These experiments found that overexpression of A20 induced apoptosis and silencing of A20 was associated with resistance to apoptosis and enhanced clonogenicity. The cells with down-regulated A20 exhibited enhanced NF-κB activities, which may account for the observed effects. These results indicate that our study provides a novel insight into molecular mechanisms leading to lymphoma and that specific targeting of NF-κB pathways may be advantageous for treatment.
Introduction
Chromosome band 6q deletion is one of the most common genomic alterations in non-Hodgkin lymphomas. Because the deleted regions are highly heterogeneous for different types of lymphoma, however, the question of which genes are responsible for oncogenesis remains unanswered. We previously demonstrated that chromosome band 6q23.3-q24.1 was deleted in 37.5% of ocular adnexal mucosa-associated lymphoid tissue (MALT) lymphomas, and these cases exhibited no MALT1 gene translocations, suggesting that the gene in the deleted region may play a pivotal role in lymphomagenesis.1 Among the genes mapped to the region, we have identified the tumor necrosis factor, alpha-induced protein 3 (TNFAIP3) gene (also known as A20) as the primary target in these cases.2
Although functions of A20 have not yet been fully explored, it is known to negatively regulate the functions of the transcription factor, nuclear factor-κB (NF-κB). A20 was originally identified as a tumor necrosis factor (TNF)–induced gene in human umbilical vein endothelial cells,3 and is capable of inhibiting both TNF-induced apoptosis and NF-κB activation via inhibiting IκB phosphorylation in response to TNF.4 Moreover, A20 knockout mice fail to terminate TNF-induced NF-κB activation, develop severe inflammation and cachexia, and die during the neonatal period.5 The fact that A20 controls NF-κB–activating pathways has led to the hypothesis that it could be involved in lymphomagenesis.
Constitutive aberrant activation of NF-κB signaling has been detected in various lymphoid malignancies and plays a key role in the development of these malignancies. However, the genetic and molecular mechanisms underlying this activation are largely unknown. Oncogenic gene mutations or chromosomal translocations in NF-κB signaling could lead to deregulation of NF-κB, and/or inactivation of negative regulators may also affect aberrant NF-κB activation. Because deletion of chromosome 6q is a common genomic alteration in non-Hodgkin lymphomas, A20 would be such a candidate gene for other subtypes of non-Hodgkin lymphomas, as it does in the case of MALT lymphoma.
In this study, we first investigated the frequency of A20 gene inactivation in different subtypes of non-Hodgkin lymphomas and then analyzed the effects of the silencing of A20 expression in Epstein-Barr virus–infected lymphoblastoid cell lines (EB-LCLs). The results present evidence that A20 is a putative tumor suppressor gene.
Methods
Patient samples, cell culture, and reagents
All samples were obtained from patients with the approval of the Institutional Review Board at Aichi Cancer Center and informed consent obtained in accordance with the Declaration of Helsinki. All cell lines were maintained in Iscove modified Dulbecco medium with 10% fetal calf serum, penicillin (50 U/mL), streptomycin (50 μg/mL), and β-mercaptoethanol (50 μM) at 37°C in 5% of carbon dioxide. EB-LCLs, UH3, and CB33 were derived from normal human peripheral blood B cells6 and kindly provided by R. Dalla-Fabera (Columbia University). Peripheral blood B cells were obtained from healthy volunteers by the human CD19 multisort kit (Miltenyi Biotec), in accordance with the manufacturer's protocol. CD19-positive B cells were stimulated with 50 μg/μL anti-IgM for 48 hours. The goat anti–human IgM was purchased from Southern Biotechnology Associates, and the IκB kinase (IKK) specific inhibitor, BMS-345541, was purchased from Calbiochem.
Array CGH analysis and microarray analysis
We reexamined our published array comparative genomic hybridization (CGH) analysis data for 383 cases of non-Hodgkin lymphoma by focusing on chromosome band 6q deletion. The definition of copy number imbalance was given in a previous study. These data are composed of 29 cases of mantle cell lymphoma (MCL),7 64 cases of MALT lymphoma,1,8 102 cases of diffuse large B-cell lymphoma (DLBCL),9 23 cases of follicular lymphoma,10,11 19 cases of sporadic Burkitt lymphoma,12 27 cases of natural killer (NK) cell lymphoma,13 68 cases of adult T-cell leukemia/lymphoma (ATLL),14 and 51 cases of peripheral T-cell lymphoma unspecified (PTCL-u).15 In addition, we reexamined our published microarray expression analysis data of DLBCL samples from 46 cases by focusing on correlation with A20 genomic status. DLBCL cases were divided into germinal center B cell–like (GCB) and activated B cell–like (ABC) subtypes as defined in a previous study.9 The microarray data can be found in the GEO public database under accession no. GSE16920.
FISH analysis for detection of A20 deletion
Dual-color fluorescence in situ hybridization (FISH) was performed for DLBCL and MCL samples to confirm the deletion of A20 detected by array CGH analysis according to a previously described method.1 For hybridization, a spectrum green–labeled RP11-277K14 probe at chromosome 6q centromere and a spectrum red–labeled RP11-356I2 probe were used for detection of the A20 gene at chromosome band 6q23.3.
Quantitative real-time RT-PCR analysis
SYBR premix ExTaq (Takara Bion) was used for polymerase chain reaction (PCR) while the β-actin gene was simultaneously run as a control and used for normalization. The relative abundance of mRNA levels was determined from standard curves generated from a serially diluted plasmid prepared as previously described.2 All experiments were performed in duplicate. The healthy male peripheral activated B-cell cDNA was used as control for determining the relative expression level of A20/β-actin. Specific primers used in this study are described in supplemental Table 2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sodium bisulfate modification and MSP analysis
The genomic DNA obtained from peripheral B cells was methylated with SssI methylase (New England Biolabs) and served as a methylated control DNA. For bisulfite treatment, 1 μg genomic DNA was used, digested with BamHI as previously described.16 The primer sequences for methylation-specific PCR (MSP) are described in supplemental Table 3. The PCR conditions consisted of 10 minutes at 95°C for initial denaturation, followed by 35 cycles at 95°C (30 seconds), 54°C (30 seconds), and 72°C (30 seconds) and a final extension of 10 minutes at 72°C using AmpliTaq Gold DNA polymerase (Applied Biosystems).
Mutational analysis of A20
All exons of A20 and CARD11 were amplified from cDNA by PCR. The direct sequencing of cDNA was performed on entire cording exons of the A20 and CARD11 coiled-coil domain. In addition, A20 genomic sequencing, which included the exon-intron junction, was conducted. Capillary electrophoresis was performed on an ABI3100 sequencer (Applied Biosystems).
Vector construction
The cDNA encoding the A20 gene was obtained from peripheral mononuclear cell cDNA by PCR amplification. The active mutant IKKβ S177E/S181E (IKKβSS/EE) cDNA was provided in the pc.DNA3.1 vector by B. Baumann (Ulm University). The lentiviral vector used in this study was CSII-EF1α-MCS-IRES2-Venus (EF1α, elongation factor; MCS, multiple cloning site; IRES, internal ribosome entry site; Venus, a variant of the yellow fluorescent protein gene), which was obtained from RIKERN BRC with permission of H. Miyoshi (RIKERN BRC) and A. Miyawaki (RIKEN Brain Science Institute). A20 was inserted upstream of the ires2-Venus element into the NotI restriction enzyme site.
For A20 silencing, short hairpin RNA (shRNA) directed against A20, firefly luciferase, and green fluorescent protein (GFP) was subcloned into CS II lentiviral vector under the control of human H1 promoter. Detail vector construction is described in the supplemental data. CSII-H1-Pgk-Puro without any shRNA probes served as a mock infection control. The sequences of small interfering RNA (siRNA) probes immediately downstream of H1 promoter are described in supplemental Table 4.
