Abstract
Deregulated c-MYC is found in a variety of cancers where it promotes proliferation as well as apoptosis. In many hematologic malignancies, enhanced NF-κB exerts prosurvival functions. Here we investigated the role of NF-κB in mouse and human c-MYC–transformed lymphomas. The NF-κB pathway is extinguished in murine lymphoma cells, and extrinsic stimuli typically inducing NF-κB activity fail to activate this pathway. Genetic activation of the NF-κB pathway induces apoptosis in these cells, whereas inhibition of NF-κB by an IκBα superrepressor provides a selective advantage in vivo. Furthermore, in human Burkitt lymphoma cells we find that NF-κB activation induces apoptosis. NF-κB up-regulates Fas and predisposes to Fas-induced cell death, which is caspase-8 mediated and can be prevented by CFLAR overexpression. We conclude that c-MYC overexpression sensitizes cells to NF-κB–induced apoptosis, and persistent inactivity of NF-κB signaling is a prerequisite for MYC-mediated tumorigenesis. We could also show that low immunogenicity and Fas insensitivity of MYC-driven lymphoma cells are reversed by activation of NF-κB. Our observations provide a molecular explanation for the described absence of the NF-κB signaling in Burkitt lymphoma and question the applicability of NF-κB inhibitors as candidates for treatment of this cancer.
Introduction
The c-MYC (MYC) transcription factor has been implicated in the control of many aspects of tumor cell biology. It promotes cell proliferation and restrains differentiation. To do so, MYC controls transcription of multiple genes involved in cell growth and metabolism, vasculogenesis, cell adhesion, and genomic stability.1 MYC is a basic helix-loop-helix-leucine zipper transcription factor that dimerizes with the related protein MAX. MYC/MAX heterodimers bind to specific DNA elements, designated as E-boxes, located in the promoter regions of target genes mediating either activation or repression of transcription.2 Complex regulatory networks control MYC activity leading to up-regulation of its expression in response to mitogens or suppression upon growth inhibitory signals.3 Increased amounts of MYC protein are found in many types of human cancer because control mechanisms keeping MYC in check are inactivated during malignant transformation of cells.1,3 In Burkitt lymphoma (BL), a highly aggressive non-Hodgkin lymphoma, overexpression of MYC is invariably connected to chromosomal translocations of the MYC proto-oncogene to immunoglobulin loci. Elevated expression of MYC likewise induces malignant transformation in mouse models and persistent MYC expression is required for tumor growth.4
MYC induces proliferation, but at the same time it can induce cell death. Several MYC-induced pathways, such as activation of the p53/ARF pathway, changes in expression, and activity of BCL2 proteins or alterations in death receptor signaling have been linked to apoptosis.3 These most likely serve as safeguard mechanisms against unrestricted growth and malignant transformation. Inactivation of the MYC-induced apoptotic program is therefore a prerequisite for tumorigenesis.
The NF-κB transcription factor is also linked to tumor initiation and progression.5,6 NF-κB binds to the DNA as homodimer or heterodimer composed of 2 of 5 related subunits: RelA(p65), RelB, c-Rel, p50/105 (NF-κB1), and p52/100 (NF-κB2).7 Inactive NF-κB complexes are sequestered in the cytosol bound to inhibitors, the IκB proteins. Activation of NF-κB involves phosphorylation, ubiquitination, and proteasomal degradation of the IκB proteins allowing nuclear translocation of the NF-κB dimer. Initiation of transcription relies on NF-κB DNA binding that may involve multimerization with additional proteins such as RPS3.8 The IκB kinase (IKK) complex consisting of IKKα/IKK1, IKKβ/IKK2, and the regulatory subunit IKKγ/NEMO phosphorylates the IκB proteins. In the classical pathway predominantly IKK2 is active. Activation of the IKK complex occurs downstream of many oncoproteins, and NF-κB target genes involved in cell proliferation, growth, and survival are induced in many different human tumors.9 Mutations leading to enhanced NF-κB activity are a recurrent theme in hematologic malignancies.10 NF-κB initiates expression of various antiapoptotic proteins, such as BCL-2, BCL-XL, cIAPs, and CFLAR (c-FLIP). Thus, apoptosis evasion is an important contribution of NF-κB in the pathogenesis of human tumors.9
Surprisingly, in the EμMyc-mouse model NF-κB activity was found to be dispensable for lymphomagenesis.11 Low NF-κB DNA-binding activities were also reported for a conditional MYC-driven human B-cell lymphoma line.12 Moreover, in 2 recent studies low expression of NF-κB target genes was reported as hallmarks of BL.13,14 We therefore investigated the role of NF-κB in MYC-driven lymphomas using genetic approaches to either induce or block the NF-κB pathway. Both in conditionally MYC-transformed murine lymphomas as well as in human BL we find that NF-κB functions as a tumor suppressor.
Methods
Cell lines and cell culture
Lymphoma cells were cultured in RPMI medium supplemented with 10% FCS, l-glutamine (2 mM), penicillin (100 U/mL), streptomycin (100 μg/mL), nonessential amino acids, and β-mercaptoethanol (50 μM). In murine lymphoma cell lines, MYC expression was abrogated by doxycycline (Dox) treatment (2 μg/mL) for the indicated time periods. For NF-κB activation, cells were stimulated with TNFα (40 ng/mL), PMA (5 ng/mL) and ionomycin (1 μg/mL), LPS (1 μg/mL), daunorubicin (5 μM), or okadaic acid (200 nM) for the indicated times.
Western blot analysis and electrophoretic mobility shift assay
Preparation of nuclear and whole-cell extracts was performed as described earlier.15 For Western blot analysis 40 μg whole-cell protein extracts per lane were separated on 12.5% polyacrylamide gels and transferred onto polyvinylene difluoride membranes (Millipore). Membranes were blocked with 7.5% dry milk in TBS containing 0.2% Tween 20. For subsequent washes, 0.2% Tween 20 in TBS was used. Affinity-purified rabbit antibodies against IKK2 (SC-7607), IκBα (SC-371), RelA/p65 (SC-372) or MYC (SC-764; Santa Cruz Biotechnology), and horseradish peroxidase–coupled goat anti–rabbit IgG secondary antibody (Thermo Scientific) were used. For electrophoretic mobility shift assays (EMSA), 5 μg nuclear or whole-cell extracts was incubated for 20 minutes at room temperature with 3 μg poly(dI/dC), 10 μg bovine serum albumin in binding buffer (50 mM NaCl, 1 mM dithiothreitol, 10 mM Tris-HCl, 1 mM EDTA, 5% glycerol), and radiolabeled oligonucleotides containing specific sites for binding of NF-κB (5′-GCC TGG GAA AGT CCC CTC AA-3′), Oct (5′-ACC TGG GTA ATT TGC ATT TCT AAA AT-3′), or SP-1 (5′-ATT CGA TCG GGG CGG GGC GAG C-3′). DNA-protein complexes were separated on native 6% polyacrylamide gels.
Retroviral vectors and stable producer cell line
The pCFG-IEGZ retroviral vectors containing TD-IκBα or CA-IKK2 constructs have been described earlier.16 φNX producer cells (2 × 106/10-cm plate) were grown in DMEM supplemented with FCS (10%), l-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL) and transfected with 10 μg plasmid DNA using calcium phosphate. Forty-eight hours after transfection, efficiency was determined by flow cytometric analysis of GFP expression (typically 70%-80% positive). Supernatants were supplemented with 8 μg/mL polybrene, and cells (5 × 105/well in 24-well plates) were infected with 750 μL/well by spin infection (90 minutes at 1200g and 37°C). Twenty-four hours after infection, efficiency was determined by flow cytometric GFP analysis. Infected cells were selected with 100 μg/mL Zeocin for 10 to 14 days and GFP expression was checked by flow cytometry.
Tumor transplantation
Murine B-lymphoma line 5522 transduced with retroviral IEGZ-empty vector or IEGZ-TD-IκBα vector were mixed with untransduced parental cells at a ratio of 1:10. Lymphoma cells (107) of the mixed populations were injected intraperitoneally into syngeneic mice. After 1 week, recipient mice were killed and tumor cells were isolated from ascites. Percentages of GFP+ cells in the isolated populations were determined by flow cytometry. Approval for the use of mice in this study was obtained from the Regierungspraesidium (TV-709).