Lentiviral pseudotyping and infection
The lentiviral vectors were generated by cotransfection into HEK293T cells by FuGENE 6 (Roche Applied Science) with pCAG-HIVgp (packaging plasmid) and pCMV-VSV-G-RSV-Rev (Rev and VSV-G expression plasmid), which were provided by Dr Miyoshi. Infectious lentiviruses in the culture supernatants were harvested at 48 hours after transfection. Transductions were performed by spin infection, and 48 hours later the infected cells expressing Venus were enriched by fluorescence-activated cell sorter (FACS) sorting using a cell sorter (JSAN; Bay Bioscience), whereas the cells expressing the puromycin-resistant gene were kept under puromycin (2 μg/mL) culturing conditions for 4 days.
Western blot analysis
Total cell lysates were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted onto a nylon membrane. The individual blots were detected with specific antibodies, anti-A20 antibody (59A426; Abcam), anti-p50 antibody (sc-114; Santa Cruz Biotechnology), anti-p52/p100 (NFKB1) antibody (sc-7386; Santa Cruz Biotechnology), and anti-p65 antibody (sc-372; Santa Cruz Biotechnology). Anti–α tubulin antibody (T9026; Sigma-Aldrich) or antilamin B1 (LMNB1) antibody (33-2000; Zymed Laboratories) was used for comparison of loaded protein amounts. Primary staining was visualized with a goat anti–mouse Ig-horseradish peroxidase–conjugated secondary antibody using ECL Western blotting detection reagents (GE Healthcare).
NF-κB reporter assay
The luciferase assay was performed by lentiviral transduction of the NF-κB reporter vector CSII-5κB-mCMV-fLuc+. CSII-mCMV-fLuc+ served as a negative control of NF-κB activation. CSII-TK-RhL-expressing Renilla luciferase was coinfected for normalization of infection efficiency. The construction of lentiviral vectors for reporter assay was described in the supplemental data. Luciferase activity was determined with a luciferase assay system (Promega) 48 hours after infection.
NF-κB DNA-binding assay
DNA-binding activities of NF-κB, p50, and p52 were examined in nuclear extracts according to the protocols provided with the TransAM kit (Active Motif); 10 μg nuclear extraction protein was used for assessing the nuclear translocation of active p50 and p52 according to the manufacturer's instructions.
Apoptosis assay
To induce apoptosis, 106 cells of LCL were treated with H2O2 (50 μM; Wako) or etoposide (50 μM; Sigma-Aldrich) for 24 hours. Apoptotic cell death was determined with the Annexin V Cy3 Apoptosis Detection Kit (Abcam) according to the manufacturer's instructions. Briefly, the cells were stained for 15 minutes at room temperature in the dark and analyzed on a FACSCalibur flow cytometer using BD-CellQuest software (BD Biosciences).
Clonogenicity in methylcellulose-based media
Colony assays were performed 3 times in independent experiments, and each assay was performed in triplicate by embedding 104 cells in 1 mL Iscove modified Dulbecco medium containing 25% fetal calf serum and 0.75% methylcellulose (Wako) in 60-mm plates. Colonies were scored on day 14.
Results
Frequent deletion of A20 found in subgroups of non-Hodgkin lymphomas
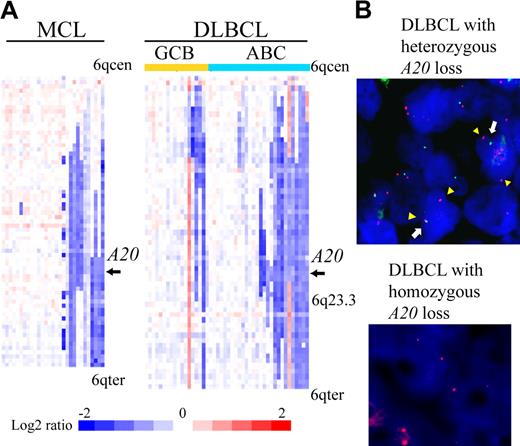
We examined our own array-CGH data1,7-15 for 383 cases to determine frequencies of A20 gene deletion. The results are summarized in Table 1. A total of 84 of 383 non-Hodgkin lymphoma cases (21.9%) were found to have the deletion (heterozygous loss in 81 cases, homozygous loss in 3). An analysis of lymphoma subtypes showed that deletion occurred most frequently in MCLs (31%) and DLBCLs (38%). Previous microarray studies of gene expression divided DLBCL into 2 subtypes, ABC DLBCL and GCB DLBCL.17,18 The analysis also revealed that the ABC subtype of DLBCL (14 of 28 cases, 50%) is more frequently associated with the deletion of A20 than GCB subtype (4 of 18 cases, 22.2%). Detailed heat map images of the deletion in MCL and DLBCL showed that the A20 locus is located within the minimal common region of deletion of chromosome band 6q both in MCL and in ABC DLBCL (Figure 1A). Our dataset did not show any specific correlation between A20 genomic deletion and pathologic characteristics, such as the blastoid variant, in MCL cases (Table 1).
A20 deletion in MCLs and DLBCLs. (A) The heat map represents log2 ratios of bacterial artificial chromosome (BAC)/P-1 phage-derived chromosome (PAC) clones, which are shown in order from chromosome band 6q centromere (6qcen) to 6q telomere (6qter). The genomic profiles of the chromosome band 6q are depicted in 29 cases of MCL and 46 cases of DLBCL. DLBCL cases are divided into 18 cases of GCB and 28 cases of ABC subtypes as defined.12 Relative copy number log2 ratio is displayed according to the color scale.  represents the location of A20. (B) Dual-color FISH analysis of DLBCL and MCL cases was performed with a combination of the BAC RP11-356I2 (A20 gene probe) green probe and RP11-277K14 (6q centromere probe) red probe to confirm array CGH results. Representative results of FISH analysis are shown. Two red signals (arrowheads) and one green signal (white arrow) are seen in one cell, indicating heterozygous deletion of the A20 gene, and 2 red signals but no green signal are seen in another cell, indicating homozygous deletion of the A20 gene.
represents the location of A20. (B) Dual-color FISH analysis of DLBCL and MCL cases was performed with a combination of the BAC RP11-356I2 (A20 gene probe) green probe and RP11-277K14 (6q centromere probe) red probe to confirm array CGH results. Representative results of FISH analysis are shown. Two red signals (arrowheads) and one green signal (white arrow) are seen in one cell, indicating heterozygous deletion of the A20 gene, and 2 red signals but no green signal are seen in another cell, indicating homozygous deletion of the A20 gene.
A20 deletion in MCLs and DLBCLs. (A) The heat map represents log2 ratios of bacterial artificial chromosome (BAC)/P-1 phage-derived chromosome (PAC) clones, which are shown in order from chromosome band 6q centromere (6qcen) to 6q telomere (6qter). The genomic profiles of the chromosome band 6q are depicted in 29 cases of MCL and 46 cases of DLBCL. DLBCL cases are divided into 18 cases of GCB and 28 cases of ABC subtypes as defined.12 Relative copy number log2 ratio is displayed according to the color scale.  represents the location of A20. (B) Dual-color FISH analysis of DLBCL and MCL cases was performed with a combination of the BAC RP11-356I2 (A20 gene probe) green probe and RP11-277K14 (6q centromere probe) red probe to confirm array CGH results. Representative results of FISH analysis are shown. Two red signals (arrowheads) and one green signal (white arrow) are seen in one cell, indicating heterozygous deletion of the A20 gene, and 2 red signals but no green signal are seen in another cell, indicating homozygous deletion of the A20 gene.
represents the location of A20. (B) Dual-color FISH analysis of DLBCL and MCL cases was performed with a combination of the BAC RP11-356I2 (A20 gene probe) green probe and RP11-277K14 (6q centromere probe) red probe to confirm array CGH results. Representative results of FISH analysis are shown. Two red signals (arrowheads) and one green signal (white arrow) are seen in one cell, indicating heterozygous deletion of the A20 gene, and 2 red signals but no green signal are seen in another cell, indicating homozygous deletion of the A20 gene.
Frequency of A20 deletion in non-Hodgkin lymphomas
| Histologic subgroup . | n (%) . |
|---|---|
| Mantle cell lymphoma | 9/29 (31) |
| Blastoid variant | 0/2 (0) |
| Pleomorphic variant | 1/3 (33.3) |
| MALT lymphoma | 11/64 (17.2) |
| Ocula adnexa | 9/24 (37.5) |
| Lung | 0/11 (0) |
| Stomach | 2/29 (6.9) |
| DLBCL | 39/102 (38) |
| GCB type | 4/18 (22.2) |
| ABC type | 14/28 (50) |
| Follicular lymphoma | 6/23 (26) |
| Grade 1 | 1/7 (14.2) |
| Grade 2 | 4/14 (28.5) |
| Grade 3A | 1/2 (50) |
| Burkitt lymphoma | 2/19 (10.5) |
| NK-cell lymphoma | 5/27 (18.5) |
| Aggressive NK-cell leukemia | 2/10 (20) |
| ENKL | 3/17 (17.6) |
| ATLL | 7/68 (10.3) |
| Acute type | 0/19 (0) |
| Lymphoma type | 7/49 (14.3) |
| PTCL-u | 5/51 (9.8) |
| Total | 84/383 (21.9) |
| Histologic subgroup . | n (%) . |
|---|---|
| Mantle cell lymphoma | 9/29 (31) |
| Blastoid variant | 0/2 (0) |
| Pleomorphic variant | 1/3 (33.3) |
| MALT lymphoma | 11/64 (17.2) |
| Ocula adnexa | 9/24 (37.5) |
| Lung | 0/11 (0) |
| Stomach | 2/29 (6.9) |
| DLBCL | 39/102 (38) |
| GCB type | 4/18 (22.2) |
| ABC type | 14/28 (50) |
| Follicular lymphoma | 6/23 (26) |
| Grade 1 | 1/7 (14.2) |
| Grade 2 | 4/14 (28.5) |
| Grade 3A | 1/2 (50) |
| Burkitt lymphoma | 2/19 (10.5) |
| NK-cell lymphoma | 5/27 (18.5) |
| Aggressive NK-cell leukemia | 2/10 (20) |
| ENKL | 3/17 (17.6) |
| ATLL | 7/68 (10.3) |
| Acute type | 0/19 (0) |
| Lymphoma type | 7/49 (14.3) |
| PTCL-u | 5/51 (9.8) |
| Total | 84/383 (21.9) |
ENKL indicates extranodal NK-cell lymphoma.
Interestingly, our data showed that the A20 deletion tends to correlate with histologic grade in follicular lymphoma cases (Table 1), suggesting that the A20 deletion may have relevance to disease progression. The analysis of subtypes of MALT lymphomas demonstrated that the A20 deletion was strongly associated with ocular adnexa subtype but rarely detected in other subtypes of MALT lymphomas (supplemental Figure 1). Among subtypes of gastric MALT lymphomas, A20 deletion appears selectively in those not associated with Helicobacter pylori infection (supplemental Table 1).
Dual-color FISH analysis using spectrum red-labeled BAC RP11-277K14 for the chromosome 6q centromer probe and spectrum green-labeled BAC RP11-375I2 for the A20 probe was performed to confirm the results of array CGH analysis. Representative results are shown in Figure 1B. These FISH results correlated well with the array CGH data.
Expression level of A20 correlates with genome status of ABC DLBCL, but not GCB DLBCL or MCL
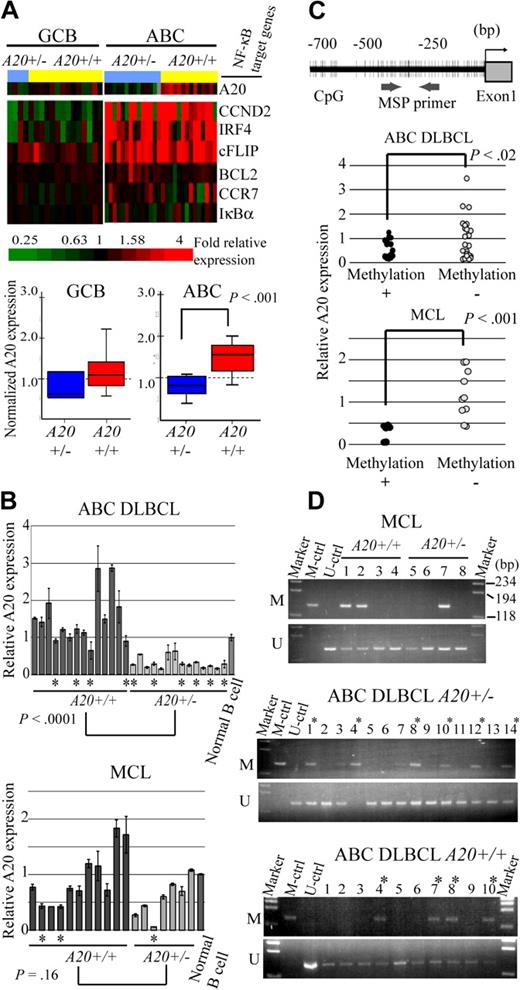
Because A20 was located in the minimal common deleted region of chromosome band 6q of ABC DLBCL and most frequently deleted in the ABC DLBCL subtypes of the analyzed lymphoma, we next examined whether the A20 genome status correlates with the expression level of the gene (Figure 2A). The microarray analysis of gene expressions disclosed that the expression level of A20 correlated well with the genome status of ABC DLBCL with statistical significance as determined by the Student t test. Interestingly, however, NF-κB target genes (CCND2, IRF4, CFLAR, BCL2, CCR7, and NFKBIA) were highly expressed in ABC DLBCL regardless of the genome status of A20 (A20+/− vs A20+/+). In GCB DLBCL, expression of A20 was lower than in ABC DLBCL, as was that of the NF-κB target genes. Because the correlation between A20 deletion and its expression had not yet been determined for MCL cases, quantitative real-time reverse-transcribed (RT)–PCR analysis for the ABC DLBCL and MCL samples was conducted for validation of the correlation between genomic status and expression of A20 (Figure 2B). In line with the microarray analysis findings, the results indicated that the expression level of A20 correlates well with genomic loss in ABC DLBCL samples, but that it is unrelated to genomic status of MCL cases.
A20 gene inactivation in the genomic subgroups of DLBCL and MCL. (A) Differential A20 expression in the genomic subgroups of DLBCL and MCL. GCB and ABC DLBCL were divided into subgroups with heterozygous A20 deletion and without A20 deletion. (Top panel) Data from DNA microarray analysis of A20 and NF-κB target gene expression in DLBCL clinical samples. Multiples of relative expression are shown according to the color scale. A20+/− indicates the cases in the heterozygous A20 deletion subgroup; A20+/+, the cases in the subgroup without A20 deletion. The NF-κB target gene set used in this study corresponds to Davis et al.19 The box-and-whisker diagrams depicted in the bottom panel represent median expression values and the interquartile range of A20 in the GCB and ABC DLBCL subgroups. Scale bar represents 5 percentile ranges of A20 expression. (B) Real-time RT-PCR analysis of A20 expression in DLBCL and MCL cases. Among ABC DLBCL cases, 14 patients with A20 loss and 14 patients without A20 loss were analyzed, and analysis of 7 patients with A20 loss and 11 patients without A20 loss was performed among MCL cases. Expressions were normalized on the basis of the corresponding β-actin content, and relative expression levels were established by comparing them with those of healthy male activated B-cell control. *Cases with A20 promoter methylation. (C) Correlation between promoter methylation and expression of A20 in DLBCL and MCL. A20 promoter methylation was subjected to MSP analysis. The location of the CpG and MSP primer regions analyzed in this study is shown in the top panel. (Bottom panel) The inverse correlation between A20 promoter methylation and gene expression analyzed by MSP. A20+/− indicates the cases in the heterozygous A20 deletion subgroup; A20+/+, those in the subgroup without A20 deletion. (D) A20 promoter methylation status in DLBCL and MCL tissues. The A20 gene promoter was methylated in colorectal cancer tissues. M denotes the PCR products of the amplified methylated DNA; and U, those of the unmethylated DNA.