Vectors and transfections
pSFI and pRTS vectors were kindly provided by D. Eick and G. W. Bornkamm.17 TD-IκBα and CA-IKK2 cDNAs15 were first inserted into blunt-end EcoRV sites of the pSFI subcloning vector and then recloned into the pRTS-1 vector. pRETRO-SUPER vectors containing anti–caspase-8 shRNA had the following sequence: forward, 5′-gatctcgggtcatgctctatcagatttcaagagaatctgatagagcatgaccctttttggaaa-3′ and reverse, 5′-agcttttccaaaaagggtcatgctctatcagattctcttgaaatctgatagagcatgacccga-3′. pcDNA-FLIPL was kindly provided by P. Krammer.18 Transfections were performed by Nucleofection (Amaxa). Conditional transgene expression was induced with doxycycline (0.5 μg/mL) and GFP expression was analyzed by flow cytometer.
Cell counts and fluorescence-activated cell sorting analyses
Proliferation of cells was determined by viable cell counts (trypan blue staining) or by flow cytometer. Cell counts were performed in triplicates. Apoptosis was determined by annexin-V and 7-AAD stainings (Apoptosis detection kit; BD Pharmingen) and flow cytometric analysis. Apoptosis was inhibited by addition of pan-caspase inhibitor z-VAD (20 μM). Surface expression of Fas was determined by staining with APO-I and Alexa-647–conjugated secondary antibody. GFP+ and GFP− fractions were analyzed separately for Alexa-647 fluorescence.
Gene expression profiling
Ramos cells were transfected in triplicates with pRTS-GFP or pRTS-CA-IKK2, selected with hygromycin, and transgene expression was induced with doxycycline (0.5 μg/mL) for 48 hours. RNA was isolated with RNeasy mini kit (QIAGEN), and gene expression profiling (GEP) was performed using Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix). Total RNA (2 μg) was labeled using the GeneChip One-Cycle Target Labeling assay kit (Affymetrix). After hybridization, arrays were stained and washed in a FS 450 Fluidics station (Affymetrix) before imaging on an Affymetrix GeneChip (3000) scanner. Raw data were generated using the GCOS 1.4 software (Affymetrix). Probe level data were obtained using the robust multichip average (RMA) normalization algorithm and CEL files were loaded into Genesifter (http://genesifter.net; VizX Laboratories). Genes were identified as differentially expressed among the 2 classes if a 2-sample t test revealed a nominal significance level of .05 and the ratio between the 2 classes was at least 2-fold. Calculation of false discovery rate was done according to the method of Benjamini and Hochberg.19 Biologic significance was determined using Gene Ontology reports.20 Microarray data have been deposited with Gene Expression Omnibus (GEO) under accession number GSE17129.21
RNA-isolation and reverse-transcription–PCR
Total RNA was isolated with QIAzol lysis reagent (QIAGEN). Total RNA (2 μg) was reverse transcribed with AMV reverse transcriptase (Roche) and polymerase chain reaction (PCR) was performed using Taq DNA polymerase (Amersham Pharmacia Biotech). The primers used were hFas_fwd (5′-CAA GTG ACT GAC ATC AAC TCC-3′), hFas_rev (5′-CCT TGG TTT TCC TTT CTG TGC-3′), hb-actin_fwd (ATC TGG CAC CAC ACC TTC TAC AAT GAG CTG CG-3′), and hb-actin_rev (5′-CGT CAT ACT CCT GCT TGC TGA TCC ACA TCT GC-3′). Conditions for Fas PCR were as follows: 30 cycles at 56°C for 40 seconds, 72°C for 1 minute, and 94°C for 40 seconds. Conditions for β-actin were as follows: 20 cycles at 58°C for 40 seconds, 72°C for 70 seconds, and 94°C for 40 seconds.
Apoptosis induction
After transfection with pRTS-GFP or pRTS-CA-IKK2, Ramos cells were treated with doxycycline (0.5 μg/mL) for 48 hours to induce transgene expression. APO-I containing supernatant (5% final concentration [f.c.]) was added and apoptosis was determined by trypan blue stainings.
Results
MYC-dependent murine lymphoma cells lack NF-κB activity
To evaluate a potential role of the NF-κB transcription factor for MYC-driven lymphoma development, we determined its activity in conditionally MYC-transformed mouse lymphoma cell lines.4,22 Basal NF-κB activity was measured by EMSA and revealed low to undetectable NF-κB in these cells (Figure 1A). We investigated the effects of typical NF-κB–inducing stimulators on NF-κB binding activity. Most T- and B-cell lymphomas showed either no or very low induction of NF-κB activity in response to LPS, TNFα, or PMA/ionomycin (Figure 1B). To test whether alternative activation pathways are functional in these cells, we checked for NF-κB activity after stimulation with daunorubicin and okadaic acid. Cells showed robust increases in NF-κB binding, indicating the presence of functional NF-κB proteins in the cytosol (Figure 1C).
NF-κB activity is impaired in the MYC-transformed cells. (A) NF-κB EMSA using nuclear extracts of different murine lymphoma cells. WEHI231 cells show constitutive NF-κB activity and were used as control. Oct1 DNA binding served as a control for the integrity of the nuclear extracts. (B) EMSA using protein extracts from lymphoma cells treated with the standard NF-κB inducers TNFα (40 ng/mL), PMA (5 ng/mL) and ionomycin (1 μg/mL), or LPS (1 μg/mL) for the indicated time points. Jurkat cells treated with PMA/ionomycin for 1 hour were used as control for NF-κB binding. Sp1 DNA was used as a control for the integrity of the cell extracts. (C) NF-κB EMSA with nuclear extracts of B-cell lymphoma cell line 5522 treated for 2 hours with daunorubicin (5 μM) or okadaic acid (200 nM). Oct1 was used as control for the integrity of the extracts. (D) Immunoblot with cell extracts of B-cell lymphoma cell lines. Doxycycline treatment abrogates transgenic MYC expression. Cells were treated with doxycycline (2 μg/mL) for 12 hours or were left untreated. Expression of p65 was used as loading control. (E) EMSA using cell extracts of 2 lymphoma cell lines demonstrates induction of NF-κB activity in the absence of MYC expression. Immunoblot shows transient loss of MYC expression after doxycycline treatment. Cells were left untreated (lanes 1-2), pulse treated with doxycycline for 12 hours (lanes 3-4), and again left untreated for the consecutive days (lanes 5-6).
NF-κB activity is impaired in the MYC-transformed cells. (A) NF-κB EMSA using nuclear extracts of different murine lymphoma cells. WEHI231 cells show constitutive NF-κB activity and were used as control. Oct1 DNA binding served as a control for the integrity of the nuclear extracts. (B) EMSA using protein extracts from lymphoma cells treated with the standard NF-κB inducers TNFα (40 ng/mL), PMA (5 ng/mL) and ionomycin (1 μg/mL), or LPS (1 μg/mL) for the indicated time points. Jurkat cells treated with PMA/ionomycin for 1 hour were used as control for NF-κB binding. Sp1 DNA was used as a control for the integrity of the cell extracts. (C) NF-κB EMSA with nuclear extracts of B-cell lymphoma cell line 5522 treated for 2 hours with daunorubicin (5 μM) or okadaic acid (200 nM). Oct1 was used as control for the integrity of the extracts. (D) Immunoblot with cell extracts of B-cell lymphoma cell lines. Doxycycline treatment abrogates transgenic MYC expression. Cells were treated with doxycycline (2 μg/mL) for 12 hours or were left untreated. Expression of p65 was used as loading control. (E) EMSA using cell extracts of 2 lymphoma cell lines demonstrates induction of NF-κB activity in the absence of MYC expression. Immunoblot shows transient loss of MYC expression after doxycycline treatment. Cells were left untreated (lanes 1-2), pulse treated with doxycycline for 12 hours (lanes 3-4), and again left untreated for the consecutive days (lanes 5-6).
Growth and survival of cell lines established from tumor-bearing mice have been shown to be dependent on constant MYC overexpression. This oncogene addiction is a common attribute of MYC-induced tumors. To test whether MYC itself has an influence on NF-κB activity, we turned off its expression by addition of doxycycline. This treatment abrogated transgenic MYC expression in the lymphoma cells (Figure 1D). After washing out doxycycline, MYC expression was recovered (Figure 1E bottom panel). In cells lacking MYC, we detected NF-κB activity by EMSA and nuclear translocation of p65 (Figure 1Eiii-iv, supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). However, upon re-expression of MYC, NF-κB activity was again abolished (Figure 1Ev-vi).