A20 gene inactivation in the genomic subgroups of DLBCL and MCL. (A) Differential A20 expression in the genomic subgroups of DLBCL and MCL. GCB and ABC DLBCL were divided into subgroups with heterozygous A20 deletion and without A20 deletion. (Top panel) Data from DNA microarray analysis of A20 and NF-κB target gene expression in DLBCL clinical samples. Multiples of relative expression are shown according to the color scale. A20+/− indicates the cases in the heterozygous A20 deletion subgroup; A20+/+, the cases in the subgroup without A20 deletion. The NF-κB target gene set used in this study corresponds to Davis et al.19 The box-and-whisker diagrams depicted in the bottom panel represent median expression values and the interquartile range of A20 in the GCB and ABC DLBCL subgroups. Scale bar represents 5 percentile ranges of A20 expression. (B) Real-time RT-PCR analysis of A20 expression in DLBCL and MCL cases. Among ABC DLBCL cases, 14 patients with A20 loss and 14 patients without A20 loss were analyzed, and analysis of 7 patients with A20 loss and 11 patients without A20 loss was performed among MCL cases. Expressions were normalized on the basis of the corresponding β-actin content, and relative expression levels were established by comparing them with those of healthy male activated B-cell control. *Cases with A20 promoter methylation. (C) Correlation between promoter methylation and expression of A20 in DLBCL and MCL. A20 promoter methylation was subjected to MSP analysis. The location of the CpG and MSP primer regions analyzed in this study is shown in the top panel. (Bottom panel) The inverse correlation between A20 promoter methylation and gene expression analyzed by MSP. A20+/− indicates the cases in the heterozygous A20 deletion subgroup; A20+/+, those in the subgroup without A20 deletion. (D) A20 promoter methylation status in DLBCL and MCL tissues. The A20 gene promoter was methylated in colorectal cancer tissues. M denotes the PCR products of the amplified methylated DNA; and U, those of the unmethylated DNA.
A20 is frequently inactivated as a result of promoter methylation or gene mutation in ABC DLBCL and MCL
Because inactivation mechanisms other than genomic deletion may also affect lymphoma development, we next analyzed A20 promoter CpG methylation and performed direct sequence analysis of the coding region of the A20 transcript in ABC DLBCL and MCL samples, which correspond to normal peripheral B cells. In normal peripheral B-cell samples, unmethylated DNA was amplified, but methylated DNA was not, thus demonstrating that peripheral B-cell DNA served as unmethylated control (Figure 2D). The A20 promoter region examined for methylation was also located within a CpG island (CpG island information at http://genome.ucsc.edu/index.html). The A20 promoter was methylated in 10 of 24 ABC DLBCL samples (41.6%) and 3 of 8 MCL samples (37.5%; Figure 2D; Table 2). Furthermore, the expression level of A20 was found to correlate with A20 promoter methylation status both in ABC DLBCL and MCL samples (Figure 2C). Although these results require further verification by bisulfate-modified DNA sequencing analysis, they suggest that A20 expression might be repressed as a result of promoter methylation during lymphoma development.
Features of A20 inactivation in ABC DLBCL and MCL
| . | ABC DLBCL . | MCL . |
|---|---|---|
| A20 mutation, n (%) | ||
| Total | 15/28 (53.5) | 3/18 (16.6) |
| With A20 deletion | 8/14 (57.1) | 1/7 (14.2) |
| Without A20 deletion | 7/14 (50) | 2/11 (18.1) |
| Promoter methylation, total, n (%) | 10/24 (41.6) | 3/8 (37.5) |
| A20 status, n (%) | ||
| Biallelic inactivation | 9/28 (32.1) | 3/18 (16.6) |
| Monoallelic inactivation | 13/28 (46.4) | 8/18 (44.4) |
| Intact | 6/28 (21.4) | 6/28 (21.4) |
| CARD11 mutation, n (%) | ||
| Total | 4/28 (14.2) | N.D. |
| With biallelic A20 inactivation | 0/9 (0) N.D. | |
| With monoallelic A20 inactivation | 2/13 (15.3) | N.D. |
| Without A20 inactivation | 2/6 (33.3) | N.D. |
| . | ABC DLBCL . | MCL . |
|---|---|---|
| A20 mutation, n (%) | ||
| Total | 15/28 (53.5) | 3/18 (16.6) |
| With A20 deletion | 8/14 (57.1) | 1/7 (14.2) |
| Without A20 deletion | 7/14 (50) | 2/11 (18.1) |
| Promoter methylation, total, n (%) | 10/24 (41.6) | 3/8 (37.5) |
| A20 status, n (%) | ||
| Biallelic inactivation | 9/28 (32.1) | 3/18 (16.6) |
| Monoallelic inactivation | 13/28 (46.4) | 8/18 (44.4) |
| Intact | 6/28 (21.4) | 6/28 (21.4) |
| CARD11 mutation, n (%) | ||
| Total | 4/28 (14.2) | N.D. |
| With biallelic A20 inactivation | 0/9 (0) N.D. | |
| With monoallelic A20 inactivation | 2/13 (15.3) | N.D. |
| Without A20 inactivation | 2/6 (33.3) | N.D. |
ND indicates not done.
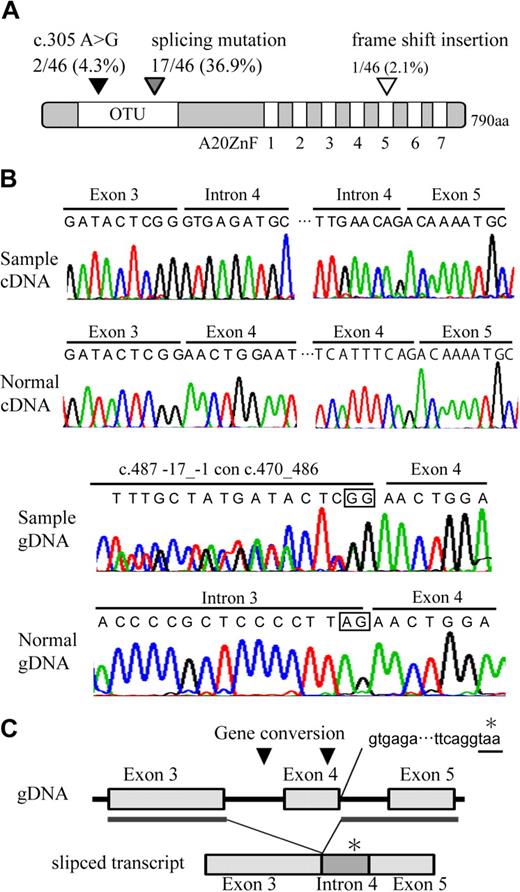
Direct sequence analysis of the A20 transcript identified one missense mutation, c.371 A > G, leading to the presence of N102S in 2 of the 46 samples, a frame shift insertion in one of the samples, and an aberrantly spliced transcript, which is associated with exon 4 skipping, as well as the introduction of termination codon in 17 of the samples (Figure 3A-B, supplemental Figure 6; Table 2). The truncated product of A20 lacks the 579 C terminal amino acids, including the OTU domain and A20 zinc finger domains. As spliced mutation was seen to occur frequently in our search, genomic sequence was examined including the exon-intron junction. We found 2 interallelic gene conversion, c.487-17_-1 con c.470_486, replacing the splicing acceptor site of intron 3 to a part of exon 3 sequence in 10 of the samples, and c.582_634 con c.434_486, replacing a part of exon 4 sequence to exon 3 in 12 of the samples. The conversion disrupting splicing acceptor site may activate cryptic splicing acceptor site and may result in splicing mutation. The positions of the A20 mutations at genomic DNA, cDNA, and protein levels are based on NC_000006.10, NM_006290.2, and NP_006281.1, respectively. Nomenclature for the description of gene mutations is based on a previous report20 (http://www.hgvs.org/mutnomen/).
Distribution and features of A20 mutation in ABC DLBCL and MCL. (A) Schematic representation of human A20 with its functional domains. OTU is a deubiquitinating enzyme domain. A20 ZnF indicates the A20 zinc finger domain, which exerts E3 ligase activity. The location of A20 mutations is represented on the map with triangles. (B) Electrofluorograms showing partial nucleotide sequence of the A20 cDNA and genomic DNA (gDNA) of the patient sample and normal control. The splicing mutation in the patient results in exon 4 skipping and inclusion of whole intron 4 sequence. The intrallelic gene conversion in the patient disrupts a consensus sequence of acceptor splicing site of intron 3 (c.582_634 con c.434_486). Square in the sequence represents consensus sequence of splicing acceptor site. (C) Schematic representation of exon 4 skipping is shown. The location of gene conversions is represented on the map in triangles. The gene conversion may induce activation of cryptic splicing acceptor site of exon 4. Inclusion of whole intron 4 sequence in aberrantly spliced product adds 90 nucleotides followed by a stop codon (*).
Distribution and features of A20 mutation in ABC DLBCL and MCL. (A) Schematic representation of human A20 with its functional domains. OTU is a deubiquitinating enzyme domain. A20 ZnF indicates the A20 zinc finger domain, which exerts E3 ligase activity. The location of A20 mutations is represented on the map with triangles. (B) Electrofluorograms showing partial nucleotide sequence of the A20 cDNA and genomic DNA (gDNA) of the patient sample and normal control. The splicing mutation in the patient results in exon 4 skipping and inclusion of whole intron 4 sequence. The intrallelic gene conversion in the patient disrupts a consensus sequence of acceptor splicing site of intron 3 (c.582_634 con c.434_486). Square in the sequence represents consensus sequence of splicing acceptor site. (C) Schematic representation of exon 4 skipping is shown. The location of gene conversions is represented on the map in triangles. The gene conversion may induce activation of cryptic splicing acceptor site of exon 4. Inclusion of whole intron 4 sequence in aberrantly spliced product adds 90 nucleotides followed by a stop codon (*).
We defined the cases with both A20 monoallelic deletion and gene mutation or the cases whose A20 transcript was not amplified by PCR as biallelic A20 inactivation cases. Based on this definition, biallelic A20 inactivation occurred in 9 of 28 (41.6%) ABC DLBCL and 3 of 18 (37.5%) MCL cases (Table 2). We also defined the cases with A20 monoallelic deletion or gene mutation alone as A20 monoallelic inactivation cases, which accounted for 13 (46.4%) ABC DLBCL and 8 (44.4%) MCL cases. Because the CARD11 mutation has been reported to induce aberrant NF-κB activation in ABC DLBCL,21 we analyzed the relationship between A20 inactivation and CARD11 mutation. CARD11 mutation was found to occur in 4 (14.2%) of ABC DLBCL cases (Table 2; supplemental Figure 5). Interestingly, the CARD11 mutation was not observed in the cases with biallelic A20 inactivation.
Reintroduction of A20 induces apoptosis in lymphoma cell lines with A20 deletion
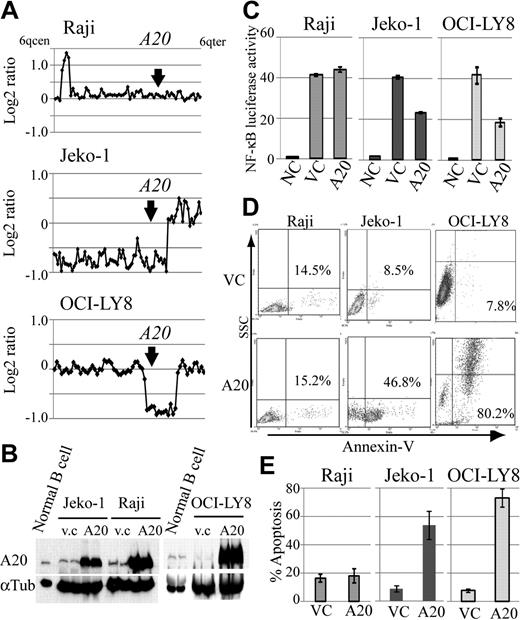
Because A20 inactivation was frequent genomic aberrations in DLBCLs and MCLs, we also investigated the role of A20 as a potential tumor suppressor in lymphoma cell lines with different A20 genomic status, and compared the expression levels of A20 in human peripheral B-cell and lymphoma cell lines. For this purpose, we constructed a lentivirus A20 expression vector, which is capable of highly efficient infection, and used it to introduce A20 into the MCL cell line Jeko-1 and the DLBCL cell line OCI-LY8 with A20 monoallelic deletion, which corresponds to the Burkitt lymphoma cell line Raji without the A20 deletion (Figure 4A). Western blot analysis showed that A20 protein expression in human peripheral B cells stimulated by anti-IgM is higher than that in lymphoma cell lines with A20 deletion (Figure 4B). The reintroduction of A20 induced cellular apoptosis in the cell lines with A20 deletion, whereas the control lymphoma cell line without A20 deletion, Raji, was not affected by A20 overexpression (Figure 4D-E). Because we assume that this finding was the result of the role of A20 as a negative regulator of NF-κB signaling, we used a luciferase assay to evaluate its effect on NF-κB. Similar to findings obtained with the induction of apoptosis, the reintroduction of A20 was associated with suppression of NF-κB activity in the cell lines with A20 deletion, but not in Raji (Figure 4C).
Reintroduction of A20 induces apoptosis in the lymphoma cell lines with A20 deletion. The oncosuppressive effects of A20 on MCL and DLBCL lymphoma cell lines infected with a lentivirus, expressing both A20 and Venus or Venus only as control. (A) Genomic profiles of analyzed lymphoma cell lines. Dots represent the log2 ratios of BAC/PAC clones, which are shown in order from chromosome band 6q centromere to 6q telomere. The location of A20 is indicated with an arrow. (B) Western blot analysis for comparison of A20 expression in human peripheral B cells treated with anti-IgM and in lymphoma cell lines. All cell lines with A20 transduction by a lentiviral vector showed high expression levels for the A20 protein. Infection efficiency of each cell line is shown in supplemental Figure 7. (C) Overexpression of A20 induced repression of NF-κB activity in lymphoma cell lines with A20 deletion as measured with a lentiviral reporter assay. In contrast, NF-κB activity is not affected by overexpression of A20 in Raji. The findings of triplicate experiments are presented as mean ± SD. NC indicates negative control; VC, vector-only control. Effect of A20 overexpression on apoptosis correlates well with NF-κB-inhibiting effect. (D) The findings of representative apoptosis assay measured by annexin V staining. (E) Triplicate experiments are presented as mean ± SD.