Murine lymphoma cells are sensitive to IKK2 activation
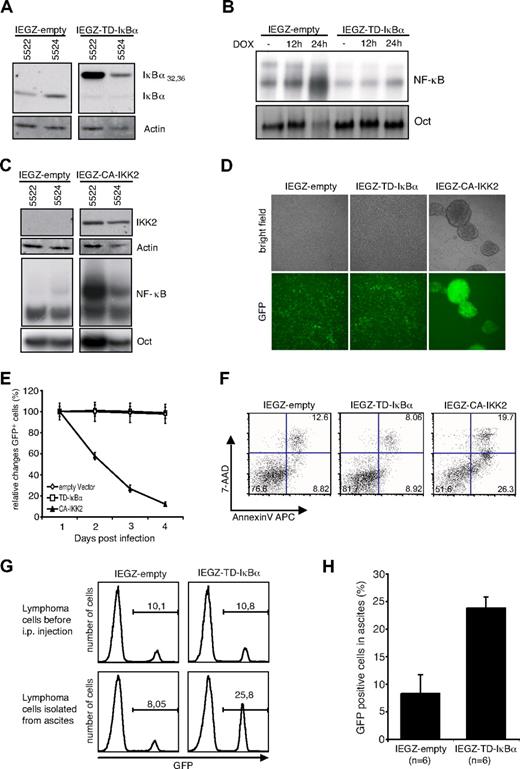
To determine whether absence of NF-κB signaling is critical for MYC-induced tumorigenesis, we modulated NF-κB activity by retroviral introduction of dominant mutants interfering with the NF-κB pathway. Inhibition of NF-κB activity was achieved by a transdominant IκBα (S32,36A) superrepressor (IEGZ-TD-IκBα), which cannot be phosphorylated by IKK. A constitutively active allele of the IκB kinase 2 (IEGZ-CA-IKK2) was used to activate the NF-κB pathway. Internal ribosomal entry sites couple expression of NF-κB modulators with a GFP-Zeocin fusion protein, allowing identification and selection of infected cells.23,24
Several MYC-dependent lymphoma B-cell lines were infected with either the TD-IκBα, the CA-IKK2 mutant, or a control vector (IEGZ-empty) containing only the GFP-Zeocin fusion gene. Twenty-four hours later, frequencies of GFP-expressing cells were evaluated by flow cytometry. Infection efficiencies were similar for the different viruses (supplemental Figure 2A). Expression of TD-IκBα was detectable 24 hours after infection (Figure 2A), accompanied by a reduction of endogenous IκBα levels. The activation of NF-κB after MYC down-regulation was strongly reduced in the presence of the superrepressor (Figure 2B). Expression of CA-IKK2 was also detectable 24 hours after infection and resulted in a strong induction of NF-κB (Figure 2C).
Modulation of NF-κB signaling pathway in murine lymphoma cells. (A) Immunoblot analysis of TD-IκBα expression in transduced murine lymphoma cell line. Cells were spin-infected with retroviral vectors IEGZ-empty or IEGZ-TD-IκBα and protein extracts were isolated 24 hours after infection. (B) EMSA of NF-κB binding activity in transduced lymphoma cell lines. NF-κB activation after doxycycline-mediated MYC inactivation was detectable only in IEGZ-empty– but not in IEGZ-TD-IκBα–transduced cells. Doxycycline (2 μg/mL) was added for 12 or 24 hours, respectively. (C) Immunoblot analysis of CA-IKK2 expression in transduced lymphoma lines. Protein extracts of transduced cells were isolated 24 hours after the infection. EMSA of the extracts displays induction of NF-κB binding in CA-IKK2–expressing cells. (D) Morphologic examination of transduced cell line 5522. Bright field and fluorescence microscopy of transduced cells 2 days after infection. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (E) Flow cytometric determination of changes in the GFP-positive fractions in transduced cultures. Murine lymphoma cells were transduced with IEGZ-empty (◇), IEGZ-TD-IκBα (□), or IEGZ-CA-IKK2 (▲), and percentage of GFP+ cells was measured by flow cytometer at the indicated time points after retroviral infection. Percentage of GFP+ cells at day 1 after infection was set to 100% to facilitate comparisons. (F) Apoptosis detection of murine lymphoma cell line transduced with IEGZ-empty, IEGZ-TD-IκBα, or IEGZ-CA-IKK2. At day 3 after the infection the cells were stained and analyzed for annexin-V–APC and 7-AAD binding. (G) In vivo tumor competition assay. Murine B-lymphoma cell line 5522 was transduced with either IEGZ-empty or TD-IκBα. Transduced GFP+ cells were then mixed with parental nontransduced cells at a ratio of 1:10. Cells of mixed populations (107) were injected intraperitoneally into syngeneic recipient mice and lymphocytes were isolated from ascites 1 week later. Flow cytometric determination of the GFP+ fractions is shown for the cells before transplantation (top panels) and after isolation of ascites of recipient mice (bottom panels). (H) Statistical analyses of competitive tumor transplantation. Mean values of the percentage of the GFP+ fraction of cells isolated from recipient mice (n = 6 for each group) detected by flow cytometry.
Modulation of NF-κB signaling pathway in murine lymphoma cells. (A) Immunoblot analysis of TD-IκBα expression in transduced murine lymphoma cell line. Cells were spin-infected with retroviral vectors IEGZ-empty or IEGZ-TD-IκBα and protein extracts were isolated 24 hours after infection. (B) EMSA of NF-κB binding activity in transduced lymphoma cell lines. NF-κB activation after doxycycline-mediated MYC inactivation was detectable only in IEGZ-empty– but not in IEGZ-TD-IκBα–transduced cells. Doxycycline (2 μg/mL) was added for 12 or 24 hours, respectively. (C) Immunoblot analysis of CA-IKK2 expression in transduced lymphoma lines. Protein extracts of transduced cells were isolated 24 hours after the infection. EMSA of the extracts displays induction of NF-κB binding in CA-IKK2–expressing cells. (D) Morphologic examination of transduced cell line 5522. Bright field and fluorescence microscopy of transduced cells 2 days after infection. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (E) Flow cytometric determination of changes in the GFP-positive fractions in transduced cultures. Murine lymphoma cells were transduced with IEGZ-empty (◇), IEGZ-TD-IκBα (□), or IEGZ-CA-IKK2 (▲), and percentage of GFP+ cells was measured by flow cytometer at the indicated time points after retroviral infection. Percentage of GFP+ cells at day 1 after infection was set to 100% to facilitate comparisons. (F) Apoptosis detection of murine lymphoma cell line transduced with IEGZ-empty, IEGZ-TD-IκBα, or IEGZ-CA-IKK2. At day 3 after the infection the cells were stained and analyzed for annexin-V–APC and 7-AAD binding. (G) In vivo tumor competition assay. Murine B-lymphoma cell line 5522 was transduced with either IEGZ-empty or TD-IκBα. Transduced GFP+ cells were then mixed with parental nontransduced cells at a ratio of 1:10. Cells of mixed populations (107) were injected intraperitoneally into syngeneic recipient mice and lymphocytes were isolated from ascites 1 week later. Flow cytometric determination of the GFP+ fractions is shown for the cells before transplantation (top panels) and after isolation of ascites of recipient mice (bottom panels). (H) Statistical analyses of competitive tumor transplantation. Mean values of the percentage of the GFP+ fraction of cells isolated from recipient mice (n = 6 for each group) detected by flow cytometry.
When we analyzed the infected cells it became apparent that CA-IKK2–expressing cells displayed a dramatically altered phenotype. Whereas IEGZ-empty– and IEGZ-TD-IκBα–infected cells showed single-cell distribution, expression of CA-IKK2 induced pronounced aggregation leading to large clusters of cells (Figure 2D). Selection for Zeocin resistance was feasible for TD-IκBα or control vector, but failed for the CA-IKK2 infections. These results implicate that CA-IKK2 expression interferes with proliferation and/or viability of the lymphoma cells. To investigate this issue we monitored changes in the infected GFP+ compartment in the absence of selective pressure. No changes in the percentages of GFP+ populations were seen in empty vector and TD-IκBα–infected cultures. In contrast, GFP+ cells rapidly disappeared from CA-IKK2–infected cultures (Figure 2E; supplemental Figure 2B). Annexin-V and 7-AAD staining revealed increased levels of apoptosis after expression of CA-IKK2 (Figure 2F; supplemental Figure 2C). We therefore conclude that the reduction of CA-IKK2–infected cells is at least partially the result of apoptosis induction.