Reintroduction of A20 induces apoptosis in the lymphoma cell lines with A20 deletion. The oncosuppressive effects of A20 on MCL and DLBCL lymphoma cell lines infected with a lentivirus, expressing both A20 and Venus or Venus only as control. (A) Genomic profiles of analyzed lymphoma cell lines. Dots represent the log2 ratios of BAC/PAC clones, which are shown in order from chromosome band 6q centromere to 6q telomere. The location of A20 is indicated with an arrow. (B) Western blot analysis for comparison of A20 expression in human peripheral B cells treated with anti-IgM and in lymphoma cell lines. All cell lines with A20 transduction by a lentiviral vector showed high expression levels for the A20 protein. Infection efficiency of each cell line is shown in supplemental Figure 7. (C) Overexpression of A20 induced repression of NF-κB activity in lymphoma cell lines with A20 deletion as measured with a lentiviral reporter assay. In contrast, NF-κB activity is not affected by overexpression of A20 in Raji. The findings of triplicate experiments are presented as mean ± SD. NC indicates negative control; VC, vector-only control. Effect of A20 overexpression on apoptosis correlates well with NF-κB-inhibiting effect. (D) The findings of representative apoptosis assay measured by annexin V staining. (E) Triplicate experiments are presented as mean ± SD.
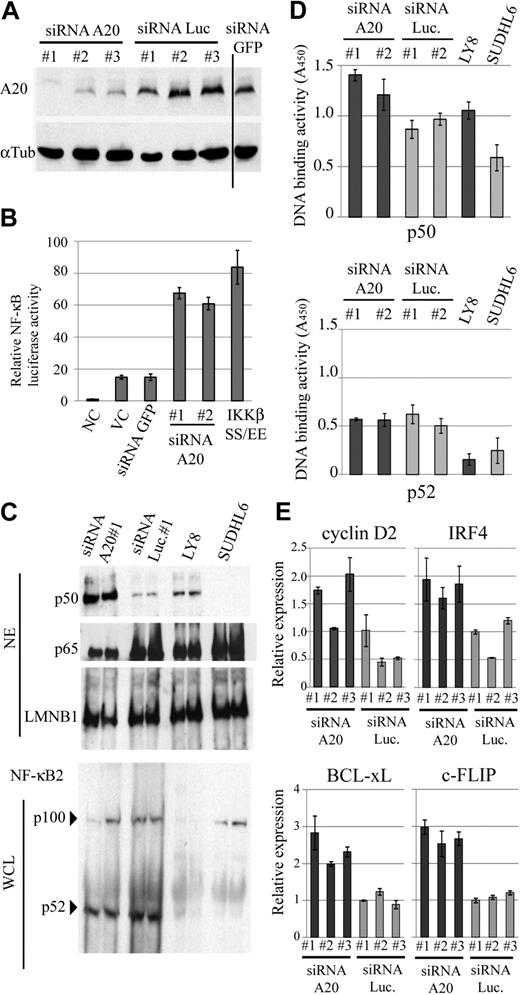
A20 silencing induces constitutive activation of NF-κB
Although the genome status of A20 appeared to be only tenuously related to NF-κB activation in the clinical DLBCL samples, it nevertheless would be possible that A20 could contribute to deregulation of NF-κB pathways in pathogenesis. To experimentally test this possibility, we first examined whether down-regulation of A20 could contribute to NF-κB activation. To investigate the cellular effect of A20 silencing, we used well-characterized EB-LCLs, CB33, and UH3 because A20 is highly expressed in these cell lines. We used the siRNA strategy to silence A20 expression and monitored the resulting NF-κB status in siRNA-transduced CB33. For this assay, we selected a lentiviral reporter system, as in this system the reporter is integrated into the genome and thus reflects changes in the activities of the NF-κB in a natural chromatin environment more faithfully than is possible with the more commonly used plasmid transfection assay system.22 Three different siRNAs, targeting distinctive sequences in A20, effectively down-regulated expression of A20 in an EB-LCL CB33 (Figure 5A), whereas control siRNAs directed against luciferase and GFP had no such effect. In parallel with the silencing of A20, NF-κB activity was found to be increased by a factor of 3 to 5 compared with mock infection or control shRNA directed against GFP (Figure 5B). A constitutive active IKKβ (IKKβSS/EE) served as a positive control of the assay. In addition, we performed real-time PCR for a comparison of the expression of cancer-related NF-κB target genes (Cyclin D2, c-FLIP, BCL-xL, and IRF4) in LCLs expressing siRNA for A20 and for luciferase. The expression of each of the 4 NF-κB target genes was found to correlate with A20 silencing (Figure 5E). We next examined the effect of A20 knockdown on canonical or noncanonical NF-κB signaling in LCLs and lymphoma cell lines with (OCI-LY8) and without (SUDHL6) A20 deletion2 by enzyme-linked immunosorbent assay-based DNA binding assay and immunobloting assay. We found that nuclear translocation and NF-κB motif DNA-binding activity of p50 increased as a result of A20 silencing, whereas p100 processing and p52-binding activity were not affected (Figure 5C-D). Similar to findings obtained with the LCLs with A20 knockdown, A20 deletion was associated with activation of NF-κB signaling in the lymphoma cell lines. These findings suggest that A20 is associated with the canonical NF-κB pathway.
A20 silencing in EB-LCLs induces constitutive NF-κB activation. (A) Repression efficiency of A20 when using siRNA was determined by immunobloting in whole-cell extracts of CB33 expressing A20, luciferase, and GFP siRNA. Vertical line represents a repositioned gel lane. (B) CB33 cells were cotransduced with a lentiviral vecter encoding κB sequence–firefly luciferase and Renilla luciferase reporter vector. The data shown represent the mean relative luciferase activity normalized against Renilla luciferase activity ± SD. The experiments were performed in triplicate. IKKβSS/EE served as positive control for NF-κB activation. (C) Immunoblots for the canonical and noncanonical NF-κB protein from the indicated cell lines. DLBCL cell lines with (OCI-LY8) and without (SUDHL6) A20 deletion were included in each blot. A20 deletion and A20 silencing are involved in p50 nuclear translocation, whereas A20 had little effect on p100 processing. NE indicates nuclear extract; and WCL, whole-cell lysate. (D) Effect of A20 knockdown on the nuclear DNA-binding activity of the NF-κB p50 and p52 subunits. Binding to an oligonucleotide containing the NF-κB consensus sequence was measured in nuclear extracts from LCLs expressing siRNA for A20 and luciferase. DNA-binding activity was quantified by colorimetry. The findings of triplicate experiments are presented as mean ± SD. NC indicates negative control; VC, vector-only control; and Luc, firefly luciferase. (E) Expression of cancer-related NF-κB target genes in A20 knockdown CB33. Relative expression level of the 4 NF-κB target genes (Cyclin D2, IRF4, BCL-xL, and c-FLIP) is determined by real-time PCR and normalized on the basis of the corresponding β-actin content. The findings of triplicate experiments are presented as mean ± SD.
A20 silencing in EB-LCLs induces constitutive NF-κB activation. (A) Repression efficiency of A20 when using siRNA was determined by immunobloting in whole-cell extracts of CB33 expressing A20, luciferase, and GFP siRNA. Vertical line represents a repositioned gel lane. (B) CB33 cells were cotransduced with a lentiviral vecter encoding κB sequence–firefly luciferase and Renilla luciferase reporter vector. The data shown represent the mean relative luciferase activity normalized against Renilla luciferase activity ± SD. The experiments were performed in triplicate. IKKβSS/EE served as positive control for NF-κB activation. (C) Immunoblots for the canonical and noncanonical NF-κB protein from the indicated cell lines. DLBCL cell lines with (OCI-LY8) and without (SUDHL6) A20 deletion were included in each blot. A20 deletion and A20 silencing are involved in p50 nuclear translocation, whereas A20 had little effect on p100 processing. NE indicates nuclear extract; and WCL, whole-cell lysate. (D) Effect of A20 knockdown on the nuclear DNA-binding activity of the NF-κB p50 and p52 subunits. Binding to an oligonucleotide containing the NF-κB consensus sequence was measured in nuclear extracts from LCLs expressing siRNA for A20 and luciferase. DNA-binding activity was quantified by colorimetry. The findings of triplicate experiments are presented as mean ± SD. NC indicates negative control; VC, vector-only control; and Luc, firefly luciferase. (E) Expression of cancer-related NF-κB target genes in A20 knockdown CB33. Relative expression level of the 4 NF-κB target genes (Cyclin D2, IRF4, BCL-xL, and c-FLIP) is determined by real-time PCR and normalized on the basis of the corresponding β-actin content. The findings of triplicate experiments are presented as mean ± SD.