To test whether blocking of NF-κB might have a selective advantage for lymphoma cells, we performed in vivo competition assays. MYC-driven lymphoma cells transduced with retroviral IEGZ-empty or IEGZ-TD-IκBα were combined with parental cells at a ratio of 1:10. Flow cytometric analysis confirmed that approximately 10% of these mixed populations were GFP positive (Figure 2G). Murine lymphoma cells (107) of the mixed populations were injected intraperitoneally into syngeneic mice. Recipient mice developed ascites and lymphocytes, isolated from ascites, were analyzed for GFP (Figure 2G). The fraction of GFP-positive cells within the group carrying the TD-IκBα–transduced cells was increased to 23.8 plus or minus 2.0, whereas it stayed at roughly 10% when empty virus–infected cells were mixed (Figure 2H). This indicates a selective advantage for lymphoma cells with blocked NF-κB activation.
IKK2 activation induces growth inhibition and apoptosis in human Burkitt lymphoma cells
NF-κB activity has been described in human MYC-expressing lymphomas,25,26 arguing against a general incompatibility of these transcription factors in human cancer cells. We therefore analyzed consequences of NF-κB modulation in human MYC-driven lymphomas. The best model is the highly aggressive human Burkitt lymphoma (BL), where transcriptional deregulation of MYC due to chromosomal translocation is the pivotal event in lymphomagenesis.
We modulated NF-κB activity in the Ramos BL cell line using an episomal conditional expression system (pRTS) allowing high expression of the transgene upon doxycycline addition without any background activity in the absence of inducer.17 Ramos cells were transfected with a control plasmid containing only GFP (pRTS-GFP), the IκBα superrepressor (pRTS-TD-IκBα), or the constitutive active IKK2 (pRTS-CA-IKK2). Selection for hygromycin resistance and induction with doxycycline resulted in more than 90% GFP-positive cells (supplemental Figure 3) and transgene expression (Figure 3A). Ramos cells show low basal NF-κB levels (Figure 3B) consisting of RelB-containing heterodimers (supplemental Figure 4),15,27 which were reduced in TD-IκBα–expressing cells (Figure 3B). CA-IKK2 mediated an induction of NF-κB activity (Figure 3B), which consists of p50 and p65/RelA subunits (supplemental Figure 4).
Transfection of Ramos cells with inducible NF-κB modulators. (A) Immunoblot analysis of transfected Ramos cells untreated or treated with doxycycline (0.5 μg/m) for 48 hours. Cell extracts were analyzed for CA-IKK2, for TD-IκBα, and as a control for actin expression. (B) EMSA of cell extracts shows induction of NF-κB activity after doxycycline addition. Oct1 DNA was used as control for the integrity of the extracts. (C) Morphologic examination of transfected Ramos. Bright field and fluorescence microscopy of cells treated with doxycycline for 5 days. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (D) Proliferation assay of Ramos expressing pRTS-GFP (◇), pRTS-TD-IκBα (□), or pRTS-CA-IKK2 (▲). GFP-positive cells were counted by flow cytometer at the indicated time points after doxycycline addition. Error bars represent SDs from triplicate experiments.
Transfection of Ramos cells with inducible NF-κB modulators. (A) Immunoblot analysis of transfected Ramos cells untreated or treated with doxycycline (0.5 μg/m) for 48 hours. Cell extracts were analyzed for CA-IKK2, for TD-IκBα, and as a control for actin expression. (B) EMSA of cell extracts shows induction of NF-κB activity after doxycycline addition. Oct1 DNA was used as control for the integrity of the extracts. (C) Morphologic examination of transfected Ramos. Bright field and fluorescence microscopy of cells treated with doxycycline for 5 days. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (D) Proliferation assay of Ramos expressing pRTS-GFP (◇), pRTS-TD-IκBα (□), or pRTS-CA-IKK2 (▲). GFP-positive cells were counted by flow cytometer at the indicated time points after doxycycline addition. Error bars represent SDs from triplicate experiments.
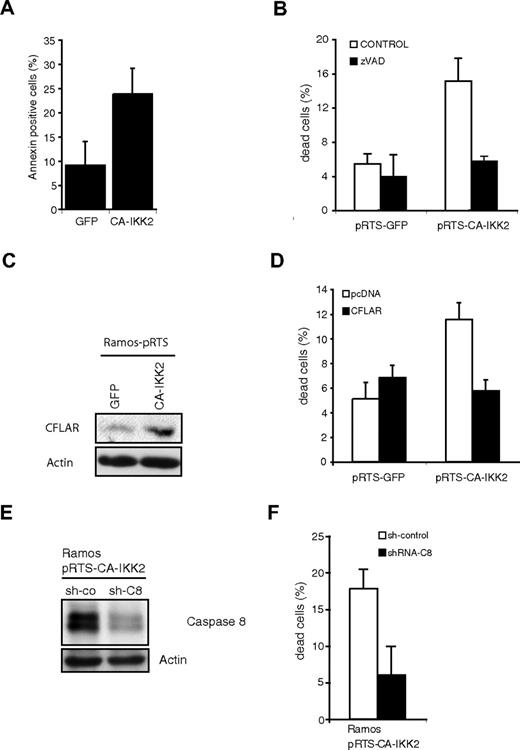
Concerning morphologic appearance of the infected cultures, no differences between TD-IκBα– and control-transfected cells or nontransfected cells were seen (Figure 3C and data not shown). However, upon CA-IKK2 induction Ramos cells showed rapid clumping, highly reminiscent of the phenotype induced in murine lymphoma lines (Figure 3C; Figure 2E). This phenotype was also observed in the transfected BL cell lines Raji, Namalwa, and BL-30 upon transgene induction (supplemental Figure 5). We analyzed the proliferation by counting GFP-positive cells over a time course of 1 week. Whereas no difference was seen between TD-IκBα–expressing cells and controls, the CA-IKK2–expressing cells showed reduced proliferation (Figure 3D). CA-IKK2–transfected cultures showed a higher percentage of annexin-V–positive cells (Figure 4A). In addition, we found increased percentages of dead cells in Ramos-expressing CA-IKK2 (Figure 4B). Also in Raji, Namalwa, and BL-30 cells expressing CA-IKK2 we observed 2- to 4-fold increases in dead cell numbers compared with pRTS-GFP– or pRTS-TD-IκBα–transfected cells (supplemental Figure 6). Increased cell death after CA-IKK2 expression was prevented by addition of the pan-caspase inhibitor zVAD, suggesting that NF-κB induces apoptosis (Figure 4B).
IKK2 expression induces the extrinsic apoptotic pathway. (A) Apoptosis detection in Ramos cells transfected with empty vector or CA-IKK2. Doxycycline (0.5 μg/mL) was added to the culture medium for 5 days before cells were analyzed for annexin-V binding. (B) Percentages of dead cells after induction of transgene expression without (▭) or in the presence of caspase inhibitor zVAD (▭). Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline and in the presence or absence of zVAD (20 μM) for 4 days. (C) Expression of CFLAR in Ramos-pRTS-GFP and Ramos-pRTS-CA-IKK2. Immunoblot analysis of Ramos cells transfected with pRTS-GFP or pRTS-CA-IKK2 and treated with doxycycline (0.5 μg/m) for 48 hours. Cell extracts were analyzed for CFLAR and as a control for actin expression. (D) Percentages of dead cells after transfection of Ramos-pRTS-GFP and Ramos-pRTS-CA-IKK2 with either pcDNA (control, ▭) or a CFLAR expression vector (▬). Cells were treated with doxycycline for 7 days to induce the pRTS-system. (E) Immunoblot analysis of Ramos-pRTS-CA-IKK2 cells stably transduced with shRNA expression vectors encoding either a scrambled shRNA (sh-co) or shRNA against caspase 8 (sh-C8). (F) Percentages of dead Ramos cells expressing the nonsense shRNA (▭) or the shRNA against caspase-8 (▬) after CA-IKK2 transgene expression. Cells were treated with doxycycline (0.5 μg/m) for 6 days. All error bars represent SDs from triplicate experiments.