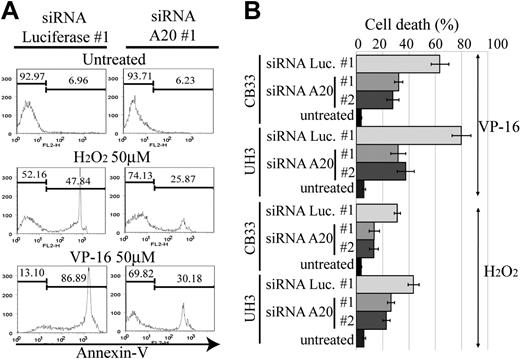
A20 silencing inhibits etoposide or H2O2-mediated apoptosis
The activation of NF-κB pathway in ABC DLBCL is thought to be associated with resistance to apoptosis, which in turn is related to poor response to conventional chemotherapy.23 Reactive oxygen species, such as H2O2 and hydroxyradicals, are generated in response to anticancer drugs and induce cellular apoptosis. NF-κB activation has been reported to lead to the elimination of reactive oxygen species,24 thus becoming the driving force for chemoresistance. We therefore next examined whether A20 silencing has an antiapoptotic effect on anticancer agents. For this purpose, we used etoposide and H2O2 to induce an intrinsic apoptosis pathway for CB33 and UH3. Apoptosis was measured by FACS with annexin V and SYTOX staining. Two independent siRNAs, each targeting a different sequence of A20 (siRNA A20 no. 1 and no. 2), were used for silencing A20. As seen in Figure 6, the resistance to apoptosis induced by etoposide or H2O2 is almost twice as high in A20 knockdown LCLs than in LCLs transduced with control siRNA directed against luciferase (siRNA luciferase no. 1).
Resistance to apoptosis in EB-LCL with silencing for A20. The cell lines shown here were treated with H2O2 or etoposide (VP-16) and then subjected to a FACS assay 24 hours later to determine the proportion of annexin V–positive cells. The effects of A20 knockdown on induction of apoptosis by H2O2 and VP-16 were examined in triplicate. (A) Representative histograms. (B) The data of triplicate experiments are presented as mean ± SD.
Resistance to apoptosis in EB-LCL with silencing for A20. The cell lines shown here were treated with H2O2 or etoposide (VP-16) and then subjected to a FACS assay 24 hours later to determine the proportion of annexin V–positive cells. The effects of A20 knockdown on induction of apoptosis by H2O2 and VP-16 were examined in triplicate. (A) Representative histograms. (B) The data of triplicate experiments are presented as mean ± SD.
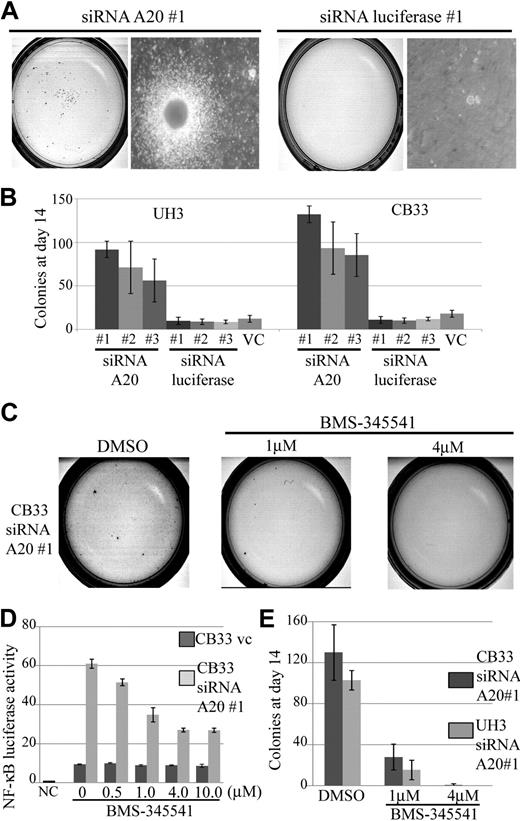
A20 silencing increases clonogenicity of EB-LCLs
The growth effect of siRNA directed against A20 compared with unrelated control directed against firefly luciferase–transduced LCLs was assessed in methylcellulose-based media colony assay. Three independent siRNAs, each targeting a different sequence of A20 (siRNA A20 nos. 1, 2, and 3), and luciferase (siRNA luciferase nos. 1, 2, and 3) were used in this assay.
The results made it clear that silencing of A20 alone was sufficient to increase the cloning efficiency of CB33 and UH3 even with a partial knockdown. The findings of a representative assay for CB33-transduced siRNA directed against A20 compared with siRNA directed against luciferase are shown in Figure 7A. A 5- to 12-fold increment in colonies on day14 was found to occur consistently in all LCLs carrying the A20 knockdown (Figure 7B). We next investigated whether blocking NF-κB activation can reduce gain colony formation ability by silencing A20. Exposure of CB33 carrying the A20 knockdown to BMS-345541, a specific IKK inhibitor contained in a liquid culture, induced inhibition of NF-κB activity even at a dose of 0.5 μM, whereas CB33-introduced vector-only control was not affected by BMS-345541 (Figure 7D). Toxicity of BMS-345541 was not observed even at a dose of 10 μM (data not shown). Exposure of CB33 carrying the A20 knockdown to methylcellulose containing BMS-345541 led to significant inhibition of colony formation, indicating that gain-of-colony formation ability by silencing A20 may depend on activation of NF-κB signaling (Figure 7E).
Colony formation assay in methylcellulose-based media of CB33 and UH3 cell lines containing siRNA directed against A20 or firefly luciferase. LCLs were embedded in 0.75% methylcellulose at 104 cells per dish. Each of the siRNAs was lentiviral vector–transduced. The assay was performed 3 times in independent infections, and each assay was performed in triplicate. The insertless vector CSII-H1-pgk-Puro served as vector control (v.c). Three kinds of siRNAs were constructed, each for a different sequence of A20 (siRNA A20 nos. 1, 2, and 3). Three kinds of luciferase siRNA (siRNA luciferase nos. 1, 2, and 3) were also generated as controls. (A) Representative colony assay of CB33. (B) The bar graph depicts the number of colonies 2 weeks after culture. The data represent the mean ± SD. CB33 carrying the A20 knockdown was plated in methylcellulose in the presence of 1 μM and 4 μM of dimethyl sulfoxide (control) or BMS-345541. (C) Representative colony assay. (D) The data represent the mean relative NF-κB luciferase activity normalized against Renilla luciferase activity ± SD after 3-day exposure of BMS-345541 in liquid culture. The experiments were performed in triplicate. NC indicates negative control; and VC, vector-only control. (E) The bar graph represents the number of colonies observed in the methylcellulose assay. Colonies were counted after 2 weeks of culturing. The data represent mean ± SD. Each assay was performed in triplicate.
Colony formation assay in methylcellulose-based media of CB33 and UH3 cell lines containing siRNA directed against A20 or firefly luciferase. LCLs were embedded in 0.75% methylcellulose at 104 cells per dish. Each of the siRNAs was lentiviral vector–transduced. The assay was performed 3 times in independent infections, and each assay was performed in triplicate. The insertless vector CSII-H1-pgk-Puro served as vector control (v.c). Three kinds of siRNAs were constructed, each for a different sequence of A20 (siRNA A20 nos. 1, 2, and 3). Three kinds of luciferase siRNA (siRNA luciferase nos. 1, 2, and 3) were also generated as controls. (A) Representative colony assay of CB33. (B) The bar graph depicts the number of colonies 2 weeks after culture. The data represent the mean ± SD. CB33 carrying the A20 knockdown was plated in methylcellulose in the presence of 1 μM and 4 μM of dimethyl sulfoxide (control) or BMS-345541. (C) Representative colony assay. (D) The data represent the mean relative NF-κB luciferase activity normalized against Renilla luciferase activity ± SD after 3-day exposure of BMS-345541 in liquid culture. The experiments were performed in triplicate. NC indicates negative control; and VC, vector-only control. (E) The bar graph represents the number of colonies observed in the methylcellulose assay. Colonies were counted after 2 weeks of culturing. The data represent mean ± SD. Each assay was performed in triplicate.