IKK2 expression induces the extrinsic apoptotic pathway. (A) Apoptosis detection in Ramos cells transfected with empty vector or CA-IKK2. Doxycycline (0.5 μg/mL) was added to the culture medium for 5 days before cells were analyzed for annexin-V binding. (B) Percentages of dead cells after induction of transgene expression without (▭) or in the presence of caspase inhibitor zVAD (▭). Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline and in the presence or absence of zVAD (20 μM) for 4 days. (C) Expression of CFLAR in Ramos-pRTS-GFP and Ramos-pRTS-CA-IKK2. Immunoblot analysis of Ramos cells transfected with pRTS-GFP or pRTS-CA-IKK2 and treated with doxycycline (0.5 μg/m) for 48 hours. Cell extracts were analyzed for CFLAR and as a control for actin expression. (D) Percentages of dead cells after transfection of Ramos-pRTS-GFP and Ramos-pRTS-CA-IKK2 with either pcDNA (control, ▭) or a CFLAR expression vector (▬). Cells were treated with doxycycline for 7 days to induce the pRTS-system. (E) Immunoblot analysis of Ramos-pRTS-CA-IKK2 cells stably transduced with shRNA expression vectors encoding either a scrambled shRNA (sh-co) or shRNA against caspase 8 (sh-C8). (F) Percentages of dead Ramos cells expressing the nonsense shRNA (▭) or the shRNA against caspase-8 (▬) after CA-IKK2 transgene expression. Cells were treated with doxycycline (0.5 μg/m) for 6 days. All error bars represent SDs from triplicate experiments.
An important antiapoptotic target gene of NF-κB is CFLAR (cFLIP), which prevents recruitment of caspase-8 to the death-inducing signaling complex. Interestingly, a direct repression of CFLAR by MYC has been reported, which renders cells more susceptible to extrinsic stimuli.28 CFLAR expression was hardly detectable by Western blot in Ramos pRTS-GFP control cells and expression increased in cells expressing CA-IKK2 (Figure 4C). However, levels were low compared with other cell lines with constitutive NF-κB activity (eg, Hodgkin cell line L428; data not shown). We tested whether ectopic CFLAR protects Ramos cells from CA-IKK2–induced apoptosis and found that transfection with a CFLAR expression vector blocked the apoptosis induced by CA-IKK2 (Figure 4D).
As CFLAR prevents caspase-8 activation, we asked whether knockdown of this protein could protect Ramos cells from CA-IKK2–induced apoptosis. Ramos cell lines stably expressing a specific shRNA showed reduced caspase-8 protein levels (Figure 4E). In these cells, CA-IKK2–induced cell death was almost completely abolished (Figure 4F). These results imply that the extrinsic death signaling pathway is responsible for the IKK2-induced apoptosis.
IKK2 effects are NF-κB dependent
To determine whether CA-IKK2–induced cell death is also seen in other lymphoma entities, we analyzed a human classical Hodgkin lymphoma cell line (KM-H2) and a cell line representing primary mediastinal B-cell lymphoma (MedB1). These cells already show considerable NF-κB activity. Conditional CA-IKK2 expression again exceeded the endogenous levels of IKK2 (Figure 5A and supplemental Figure 7A). When apoptosis in CA-IKK2–expressing KM-H2 and MedB1 cells was analyzed, no significant increase was seen (Figure 5B). As the level of NF-κB is limited by the amount of available inhibited NF-κB complexes, all observed NF-κB inductions were well within the physiologic range (supplemental Figure 7B).
Morphologic alterations and induced cell death after CA-IKK2 expression are NF-κB dependent. (A) Immunoblot of MedB1 and KM-H2 cell transfected with either pRTS-GFP or pRTS-CA-IKK2. Cells were treated with doxycycline (0.5 μg/mL) for 48 hours to induce transgene expression. Cell extracts were analyzed for CA-IKK2 and, as a control, for actin expression. (B) Percentages of dead cells transfected with pRTS-GFP (▭) or pRTS-CA-IKK2 (▬) after induction of transgene expression. Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline (0.5 μg/m) for 6 days. (C) Immunoblot analysis of Ramos cells stably transduced with IEGZ-empty or IEGZ-TD-IκBα and transfected with inducible pRTS-GFP or pRTS-CA-IKK2. Doxycycline was added to cultures (0.5 μg/mL) for 48 hours to induce transgene expression. (D) Morphologic examination of transfected Ramos cells. Fluorescence microscopy of cells treated with doxycycline for 5 days. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (E) Proliferation assay of Ramos-expressing NF-κB modulators. Ramos cells were stably transduced with IEGZ-empty and transfected with either pRTS-GFP (■) or pRTS-CA-IKK2 (○), or transduced with IEGZ-TD-IκBα transfected with either pRTS-GFP (●) or pRTS-CA-IKK2 (▵). Viable cell numbers were determined by trypan blue exclusion at the indicated time points. (F) Viable cell counts of transduced Ramos cells transfected with either pRTS-GFP (▭) or pRTS-CA-IKK2 (▬). Increased percentages of dead cells after CA-IKK2 expression were reduced in the presence of TD-IκBα. Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline (0.5 μg/mL) for 6 days. All error bars represent SDs from triplicate experiments.
Morphologic alterations and induced cell death after CA-IKK2 expression are NF-κB dependent. (A) Immunoblot of MedB1 and KM-H2 cell transfected with either pRTS-GFP or pRTS-CA-IKK2. Cells were treated with doxycycline (0.5 μg/mL) for 48 hours to induce transgene expression. Cell extracts were analyzed for CA-IKK2 and, as a control, for actin expression. (B) Percentages of dead cells transfected with pRTS-GFP (▭) or pRTS-CA-IKK2 (▬) after induction of transgene expression. Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline (0.5 μg/m) for 6 days. (C) Immunoblot analysis of Ramos cells stably transduced with IEGZ-empty or IEGZ-TD-IκBα and transfected with inducible pRTS-GFP or pRTS-CA-IKK2. Doxycycline was added to cultures (0.5 μg/mL) for 48 hours to induce transgene expression. (D) Morphologic examination of transfected Ramos cells. Fluorescence microscopy of cells treated with doxycycline for 5 days. Images were taken with a Leica DMIRBE microscope 10× NA = 0.3 PH1 acquired with a Hamamatsu digital camera C4742-95 (Hamamatsu Photonics) and Open Lab software Version 4.0.4 (Improvision). (E) Proliferation assay of Ramos-expressing NF-κB modulators. Ramos cells were stably transduced with IEGZ-empty and transfected with either pRTS-GFP (■) or pRTS-CA-IKK2 (○), or transduced with IEGZ-TD-IκBα transfected with either pRTS-GFP (●) or pRTS-CA-IKK2 (▵). Viable cell numbers were determined by trypan blue exclusion at the indicated time points. (F) Viable cell counts of transduced Ramos cells transfected with either pRTS-GFP (▭) or pRTS-CA-IKK2 (▬). Increased percentages of dead cells after CA-IKK2 expression were reduced in the presence of TD-IκBα. Viable and dead cell numbers were determined by trypan blue exclusion. Cells were treated with doxycycline (0.5 μg/mL) for 6 days. All error bars represent SDs from triplicate experiments.
Although phosphorylation of the IκB proteins is the best-established function of IKK2, other targets of this kinase have been described.29 To identify the relevant pathway, we transduced Ramos cells with a retroviral vector expressing TD-IκBα (IEGZ-TD-IκBα). Infected cells (more than 95% positive for GFP; Figure 5C bottom panels; supplemental Figure 8) were transfected with pRTS-CA-IKK2 or a control construct. EMSA revealed that TD-IκBα completely blocks CA-IKK2–induced NF-κB activity (Figure 5C). The CA-IKK2–induced massive clumping phenotype was largely attenuated (Figure 5D). Furthermore, TD-IκBα expression blocks CA-IKK2–mediated effects on proliferation (Figure 5E) and cell death, indicating that apoptosis induction is entirely NF-κB dependent (Figure 5F).
To rule out the possibility that CA-IKK2 expression leads to supraphysiologic levels of NF-κB in the transfected BL, we induced physiologic levels of endogenous NF-κB by stimulation of CD40. Treatment with anti-CD40 antibody (G28.5) induced NF-κB binding in Ramos but not in Namalwa cells (supplemental Figure 9A). Treated Ramos cells showed rapid clumping, whereas no alterations were seen for Namalwa cells (supplemental Figure 9B). Furthermore, Ramos cells showed reduced proliferation and viability after anti-CD40 treatment (supplemental Figure 9C-D).
Physiologic NF-κB stimulation had similar effects in responsive murine MYC-driven lymphoma cells. Whereas most of the tested cell lines did not induce NF-κB in response to CD40 stimulation, we identified 1 responsive cell line. In this cell line, anti-CD40–induced NF-κB was accompanied by cell clumping, reduced proliferation, and reduced viability (supplemental Figure 10A-D).
IKK2 induces proapoptotic and antiapoptotic genes
To identify genes regulated by CA-IKK2, we performed expression profiling using Affymetrix chips. By applying a threshold of 2, we identified a total of 378 genes that were differentially expressed in CA-IKK2 versus control cells. Three-hundred eight genes were up-regulated and 70 genes were down-regulated in Ramos-CA-IKK2 cells (supplemental Table 1). We were particularly interested in genes that could mediate the observed phenotypes in cell adhesion, proliferation, and apoptosis (Table 1). Several genes coding for cell adhesion molecules were up-regulated including ICAM-1, a known target of NF-κB.30 In addition, several integrins (CD11b/ITGAM, CD18/ITGB2-LFA-1, and CD58/LFA-3) and genes regulating antigen processing and presentation (CIITA, TAP, IFI30), and several chains of MHC class I and class II complexes (supplemental Table 1) were induced. We found several genes involved in cell-cycle regulation. Most prominently, we detected enhanced expression of RASSF4, a gene that has been described as a potential tumor suppressor due to its antiproliferative effects.31
Genes regulated following CA-IKK2 induction in Ramos cells
| Gene ID . | Gene name . | Fold change . |
|---|---|---|
| Cell adhesion | ||
| IL32 | Interleukin 32 | 18.4 |
| CCL5 | Chemokine (C-C motif) ligand 5 | 17.8 |
| ICAM1 | Intercellular adhesion molecule 1 (CD54) | 13.5 |
| FBLN5 | Fibulin 5 | 6.3 |
| ITGB2 | Integrin, beta 2 (CD18) | 5.7 |
| ADAM8 | ADAM metallopeptidase domain 8 | 5.2 |
| ITGAM | Integrin, alpha M (CD11b) | 3.4 |
| FCGBP | Fc fragment of IgG binding protein | 3.3 |
| CD58 | CD58 molecule | 3.0 |
| Apoptosis (proapoptotic) | ||
| FAS | Fas (TNF receptor superfamily, member 6) | 29.7 |
| BTG1 | B-cell translocation gene 1 | 4.5 |
| PLAGL1 | Pleiomorphic adenoma gene-like 1 | 4.1 |
| IFIH1 | Interferon induced with helicase C domain 1 | 3.5 |
| BID | BH3 interacting domain death agonist | 2.3 |
| RUNX3 | Runt-related transcription factor 3 | 2.0 |
| ID3 | Inhibitor of DNA binding 3 | 0.5 |
| Apoptosis (antiapoptotic) | ||
| BCL2A1 | BCL2-related protein A1 | 84.2 |
| SGK1 | Serum/glucocorticoid regulated kinase 1 | 30.9 |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | 23.6 |
| TNFAIP3 | Tumor necrosis factor alpha-induced protein 3 | 17.1 |
| CD40 | CD40 molecule, TNF receptor superfamily member 5 | 6.7 |
| IL1B | Interleukin 1 beta | 6.3 |
| NUP62 | Nucleoporin 62 kDa | 5.1 |
| ATF5 | Activating transcription factor 5 | 3.7 |
| TRAF1 | TNF receptor-associated factor 1 | 3.0 |
| PIM1 | Pim-1 oncogene | 2.9 |
| IL2RA | Interleukin 2 receptor alpha | 2.7 |
| BIR3 | baculoviral IAP repeat-containing 3 | 2.6 |
| IER3 | Immediate early response 3 | 2.5 |
| BCL2 | B-cell CLL/lymphoma 2 | 2.5 |
| BCL3 | B-cell CLL/lymphoma 3 | 2.5 |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2.4 |
| NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 2.4 |
| NEK6 | NIMA (never in mitosis gene a)–related kinase 6 | 2.3 |
| BCL2L1 | BCL2-like 1 | 2.1 |
| Cell cycle (inhibiting) | 7.8 | |
| RASSF4 | Ras association (RalGDS/AF-6) domain family 4 | 2.9 |
| PLEKHO1 | Pleckstrin homology domain containing, | 2.2 |
| WEE1 | WEE1 homolog (S. pombe) | 2.0 |
| MACF1 | Microtubule-actin cross-linking factor 1 | 2.0 |
| HGF | Hepatocyte growth factor (hepapoietin A; scatter factor) | 0.3 |
| Cell cycle (promoting) | ||
| STAG3 | Stromal antigen 3 | 3.9 |
| CYLD | Cylindromatosis (turban tumor syndrome) | 2.6 |
| TUBB2B | Tubulin, beta 2B | 2.4 |
| Gene ID . | Gene name . | Fold change . |
|---|---|---|
| Cell adhesion | ||
| IL32 | Interleukin 32 | 18.4 |
| CCL5 | Chemokine (C-C motif) ligand 5 | 17.8 |
| ICAM1 | Intercellular adhesion molecule 1 (CD54) | 13.5 |
| FBLN5 | Fibulin 5 | 6.3 |
| ITGB2 | Integrin, beta 2 (CD18) | 5.7 |
| ADAM8 | ADAM metallopeptidase domain 8 | 5.2 |
| ITGAM | Integrin, alpha M (CD11b) | 3.4 |
| FCGBP | Fc fragment of IgG binding protein | 3.3 |
| CD58 | CD58 molecule | 3.0 |
| Apoptosis (proapoptotic) | ||
| FAS | Fas (TNF receptor superfamily, member 6) | 29.7 |
| BTG1 | B-cell translocation gene 1 | 4.5 |
| PLAGL1 | Pleiomorphic adenoma gene-like 1 | 4.1 |
| IFIH1 | Interferon induced with helicase C domain 1 | 3.5 |
| BID | BH3 interacting domain death agonist | 2.3 |
| RUNX3 | Runt-related transcription factor 3 | 2.0 |
| ID3 | Inhibitor of DNA binding 3 | 0.5 |
| Apoptosis (antiapoptotic) | ||
| BCL2A1 | BCL2-related protein A1 | 84.2 |
| SGK1 | Serum/glucocorticoid regulated kinase 1 | 30.9 |
| TNFRSF9 | Tumor necrosis factor receptor superfamily, member 9 | 23.6 |
| TNFAIP3 | Tumor necrosis factor alpha-induced protein 3 | 17.1 |
| CD40 | CD40 molecule, TNF receptor superfamily member 5 | 6.7 |
| IL1B | Interleukin 1 beta | 6.3 |
| NUP62 | Nucleoporin 62 kDa | 5.1 |
| ATF5 | Activating transcription factor 5 | 3.7 |
| TRAF1 | TNF receptor-associated factor 1 | 3.0 |
| PIM1 | Pim-1 oncogene | 2.9 |
| IL2RA | Interleukin 2 receptor alpha | 2.7 |
| BIR3 | baculoviral IAP repeat-containing 3 | 2.6 |
| IER3 | Immediate early response 3 | 2.5 |
| BCL2 | B-cell CLL/lymphoma 2 | 2.5 |
| BCL3 | B-cell CLL/lymphoma 3 | 2.5 |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2.4 |
| NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 2.4 |
| NEK6 | NIMA (never in mitosis gene a)–related kinase 6 | 2.3 |
| BCL2L1 | BCL2-like 1 | 2.1 |
| Cell cycle (inhibiting) | 7.8 | |
| RASSF4 | Ras association (RalGDS/AF-6) domain family 4 | 2.9 |
| PLEKHO1 | Pleckstrin homology domain containing, | 2.2 |
| WEE1 | WEE1 homolog (S. pombe) | 2.0 |
| MACF1 | Microtubule-actin cross-linking factor 1 | 2.0 |
| HGF | Hepatocyte growth factor (hepapoietin A; scatter factor) | 0.3 |
| Cell cycle (promoting) | ||
| STAG3 | Stromal antigen 3 | 3.9 |
| CYLD | Cylindromatosis (turban tumor syndrome) | 2.6 |
| TUBB2B | Tubulin, beta 2B | 2.4 |
RNA from cells transfected with either pRTS-CA-IKK2 or pRTS-GFP and stimulated with doxycycline for 48 hours was analyzed by expression profiling. Genes were identified as differentially expressed if the ratio between the 2 classes was at least 2-fold. Biologic significance was determined using Gene Ontology.20
We also observed a large number of differently expressed genes related to cell survival and apoptosis. CA-IKK2 induced several antiapoptotic genes such as members of the antiapoptotic BCL-2 family (BCL-2, BCL2L1 /BCL-X, BCL2A1), FLIP/CFLAR, and the caspase inhibitor cIAP2/BIR3 (Table 1). However, we also detected enhanced expression of several proapoptotic genes, with expression of Fas, a death receptor–inducing apoptosis via FADD and caspase-8 activation, being strongly induced.
NF-κB signaling sensitizes BL cells to Fas-induced apoptosis
Fas-receptor signaling plays a pivotal role for lymphocyte homeostasis, and Fas induction by CA-IKK2 could render BL cells susceptible to apoptosis. We determined Fas expression on the cell surface by flow cytometry. Upon induction of CA-IKK2 expression by doxycycline, 70% to 80% of the cells became GFP positive. This allowed us to compare levels of Fas surface expression in the GFP− and the GFP+ fractions. In control-transfected Ramos cells, we found no differences in Fas expression between GFP− and GFP+ cells (Figure 6A). In contrast, cells transfected with the pRTS-CA-IKK2 showed strong up-regulation in the majority of the GFP+ cells (Figure 6A). Induction of Fas surface expression was also seen in Namalwa cells, whereas there was no significant influence of IKK2 expression in MedB1 and KM-H2 cells (supplemental Figure 11).
NF-κB activation sensitizes Ramos cells to Fas-mediated apoptosis. (A) Surface expression of Fas on Ramos cells transfected with either pRTS-GFP (top panel) or pRTS-CA-IKK2 (bottom panel). Cells were treated with doxycycline (0.5 μg/mL) for 48 hours, incubated with APO-I antibodies and a corresponding secondary antibody conjugated to Alexa-647 fluorochrome. Stained cells were subjected to flow cytometry and gates were set on GFP− (gray line) and GFP+ (black line) fractions. (B) Percentages of dead cells in Ramos transfected with pRTS-GFP or pRTS-CA-IKK2 in the absence (▭) or presence (▬) of agonistic anti-Fas antibody (APO-I). Cells were cultured in the presence of doxycycline for 4 days to induce transgene expression and as indicated treated with APO-I supernatant (5% final concentration [f.c.]) for additional 48 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments. (C) Fas mRNA expression analyzed by reverse-transcription–PCR. Total RNA of Ramos stably transduced with either IEGZ-empty or IEGZ-TD-IκBα and transfected with pRTS-GFP and pRTS-CA-IKK2, respectively, was isolated 48 hours after addition of doxycycline to the culture medium. (D) Percentages of dead cells in cultures of Ramos-IEGZ-empty (▭) or Ramos-IEGZ-TD-IκBα (▬) transfected with either pRTS-GFP or pRTS-CA-IKK2 after addition of agonistic anti-Fas antibody (APO-I). Fas sensitivity of Ramos-CA-IKK2 is abrogated by TD-IκBα expression. Doxycycline (0.5 μg/mL) was added to the culture medium for 4 days and APO-I supernatant (5% f.c.) was added for additional 24 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments.
NF-κB activation sensitizes Ramos cells to Fas-mediated apoptosis. (A) Surface expression of Fas on Ramos cells transfected with either pRTS-GFP (top panel) or pRTS-CA-IKK2 (bottom panel). Cells were treated with doxycycline (0.5 μg/mL) for 48 hours, incubated with APO-I antibodies and a corresponding secondary antibody conjugated to Alexa-647 fluorochrome. Stained cells were subjected to flow cytometry and gates were set on GFP− (gray line) and GFP+ (black line) fractions. (B) Percentages of dead cells in Ramos transfected with pRTS-GFP or pRTS-CA-IKK2 in the absence (▭) or presence (▬) of agonistic anti-Fas antibody (APO-I). Cells were cultured in the presence of doxycycline for 4 days to induce transgene expression and as indicated treated with APO-I supernatant (5% final concentration [f.c.]) for additional 48 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments. (C) Fas mRNA expression analyzed by reverse-transcription–PCR. Total RNA of Ramos stably transduced with either IEGZ-empty or IEGZ-TD-IκBα and transfected with pRTS-GFP and pRTS-CA-IKK2, respectively, was isolated 48 hours after addition of doxycycline to the culture medium. (D) Percentages of dead cells in cultures of Ramos-IEGZ-empty (▭) or Ramos-IEGZ-TD-IκBα (▬) transfected with either pRTS-GFP or pRTS-CA-IKK2 after addition of agonistic anti-Fas antibody (APO-I). Fas sensitivity of Ramos-CA-IKK2 is abrogated by TD-IκBα expression. Doxycycline (0.5 μg/mL) was added to the culture medium for 4 days and APO-I supernatant (5% f.c.) was added for additional 24 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments.
To analyze if Fas renders Ramos cells more sensitive to death induction by Fas ligation, we treated the cells with agonistic anti-Fas antibodies (APO-I). Whereas control cells were virtually resistant to APO-I treatment, CA-IKK2–expressing cells showed increased levels of dead cells (2- to 3-fold) in the absence of APO-I, which was significantly enhanced by addition of APO-I (Figure 6B). Interestingly the constitutive increase in apoptosis of CA-IKK2–expressing Ramos cells could not be prevented by addition of neutralizing anti–human FasL mAbs (supplemental Figure 12), suggesting a ligand-independent autoactivation of Fas receptors. The strong induction of Fas mRNA expression by CA-IKK2 was largely attenuated in TD-IκBα–expressing cells (Figure 6C). In addition, APO-I–induced cell death in CA-IKK2–expressing cells was completely blocked by TD-IκBα (Figure 6D).
Discussion
MYC-induced apoptosis represents a major safeguard mechanism against unrestricted growth that has to be overcome in tumorigenesis. A primary role for NF-κB in malignant transformation is prevention of apoptosis caused by certain oncogenes. NF-κB–regulated genes in lymphocytes are associated with cell-cycle progression, survival, and regulation of immune responses.32 In many hematologic malignancies such as Hodgkin lymphoma, mucosa-associated lymphatic tissue lymphoma, and diffuse large B-cell lymphoma, NF-κB is constitutively activated.33 NF-κB itself is suggested to be an oncogenic factor in lymphomagenesis or it might predispose malignant transformation by inhibiting apoptotic signals induced by other oncogenes. Indeed, the tumor-promoting role of NF-κB signaling is widely acknowledged. It therefore came as a surprise when 2 recent studies using gene expression profiling demonstrated that very low NF-κB signatures are a hallmark of BL.13,14 Our finding that MYC-transformed mouse and human lymphomas are insensitive to NF-κB inhibition extends these observations and proves that canonical NF-κB activation is dispensable for proliferation or cell survival. Apparently, in MYC-driven lymphomas NF-κB is not involved in tumor promotion. This absence of NF-κB signaling has been described for GC centroblasts, the normal counterparts of malignant BL. Therefore, the low NF-κB profile in MYC-driven lymphomas could reflect the normal differentiation program of GC-derived B cells. However, we now present evidence that NF-κB inhibition provides a selective advantage to MYC-transformed lymphomas and its activation actually interferes with both cell survival and proliferation. These observations might have implications for the development of therapies against MYC-positive tumors.
Components of the NF-κB signaling pathway are present in these cells, as NF-κB activity can be restored by turning off MYC expression. Importantly, most lymphoma cell lines were resistant to NF-κB induction by conventional extracellular stimuli. We did, however, find a few lines showing some inducible NF-κB activation upon either PMA/ionomycin or LPS treatment (data not shown). Furthermore, we found NF-κB activation after anti-CD40 stimulation in Ramos and 1 murine MYC-driven lymphoma line, whereas Namalwa and other tested murine lines showed no NF-κB activation. This suggests that although decreased inducibility of NF-κB is a common feature in these lymphoma cells, the actual pathways leading to the block in NF-κB activation in individual tumors vary. MYC-mediated NF-κB suppression has been described in several different cell systems, but it was typically associated with increased sensitivity to apoptosis induction. In mouse primary fibroblasts, inducible MYC expression impairs TNF-induced activation of NF-κB, thereby rendering them sensitive to TNF-induced cell death. Apoptosis could be inhibited by overexpression of p65/RelA.34 In TRAIL-resistant human colon cancer cell lines, expression of MYC sensitized cells to TRAIL-induced apoptosis through inactivation of NF-κB.35
Expression of CA-IKK2 in murine lymphoma cells as well as in human BL led to enhanced cell adhesion, reduced proliferation, and apoptosis. We can exclude toxic effects of CA-IKK2 overexpression, because tested Hodgkin and primary mediastinal B-cell lymphoma lines were not sensitive to CA-IKK2 expression and blocking of NF-κB in CA-IKK2–expressing Ramos prevented cell death. The physiologic activation of NF-κB in a murine MYC-driven lymphoma line or human BL Ramos by CD40 stimulation resulted in comparable effects of cell clumping and cell death.
NF-κB stimulates the expression of several antiapoptotic proteins. Among them are CFLAR, inhibitors of apoptosis (IAPs), and members of the antiapoptotic BCL-2 family.36-38 Our gene expression profiling suggests that NF-κB indeed regulates both proapoptotic and antiapoptotic genes in BL. The net consequence is, however, induction of apoptosis. Our results therefore indicate a tumor-suppressive role for NF-κB activity in MYC-induced lymphomas. Apoptosis as a safeguard mechanism against tumors operates at 2 levels. (1) It operates as an immune surveillance mechanism by instructing cytotoxic cells of the immune system to eliminate transformed cells. For this level of protection, cell-cell contact of the malignant cell with cytotoxic T cells and often presentation of tumor antigens are necessary. (2) It operates on the level of cell intrinsic pathways because deregulation of oncogenic proteins simultaneously induces apoptosis. Tumor cells escape cytotoxic T cell–mediated elimination by down-regulating accessory molecules or by blocking intracellular processing of tumor-specific antigens. BL cells show low expression of LFA-1 (CD11/CD18) and LFA-3 (CD58)39 as well as ICAM-1.40 Antigen presentation is also affected due to down-regulation of MHC molecules as well as transporters associated with antigen presentation (TAP1/TAP2).41 This nonimmunogenic phenotype is imposed by MYC expression, as it was shown in a conditional model that MYC inactivation results in up-regulation of accessory molecules and antigen presentation.42 However, the mechanism of this MYC-induced repression of immunogenicity was not clear. Our results demonstrate that IKK2/NF-κB is controlling expression of several of these such as LFA-1, LFA-3, ICAM-1, TAP1, HLA-I, and HLA-2 (supplemental Table 1). The nonimmunogenic phenotype in BL cells is largely the result of low NF-κB.
However, immune surveillance cannot account for the cell death we observed in vitro. CA-IKK2 induced several proapoptotic genes in Ramos cells. BTG1 (B-cell translocation gene 1), a Bcl-2–regulated mediator of apoptosis in breast cancer cells,43 PLAGL1, IFIH1, and RUNX3 are all considered tumor suppressors because of their apoptosis-inducing capacities.44-46 BID, a BH3-only protein promoting cytochrome c release, had been connected to MYC-induced cell-death priming.47 Most significantly, we detected strong up-regulation of Fas, a death receptor previously linked to MYC-induced apoptosis. MYC has been shown to sensitize cells to Fas-induced death.48 Insensitivity to Fas-mediated apoptosis is a common attribute of BL cells and correlates with low Fas surface expression.49,50 Ramos and Namalwa cells are completely resistant to agonistic Fas-antibody and blocking canonical NF-κB activation did not enhance sensitivity. However, NF-κB activation itself renders the cells highly susceptible to Fas-mediated apoptosis.
The high expression of Fas after NF-κB activation can contribute to apoptosis of the cells in vitro via 2 ways. (1) Apoptosis can be the result of Fas-ligand (FasL) expression by the Ramos cells that would result in Fas receptor ligation in an autocrine manner. (2) Apoptosis could be induced by a ligand-independent self-assembly of the receptors, which has been described.51 MYC-dependent FasL expression has been reported at least for some cell types.52 However, using the described method of meta-analysis of multiple cancer studies within the Oncomine database,53,54 we found no consistent pattern of FasL up-regulation in BL compared with other lymphomas, and with real-time PCR we found FasL mRNA to be below detection levels in Ramos and Namalwa cells. We observed constitutive apoptosis in response to IKK2 expression in Ramos, which was reduced by CFLAR overexpression or caspase-8 knockdown, pointing toward death receptor–mediated cell killing. CFLAR has been reported to be down-regulated by MYC, rendering cells sensitive to extrinsic death-inducing signals via caspase-8 processing.28 Indeed, we found CFLAR levels in Ramos cells to be very low and only slightly up-regulated after IKK2/NF-κB activation. Low CFLAR expression due to MYC repression could explain the toxicity of CA-IKK2, because NF-κB–mediated death receptor induction cannot be balanced by sufficient expression of the antiapoptotic adaptor molecule. NF-κB–induced apoptosis could be further enhanced by APO-I stimulation, indicating high sensitivity of the cells toward Fas signaling. Activation of self-assembling Fas receptors would explain the death-inducing activity in vitro even in the absence of enhanced FasL expression. In vivo, FasL is expressed by several cell types, especially T cells and natural killer cells. The Fas-FasL apoptosis pathway is critical for activation-induced cell death, limiting proliferation of lymphocytes in the periphery. Furthermore, Fas-mediated apoptosis is involved in negative selection of B cells in germinal centers. Fas is a known tumor suppressor also for BL. Induction of Fas expression and restoration of Fas sensitivity upon NF-κB activation impede immune escape in vivo and contribute to tumor suppression. The fact that mouse and human MYC-transformed lymphoma cells show no evidence of NF-κB activation argues for a strong selective pressure against NF-κB in lymphomagenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank K. Holzmann and the Microarray Facilities of the University of Ulm. We thank D. Eick and G. W. Bornkamm for the inducible gene expression vector system, S. Fulda and G. Strauss for providing caspase-8 shRNA and the APO-I antibody, and P. Krammer for the CFLAR expression vector.
Authorship
Contribution: K.K. designed the research, performed experiments, analyzed results, made the figures, and wrote the paper; S.S., D.M., and B.B. performed experiments; and T.W. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Wirth, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: thomas.wirth@uni-ulm.de.




![Figure 6. NF-κB activation sensitizes Ramos cells to Fas-mediated apoptosis. (A) Surface expression of Fas on Ramos cells transfected with either pRTS-GFP (top panel) or pRTS-CA-IKK2 (bottom panel). Cells were treated with doxycycline (0.5 μg/mL) for 48 hours, incubated with APO-I antibodies and a corresponding secondary antibody conjugated to Alexa-647 fluorochrome. Stained cells were subjected to flow cytometry and gates were set on GFP− (gray line) and GFP+ (black line) fractions. (B) Percentages of dead cells in Ramos transfected with pRTS-GFP or pRTS-CA-IKK2 in the absence (▭) or presence (▬) of agonistic anti-Fas antibody (APO-I). Cells were cultured in the presence of doxycycline for 4 days to induce transgene expression and as indicated treated with APO-I supernatant (5% final concentration [f.c.]) for additional 48 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments. (C) Fas mRNA expression analyzed by reverse-transcription–PCR. Total RNA of Ramos stably transduced with either IEGZ-empty or IEGZ-TD-IκBα and transfected with pRTS-GFP and pRTS-CA-IKK2, respectively, was isolated 48 hours after addition of doxycycline to the culture medium. (D) Percentages of dead cells in cultures of Ramos-IEGZ-empty (▭) or Ramos-IEGZ-TD-IκBα (▬) transfected with either pRTS-GFP or pRTS-CA-IKK2 after addition of agonistic anti-Fas antibody (APO-I). Fas sensitivity of Ramos-CA-IKK2 is abrogated by TD-IκBα expression. Doxycycline (0.5 μg/mL) was added to the culture medium for 4 days and APO-I supernatant (5% f.c.) was added for additional 24 hours. Viable and dead cell numbers were determined by trypan blue exclusion. Error bars represent SDs from triplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/12/10.1182_blood-2008-09-181008/4/m_zh89990941880006.jpeg?Expires=1765897200&Signature=T3q60duJZwhfm2cZ9bCufPsmfgq~gEPVyiUvT5em5yn9fD0NLWiglRIP5P9AGcQ3vz8eeZdmJqNxIFdIIyv08BdniAwyyfttqYylilJFmYPNQh1kM9CKdWrP15sP-IXeFOVbycHCQ80HLm2wk2ot~jLOQWJ5WRSl1VYinzcJZH3oWPN0Bntby6nr2jtbFyIe0ocgq5ihB-NbtYDKR9hqLyS7igYaFu8VsCSbQKsEcW8dXdIAOmfRQYwKuFjBK1nrx8yNjHmiTbXo8BEQh1w2uT8gvUN2iSH-TmNwdscFuiMsdb8Z7Cx~HSYrLW3PDk~yg8w2-Wjb7nEwzqkoLVvRoA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)