Finally, we demonstrated that down-regulation of A20 induced up-regulation of NF-κB activity and resistance to apoptosis, and gain-of-colony formation ability into LCLs.
Discussion
In the present study, we are able to show by array CGH analysis that genetic deletion of A20 occurred in several histologic subtypes of non-Hodgkin lymphoma. Among the cases examined, A20 deletion was more frequently found in MCL and ABC DLBCL, which are aggressive non-Hodgkin lymphoma subtypes. It has been shown that these 2 types frequently possess constitutive activation of NF-κB signaling and that the survival of MCL and ABC DLBCL lymphoma cells depends on the NF-κB pathway,19,25 suggesting that NF-κB activation constitutes a significant part of the clinical characteristics of these 2 lymphoma types. Our array CGH profiles of MCL and ABC DLBCL showed that A20 is located in the minimal common deleted region at chromosome band 6q. Moreover, gene expression analysis demonstrated that A20 genomic status correlates well with its expression level in ABC DLBCL and that A20 is frequently inactivated by promoter methylation or/and gene mutation in both ABC DLBCL and MCL. Therefore, A20 is the most likely target gene at the 6q deletion in ABC DLBCL and MCL. Although the scope of the lymphoma-analyzed scopes in our study did not cover all non-Hodgkin lymphoma subsets, our data indicate the presence of a novel genetic alteration associated with the process underlying deregulation of NF-κB pathways in lymphomagenesis. The variety of frequency of A20 loss in different lymphoma types suggests that the oncogenic role of A20 deletion also varies. The biologic significance of the oncogenic role of this variation needs to be further studied.
Activation of NF-κB target genes was observed in both ABC DLBCL cases, one with and one without A20 deletion, even though they differed in A20 genomic status. As for cases without A20 inactivation, other genetic mechanisms may play a role for activation of NF-κB signaling in ABC DLBCL development. Recent studies have demonstrated that CARD11 plays an essential role in survival of ABC DLBCL cell lines.23 The CARD11 mutation was indeed reported to be observed in 9.6% of ABC DLBCL clinical samples and oncogenic mutation of CARD11 to induce constitutive activation of NF-κB signaling.21 Our datasets showed no occurrence of CARD11 mutation in the ABC DLBCL cases with biallelic A20 inactivation, indicating that CARD11 mutation may contribute to deregulation of the NF-κB pathway in the cases without A20 biallelic inactivation, and subsequently gives rise to ABC DLBCL.
The finding that A20 is the target gene for 6q deletion in ABC DLBCL and MCL prompted us to examine the oncogenic role of A20. In addition, recent studies show that A20 deletion or gene mutations occur in MALT lymphoma,2,26,27 Waldenström macroglobulinemia,28 and MCL.29 Before our current study, however, the role of A20 in tumorigenesis was poorly understood and has remained controversial. A20 was initially characterized as an inhibitor of TNFα-induced apoptosis.3 The stable overexpression of A20 was reported to provide protection for TNFα-induced cytotoxicity in human breast carcinoma cell-line MCF-7 cells30 and for TP53 overexpression-induced apoptosis in H1299 epithelial cells.31 In contrast, the overexpression of A20 in vascular smooth muscle cells reportedly showed an antiproliferative effect,32 whereas the proapoptotic effect of A20 overexpression in human salivary adenoid cystic carcinoma cells has been also described.33 These contradictory observations may reflect the diversified role of A20 in various normal tissues and tumors. In this study, we were able to show that overexpression of A20 induced apoptosis in the MCL and DLBCL cell lines with A20 deletion. However, because these observations were made under the condition of overexpression and little is known about the oncogenic role of A20 inactivation, A20 knockdown study was required to gain an understanding of the significance of A20 deletion in lymphoma development.
No direct evidence has been shown that the inactivation of A20 by itself was sufficient to induce constitutive activation of NF-κB in human lymphocytes. Because A20 is involved in the negative feedback regulation for NF-κB activation,34 the inactivation of A20 was supposed to give rise to constitutive activation of NF-κB and lead to lymphoma development. In an effort to understand the precise molecular mechanism, a knockdown experiment was conducted as a part of our study, and it was found that the silencing of A20 expression indeed induced constitutive activation of NF-κB in human EB-LCL even with a partial knockdown. Our observation that haplo-insufficiency can result in NF-κB activation was in accordance with the report that the pro-atherosclerotic NF-κB target gene is elevated and atherosclerosis is increased in A20 haplo-insufficient mice,35 so that A20 haplo-insufficiency results in NF-κB deregulation. In our study, biallelic A20 inactivation occurred frequently in ABC DLBCL and MCL. However, monoallelic A20 inactivation was also found in these lymphomas. Because partial A20 knockdown could induce NF-κB activation of human LCLs, A20 may function in a haplo-insufficient manner, so that monoallelic A20 inactivation may also affect the tumorigenecity process in combination with other genetic alterations, such as the CARD11 or PRDM1 gene mutation,36 which were previously reported to be associated with lymphomagenesis. More importantly, we demonstrated, for the first time, that silencing of A20 induced resistance to apoptosis and increased colony formation ability in human EB-LCLs even with a partial knockdown, indicating that A20 acts as a tumor suppressor gene in B-cell lymphomagenesis. A recent report of gene expression analysis findings indicated that EB-LCLs resemble activated normal peripheral B cells more than normal germinal center B cells.37,38 Although the scope of the lymphomas analyzed in our study did not cover all non-Hodgkin lymphoma subsets or ABC DLBCL cell line expression profiles, these observations suggest that our A20 knockdown model of EB-LCLs may facilitate our understanding of ABC DLBCL development.
In conclusion, we were able to show that the A20 deletion occurs frequently in ABC DLBCL and MCL. A20 expression level correlated well with genomic status in ABC DLBCL. In addition, promoter methylation and gene mutation also occur frequently in ABC DLBCL and MCL, making A20 the most probable target gene of deletion 6q in these subtypes of lymphomas. Moreover, we demonstrated that overexpression of A20 induces apoptosis in lymphoma cell lines with A20 deletion and that silencing of A20 can induce constitutive NF-κB activation, resistance to apoptosis, and an increase in colony formation in human EB-LCLs, indicating that A20 acts as a tumor suppressor gene. We think that our novel findings constitute a key for establishing the link between inactivation of A20 and lymphomagenesis and may provide a novel approach for understanding the molecular mechanism of lymphoma development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Masuhiro Takahashi, Masutaka Higashimura, and Jun Takizawa for the discussions and encouragement throughout this study.
This work was supported in part by Grants-in-Aid from the Ministry of Health, Labor and Welfare, from the Ministry of Education, Culture, Sports, Science and Technology, from the Japan Society for the Promotion of Science and Foundation of Promotion of Cancer Research as well as Princess Takamatsu Cancer Research Fund (grant 03-23503) and Japan Leukemia Research Fund Wella Award (M.S.).
Authorship
Contribution: K.H. performed experiments, collected and analyzed data, and wrote the paper; S.T., M.N., and H.T. performed experiments and analyzed data; S.N. and Y.M. performed, collected, and analyzed data; and M.S. organized the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masao Seto, Division of Molecular Medicine, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan; e-mail: mseto@aichi-cc.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal