Abstract
BCL2 and MYC are oncogenes commonly deregulated in lymphomas. Concurrent BCL2 and MYC translocations (BCL2+/MYC+) were identified in 54 samples by karyotype and/or fluorescence in situ hybridization with the aim of correlating clinical and cytogenetic characteristics to overall survival. BCL2+/MYC+ lymphomas were diagnosed as B-cell lymphoma unclassifiable (BCLU; n = 36) with features intermediate between Burkitt lymphoma and diffuse large B-cell lymphoma (DLBCL); DLBCL (n = 17), or follicular lymphoma (n = 1). Despite the presence of a t(14;18), 5 cases were BCL2 protein–negative. Nonimmunoglobulin gene/MYC (non-IG/MYC) translocations occurred in 24 of 54 cases (44%) and were highly associated with DLBCL morphology (P < .001). Over a median follow-up of 5.3 years, 6 patients remained in remission and 32 died within 6 months of the MYC+ rearrangement, irrespective of whether MYC+ occurred at diagnosis (31 of 54) or transformation (23 of 54; P = .53). A non-IG/MYC translocation partner, absent BCL2 protein expression and treatment with rituximab-based chemotherapy, were associated with a more favorable outcome, but a low International Prognostic Index score and DLBCL morphology were independent predictors of overall survival. A comprehensive cytogenetic analysis of BCL2 and MYC status on all aggressive lymphomas may identify a group of high-risk patients who may benefit from chemotherapeutic regimens that include rituximab and/or BCL2-targeted therapy.
Introduction
BCL2 and MYC are 2 dominant acting oncogenes that are often deregulated as a result of chromosomal translocation in B-cell lymphomas. The translocation t(14;18) juxtaposes BCL2 on chromosome band 18q21 to the immunoglobulin heavy chain gene (IGH) enhancer at band 14q32, resulting in BCL2 protein overexpression and inhibition of apoptosis.1-3 It is found in 85% of follicular lymphomas (FLs) and 15% to 30% of de novo diffuse large B-cell lymphomas (DLBCLs).4 The translocation t(8;14)(q24;q32) is characterized by rapid cellular proliferation as a result of MYC deregulation driven by the IGH enhancer.5,6 This translocation is the hallmark of classic Burkitt lymphomas (BLs),5 but it is also found in a high proportion of cases previously known as atypical Burkitt/Burkitt-like lymphoma (BLL; 41%-80%)7,8 and a subset of DLBCL (∼ 5%-8%).9,10 BLLs are often associated with complex cytogenetic alterations and have an unfavorable clinical outcome compared with DLBCL and BL.7,8 In the 2008 World Health Organization criteria for the classification of lymphomas, the term BLL has been dropped in favor of B-cell lymphoma, unclassifiable, with features intermediate between BL and DLBCL.11 In this study, this lymphoma category will be referred to as B-cell lymphoma, unclassifiable (BCLU).12
Infrequently, BCL2 and MYC translocations may be found concurrently in the same specimen, so-called “double hit disease.”11 These cases may present with variable morphologies, including acute lymphoblastic leukemia/lymphoma (ALL), DLBCL, BCLU, and rarely FL.13-19 The primary event in these cases is usually the t(14;18) with the MYC translocation (MYC+) arising as a secondary genetic event.13,20 Patients with lymphomas harboring concurrent BCL2 and MYC translocations, hereafter referred to as BCL2+/MYC+ lymphomas, have been described in several small case series as having a very poor overall survival (OS).16,17 A recent study at our institution revealed that 4% (6 of 142) of patients with primary DLBCL had dual translocations involving both BCL2 and MYC.21 The clinical outcome of these patients was variable.
We conducted a comprehensive investigation of a large cohort of lymphomas with concurrent translocations of 8q24 (MYC) and 18q21 (BCL2) to identify the clinical and cytogenetic prognostic factors associated with survival.
Methods
Patient identification
Patients with non-Hodgkin lymphoma (NHL) diagnosed between 1991 and 2007 were identified in the British Columbia Cancer Agency Lymphoid Cancer and Cytogenetic Databases. We initially selected patients based on 3 requirements: accurate histologic diagnosis reassigned according to the recently published 2008 World Health Organization classification for lymphoid neoplasms, availability of cytogenetic analysis by karyotype, and detailed information on clinical outcome.12 Indications for karyotype analysis included high-grade histology, DLBCL with a high proliferation index, and FL with atypical morphologic features. A total of 1118 patients met these criteria. In addition, fluorescence in situ hybridization (FISH) analysis using commercial probes for MYC and BCL2 (“Cytogenetic analysis”) was performed on a tissue microarray constructed from duplicate 0.6-mm cores of formalin-fixed, paraffin-embedded tissue (FFPET) derived from 142 unselected diagnostic samples of DLBCL.21 Thus, information on MYC and BCL2 translocation status was available on 1260 patients. Of these, 54 patients were identified as having concurrent translocations at 18q21 and 8q24 (49 by karyotype and 5 by FISH).
Cases were considered to have BCLU if their biopsy revealed lymphoma with features intermediate between DLBCL and BL (one of the so-called gray zone lymphomas), previously called BLL. Standard diagnostic criteria were used to identify DLBCL and FL. Baseline clinical characteristics, including the International Prognostic Index (IPI) variables, were recorded.22 OS was calculated from the date of the MYC+ biopsy to the date of last follow-up or death from any cause. Progression-free survival was calculated from the date of the MYC+ biopsy to the date of last follow-up or disease progression. The chemotherapy regimens used for treatment of these patients were heterogeneous and based on treatment era. CHOP-like regimens included cyclophosphamide, doxorubicin, vincristine, and prednisone. Rituximab was added to CHOP (R-CHOP) for aggressive histology B-cell lymphomas at our institution in March 2001, and this regimen was used in 11 patients (5 BCLU and 6 DLBCL).23 Palliative regimens included observation, radiation, immunotherapy alone, and non–anthracycline-based chemotherapy. High-dose chemotherapy included ALL-type regimens (n = 2)24 and high-dose chemo-radiotherapy followed by hematopoietic stem cell transplantation (autologous n = 3; and allogeneic n = 1). Ethical approval to perform this retrospective study was granted by the University of British Columbia-British Columbia Cancer Agency Research Ethics Board and is in accordance with the Declaration of Helsinki (REB no. H08-02834).
Immunohistochemistry
Immunohistochemical analysis of archived FFPET included CD20 (L26; Dako North America), CD3 (Dako North America), CD10 (clone 56C6; Vector Laboratories), Ki-67 (Dako North America), and BCL2 (clone 124; Dako North America). Five cases had no BCL2 protein expression using clone 124 (targeting amino acids 41-54) and were subsequently analyzed using clone E17 (Epitomics; targeting amino acids 60-80). BCL2 protein or Ki-67 staining was not performed on bone marrow biopsies or peripheral blood. Cases were considered positive for CD10 and BCL2 if more than 30% of the neoplastic cells stained positively. The proliferation rate was determined in 20 samples using the monoclonal antibody Ki-67, which binds to a nuclear protein in cycling cells. A cutoff value of more than or equal to 80% was used to define a high proliferation index according to the Southwest Oncology Group study by Miller et al.25
Genomic sequencing of the BCL2 gene
The 5 cases that were BCL2 protein–negative by clone 124 were investigated for mutations in the BCL2 gene. Genomic DNA was extracted using the ALL PREP kit (QIAGEN) or the PureGene DNA purification kit (GENTRA) in 3 frozen samples and 2 FFPET samples, respectively. The BCL2 genomic sequence corresponding to amino acids 8 to 126 of the BCL2 protein was polymerase chain reaction–amplified using the following primers containing the universal −21M13F and M13R sequencing tags (italics): forward 5′-TGTAAAACGACGGCCAGTGGGTACGATAACCGGGAGAT-3′; and reverse 5′-CAGGAAACAGCTATGACCGGCGGGAGAAGTCGT-3′. The purified 334-base pair product was bidirectionally sequenced using BigDye Terminator, Version 3.1 (Applied Biosystems) and an ABI 3730 XL sequencer (Applied Biosystems). Mutations were considered present if they were observed in both forward and reverse reads.
Cytogenetic analysis
G-banded karyotype and multicolor karyotype analyses were performed as previously reported.26,27 Karyotype descriptions conform to the International System for Human Cytogenetic Nomenclature.28 MYC rearrangements were confirmed using the LSI MYC dual-color break-apart probe (Abbott Molecular) on cells fixed in methanol/acetic acid (3:1) in 48 of 54 cases (including the 5 cases identified by tissue microarray). Additional BAC probes were used to further characterize the MYC breakpoints and partner chromosomes involved in the MYC translocations (supplemental Table 2, available on the Blood website; see the Supplemental Materials link at the top of the online article). The BAC probes were directly labeled by nick translation using a commercial labeling kit according to the manufacturer's protocol (Abbott Molecular). Forty samples, including the 5 samples that were BCL2 protein–negative, were confirmed to have t(14;18) involving the BCL2 gene by FISH using the LSI IGH/BCL2 dual-color, dual-fusion translocation probe (Abbott Molecular). For probe signal scoring, a minimum of 200 interphase nuclei were examined. A cutoff threshold of more than 5% positive cells was used to confirm the presence of IGH-BCL2 and IGH-MYC translocations. The cell line Karpas 353, which contains the translocation t(8;9), was used as control material for the FISH experiments, generously provided by Dr A. Karpas.29
Statistical analysis
Statistical analyses were performed using SPSS software, Version 11.0, and the R statistical package (http://cran.r-project.org/). Fisher exact test and likelihood ratios were used to determine the significance of any differences between patients with different histologic and cytogenetic characteristics. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. Hazard ratios and 95% confidence intervals were calculated using univariate Cox proportional-hazards models. In the multivariate model, we included terms that appeared to be important on univariate analyses and then used a backward selection method to remove the nonsignificant terms from the model. A P value less than .05 (2-sided) was considered statistically significant.
Results
A total of 1260 cases of NHL were identified in the British Columbia Cancer Agency lymphoid cancer and cytogenetics databases between 1991 and 2007 as having the required clinical information, pathology review, and cytogenetic analysis available for this study. Of these, 54 cases (4%) were identified as having concurrent translocations involving MYC and BCL2 based on karyotype and/or FISH analysis. These samples were acquired from patients having different diagnoses and treatment regimens over a 16-year period.
Clinical characteristics
MYC rearrangements were found at the time of initial lymphoma diagnosis in 31 cases (57%). In 23 cases (43%), the MYC rearrangement occurred at the time of histologic transformation from a preexisting FL (n = 19) or chronic lymphocytic leukemia (n = 1). Three patients had an antecedent DLBCL. Extranodal disease was common and 59% had bone marrow and/or peripheral blood involvement at the time of detection of the MYC rearrangement. Treatments for the prior indolent lymphomas had included observation (n = 6), radiation (n = 4), single or multiagent chemotherapy with or without immunotherapy (n = 9), or this information was not available (n = 1). Treatment for the BCL2+/MYC+ lymphomas consisted of CHOP (n = 23), R-CHOP (n = 11), high-dose chemotherapy with or without stem cell transplantation (n = 6), and palliation (n = 14).
Immunohistochemical and immunophenotypic characteristics
The BCL2+/MYC+ lymphomas frequently presented as a BCLU (n = 36) or DLBCL (n = 17). One sample had a MYC+ lymphoma with FL morphology (grade 2). A high proliferation index, as determined by a Ki-67 more than or equal to 80%, was observed in 14 of 20 tested cases (including 2 of the 5 BCL2 clone 124-negative cases) and tended to correlate with a BCLU morphology, but in this limited number of samples, this observation was not statistically significant (P = .15). Immunophenotyping analysis revealed that 62% of samples coexpressed CD19, CD20, and CD10, consistent with a germinal center phenotype.
Genetic analysis of the BCL2 gene
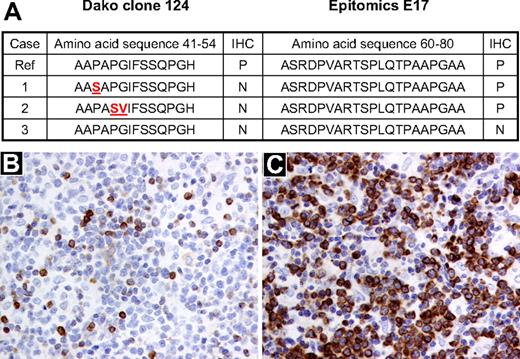
Despite the presence of t(14;18), 5 BCL2+/MYC+ lymphomas were considered to be BCL2 protein–negative by clone 124 that is used routinely in most clinical laboratories. Four of these were considered BCL2 protein–positive with the E17 antibody. Synonymous and nonsynonymous mutations in the BCL2 gene were detected in 2 of the 3 interpretable cases. These 2 cases had discrepant staining with both antibodies and harbored mutations in the flexible loop domain but not the BH3 domain (Figure 1). Both mutated cases had C → T mutations that are typical of somatic hypermutation as previously described by Tanaka et al.30 No mutations were detected in the one sample that was negative by both antibodies. However, mutations 5′ of the amplified sequence or in the promoter regions of the BCL2 gene could not be excluded. The patient with the “true” BCL2-negative biopsy is alive, free of disease more than 6 years after the MYC+ diagnosis. Two of the FFPET cases had poor quality sequence data, and mutations could not be excluded or confirmed.
Correlation between the presence of mutations in the BCL2 gene and BCL2 protein expression by immunohistochemistry using clones 124 and E17. (A) BCL2 protein expression by immunohistochemistry and corresponding amino acid sequence derived from sequencing the BCL2 gene in 3 BCL2+/MYC+ lymphoma samples. The affected amino acid changes are highlighted in red. (B) DLBCL sample (sample 2) stained with the BCL2 antibody clone 124 (Dako North America; original magnification ×200). (C) The same DLBCL sample (sample 2) stained with the BCL2 antibody E17 (Epitomics; original magnification, ×200). Ref indicates reference genome hg 19, May 2009; IHC, immunohistochemistry; P, positive; and N, negative.
Correlation between the presence of mutations in the BCL2 gene and BCL2 protein expression by immunohistochemistry using clones 124 and E17. (A) BCL2 protein expression by immunohistochemistry and corresponding amino acid sequence derived from sequencing the BCL2 gene in 3 BCL2+/MYC+ lymphoma samples. The affected amino acid changes are highlighted in red. (B) DLBCL sample (sample 2) stained with the BCL2 antibody clone 124 (Dako North America; original magnification ×200). (C) The same DLBCL sample (sample 2) stained with the BCL2 antibody E17 (Epitomics; original magnification, ×200). Ref indicates reference genome hg 19, May 2009; IHC, immunohistochemistry; P, positive; and N, negative.
Cytogenetic characteristics
By karyotype, 30 of 54 cases had MYC translocations involving the IG loci, 16 t(8;14), 11 t(8;22), and 3 t(2;8). Of the cases with t(8;14), 8 had a complex rearrangement [t(14;18)t(8,14)] where MYC was adjacent to the 3′IGH enhancer on derivative chromosome 14 and BCL2 was driven by the 5′IGH enhancer relocated to the derivative chromosome 8 as previously described.31 The remaining MYC rearrangements involved variant chromosome partners, most commonly a t(8;9)(q24;p13) (13 of 24). The other MYC partner loci included 1p36, 3p25, 3q27(BCL6), 4p13, 5q13, 12p11, and 13q31. The karyotypes and the MYC translocation partners with corresponding histology and clinical outcomes are shown in supplemental Table 1. Multiple alterations were present in almost all cases in addition to the BCL2 and MYC translocations, including breakpoints at chromosome band 3q27 (13%) and loss of 17p13 (16%) or 1p36 (36%). These results confirm previous observations that BCL2+/MYC+ lymphomas, unlike classic BL, typically have complex karyotypes.32
Given that t(8;9) was the most frequent non-IG/MYC rearrangement observed, a series of adjacent BACs were used to determine the breakpoint site at 9p13 in the cell line Karpas 353 and in 11 of 13 of the clinical cases (supplemental Figure 1). The breakpoint region covered by BACs RP11-220I1 through RP11-405L18 spans approximately 400 kb of genomic region just 5′ of the PAX5 locus as previously described by Bertrand et al.33
Characteristics associated with overall survival
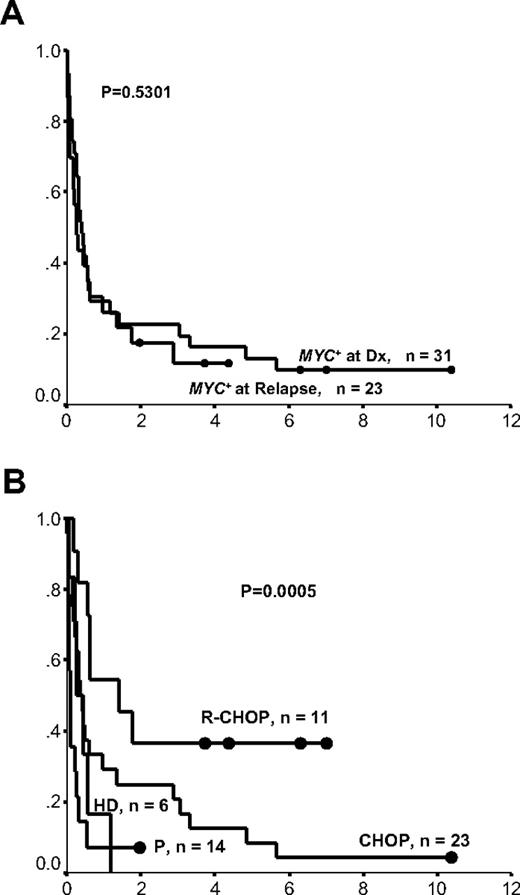
Table 1 demonstrates the effect of each clinical, immunophenotypic, and cytogenetic factor on OS for the entire cohort. Every death in this study, except for one, was attributed to lymphoma progression; thus, progression-free survival was very similar to OS (data not shown). As illustrated in Figure 2A, patients identified as having a MYC+ rearrangement at the time of their initial lymphoma diagnosis had a similar outcome to those who acquired it at the time of transformation. Only 6 of 54 patients remained alive and in remission over a median follow-up time of 5.3 years, 3 of which presented at diagnosis and 3 at relapse. Thus, samples in these 2 groups were pooled together in subsequent analyses.
Characteristics associated with overall survival in all 54 patients with BCL2+/MYC+ lymphomas
| Variable . | n (%) . | Median OS, y . | P, log-rank . |
|---|---|---|---|
| Age > 60 y at diagnosis | 28 (52) | 0.58 | .003 |
| Male sex | 32 (59) | 0.32 | .825 |
| Performance status > 1 | 19 (35) | 0.27 | .001 |
| Stage > 2 | 41 (76) | 0.30 | .065 |
| LDH > normal | 27 (50) | 0.35 | .923 |
| Extranodal sites > 1 | 19 (35) | 0.18 | < .001 |
| IPI | |||
| 0-1 | 16 (30) | 1.35 | < .001 |
| 2-3 | 24 (44) | 0.48 | |
| 4-5 | 14 (26) | 0.15 | |
| Age at MYC+ > 60 y | 32 (59) | 0.26 | .005 |
| Antecedent MYC− lymphoma | 0.41 | .530 | |
| FL | 19 (35) | ||
| CLL | 1 (2) | ||
| DLBCL | 3 (6) | ||
| Histology with MYC+ | < .001 | ||
| DLBCL | 17 (31) | 2.86 | |
| BCLU | 36 (67) | 0.26 | |
| FL | 1 (2) | ||
| Bone marrow with MYC+ | .001 | ||
| Positive | 32 (59) | 0.20 | |
| Negative | 13 (24) | 1.40 | |
| Not available | 9 (17) | ||
| BCL2 protein expression | .009 | ||
| Positive | 23 (43) | 0.48 | |
| Negative | 5 (9) | 5.66 | |
| Not available | 27 (48) | ||
| CD10 expression | .316 | ||
| Positive | 34 (63) | 0.30 | |
| Negative | 9 (17) | 0.20 | |
| Not available | 11 (20) | ||
| IG/MYC | 30 (56) | 0.26 | .017 |
| Non-IG/MYC | 24 (44) | 0.58 | |
| Treatment | |||
| HD chemo | 6 (11) | 0.26 | < .001 |
| R-CHOP | 11 (20) | 1.40 | |
| CHOP-like | 23 (43) | 0.42 | |
| Palliative | 14 (26) | 0.07 |
| Variable . | n (%) . | Median OS, y . | P, log-rank . |
|---|---|---|---|
| Age > 60 y at diagnosis | 28 (52) | 0.58 | .003 |
| Male sex | 32 (59) | 0.32 | .825 |
| Performance status > 1 | 19 (35) | 0.27 | .001 |
| Stage > 2 | 41 (76) | 0.30 | .065 |
| LDH > normal | 27 (50) | 0.35 | .923 |
| Extranodal sites > 1 | 19 (35) | 0.18 | < .001 |
| IPI | |||
| 0-1 | 16 (30) | 1.35 | < .001 |
| 2-3 | 24 (44) | 0.48 | |
| 4-5 | 14 (26) | 0.15 | |
| Age at MYC+ > 60 y | 32 (59) | 0.26 | .005 |
| Antecedent MYC− lymphoma | 0.41 | .530 | |
| FL | 19 (35) | ||
| CLL | 1 (2) | ||
| DLBCL | 3 (6) | ||
| Histology with MYC+ | < .001 | ||
| DLBCL | 17 (31) | 2.86 | |
| BCLU | 36 (67) | 0.26 | |
| FL | 1 (2) | ||
| Bone marrow with MYC+ | .001 | ||
| Positive | 32 (59) | 0.20 | |
| Negative | 13 (24) | 1.40 | |
| Not available | 9 (17) | ||
| BCL2 protein expression | .009 | ||
| Positive | 23 (43) | 0.48 | |
| Negative | 5 (9) | 5.66 | |
| Not available | 27 (48) | ||
| CD10 expression | .316 | ||
| Positive | 34 (63) | 0.30 | |
| Negative | 9 (17) | 0.20 | |
| Not available | 11 (20) | ||
| IG/MYC | 30 (56) | 0.26 | .017 |
| Non-IG/MYC | 24 (44) | 0.58 | |
| Treatment | |||
| HD chemo | 6 (11) | 0.26 | < .001 |
| R-CHOP | 11 (20) | 1.40 | |
| CHOP-like | 23 (43) | 0.42 | |
| Palliative | 14 (26) | 0.07 |
LDH indicates lactate dehydrogenase; IPI, International Prognostic Index; FL, follicular lymphoma; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; BCLU, B-cell lymphoma, unclassifiable, with features intermediate between Burkitt lymphoma and DLBCL; IG, immunoglobulin; HD chemo, high-dose chemotherapy with or without stem cell transplantation; and R-CHOP, rituximab and CHOP chemotherapy.
Survival curves of patients with BCL2+/MYC+ lymphomas according to the timing of MYC+ rearrangement and treatment regimen. (A) Timing of MYC+ rearrangement: at diagnosis or at relapse. x-axis represents cumulative survival; y-axis, time (years). (B) Treatment regimen. ● represent long-term survivors; x-axis, cumulative survival; y-axis, time (years); Dx, diagnosis; R, rituximab; HD, high-dose chemotherapy with or without stem cell transplantation; and P, palliative.
Survival curves of patients with BCL2+/MYC+ lymphomas according to the timing of MYC+ rearrangement and treatment regimen. (A) Timing of MYC+ rearrangement: at diagnosis or at relapse. x-axis represents cumulative survival; y-axis, time (years). (B) Treatment regimen. ● represent long-term survivors; x-axis, cumulative survival; y-axis, time (years); Dx, diagnosis; R, rituximab; HD, high-dose chemotherapy with or without stem cell transplantation; and P, palliative.
Figure 2B demonstrates that 32 (59%) patients died within 6 months after the diagnosis of the MYC+ rearrangement irrespective of treatment regimen. The addition of rituximab to CHOP may have improved outcome in patients with BCL2+/MYC+ lymphomas (median OS, 1.4 years vs 0.4 years for R-CHOP [n = 11] and CHOP [n = 23] treated patients, respectively, P = .05); however, too few patients were treated with R-CHOP to support any firm conclusions. The 6 patients treated with high-dose chemotherapy and stem cell transplantation, all of whom had BCLU histology, had a similar poor outcome compared with those patients treated with palliation (median survival, 3 months vs 1 month, P > .05), suggesting that even intensified therapy cannot overcome the aggressive tumor biology in these patients. Of the 10 patients with OS more than 2 years, the predominant histology was DLBCL (9 of 10; supplemental Table 1), and their treatment regimens included CHOP (n = 6) and R-CHOP (n = 4).
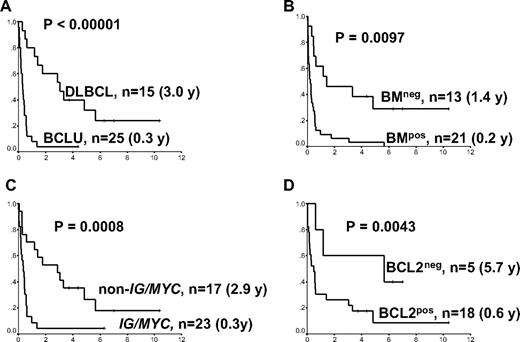
Restricting the analysis to the 40 patients treated with curative intent, which excludes the case of FL, 5 factors were associated with a more favorable outcome: a low IPI score, the absence of bone marrow involvement, DLBCL histology, the presence of a non-IG/MYC partner, and a BCL2 protein–negative biopsy (clone 124; Figure 3). There was a strong association between DLBCL morphology and the presence of a non-IG/MYC partner and a BCL2 protein–negative biopsy (P < .001 and P = .04, respectively). Conversely, BCLU morphology was associated with the presence of an IG/MYC partner and a bone marrow/leukemic presentation (P = .001). Of the 4 variables presented in Figure 3, only bone marrow/leukemic presentation of MYC+ lymphoma was associated with a high IPI score (P = .03). Other factors not associated with survival were a high proliferation index, CD10 expression, or the presence of additional cytogenetic alterations, such as a rearrangement at 3q27, a loss of 17p13 or 1p36.
Survival curves of patients with BCL2+/MYC+ lymphomas according to morphology, bone marrow involvement, MYC translocation partner, and BCL2 protein expression. Median overall survivals are shown in parentheses. (A) Morphology. (B) Bone marrow involvement. (C) MYC translocation partner. (D) BCL2 protein expression (clone 124). In all graphs, x-axis represents cumulative survival; and y-axis, time (years). BCLU indicates B-cell lymphoma unclassifiable; DLBCL, diffuse large B-cell lymphoma; BMneg, no bone marrow involvement with MYC+ lymphoma; BMpos, bone marrow involvement with MYC+ lymphoma; BCL2neg, no BCL2 protein expression by clone 124; BCL2pos, BCL2 protein expression by clone 124; and IG/MYC, MYC translocation involving one of the immunoglobulin genes.
Survival curves of patients with BCL2+/MYC+ lymphomas according to morphology, bone marrow involvement, MYC translocation partner, and BCL2 protein expression. Median overall survivals are shown in parentheses. (A) Morphology. (B) Bone marrow involvement. (C) MYC translocation partner. (D) BCL2 protein expression (clone 124). In all graphs, x-axis represents cumulative survival; and y-axis, time (years). BCLU indicates B-cell lymphoma unclassifiable; DLBCL, diffuse large B-cell lymphoma; BMneg, no bone marrow involvement with MYC+ lymphoma; BMpos, bone marrow involvement with MYC+ lymphoma; BCL2neg, no BCL2 protein expression by clone 124; BCL2pos, BCL2 protein expression by clone 124; and IG/MYC, MYC translocation involving one of the immunoglobulin genes.
Given the small sample size, multivariate Cox regression analysis was performed excluding BCL2 as a variable because this result was not available in most patients. Hazard ratios for the univariate and multivariate models are listed in Table 2. If only 3 variables are entered into the model, then morphology and IPI, not the MYC translocation partner, were the factors most predictive of OS (morphology, P < .001; IPI 2-3 vs IPI 0-1, P = .290; and IPI 4-5 vs IPI 0-1, P = .009).
Univariate and multivariate models using overall survival for 40 patients with concurrent BCL2 and MYC translocations treated with curative intent
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| BCLU vs DLBCL | 5.73 | 2.49-13.2 | < .001 | 4.04 | 1.40-11.6 | .009 |
| BM+ vs BM− | 3.27 | 1.50-7.33 | .004 | 1.98 | 0.79-5.03 | .150 |
| BCL2+ vs BCL2− | 2.86 | 0.791-10.3 | .110 | — | — | — |
| IG/MYC vs non-IG/MYC | 3.42 | 1.62-7.22 | .001 | 1.47 | 0.55-3.88 | .441 |
| IPI risk group, 2-3 vs 0-1 | 1.64 | 0.747-3.58 | .220 | 1.45 | 0.53-3.95 | .470 |
| IPI risk group, 4-5 vs 0-1 | 5.12 | 1.84-14.31 | .002 | 3.25 | 0.85-12.4 | .084 |
| Variable . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| BCLU vs DLBCL | 5.73 | 2.49-13.2 | < .001 | 4.04 | 1.40-11.6 | .009 |
| BM+ vs BM− | 3.27 | 1.50-7.33 | .004 | 1.98 | 0.79-5.03 | .150 |
| BCL2+ vs BCL2− | 2.86 | 0.791-10.3 | .110 | — | — | — |
| IG/MYC vs non-IG/MYC | 3.42 | 1.62-7.22 | .001 | 1.47 | 0.55-3.88 | .441 |
| IPI risk group, 2-3 vs 0-1 | 1.64 | 0.747-3.58 | .220 | 1.45 | 0.53-3.95 | .470 |
| IPI risk group, 4-5 vs 0-1 | 5.12 | 1.84-14.31 | .002 | 3.25 | 0.85-12.4 | .084 |
HR indicates hazard ratio; CI, confidence interval; BCLU, B-cell lymphoma, unclassifiable, with features intermediate between Burkitt lymphoma and DLBCL; DLBCL, diffuse large B-cell lymphoma; BM, bone marrow involvement; BCL2, BCL2 protein expression (clone 124); —, not applicable; IG, immunoglobulin; and IPI, International Prognostic Index.
Discussion
This study demonstrates that lymphomas with concurrent BCL2 and MYC translocations are heterogeneous in morphology, clinical presentation, and outcome. The nondescript term “double hit” lymphomas does not adequately capture the full clinical spectrum of this disease. It is more precise and clinically relevant to refer to these lymphoma subtypes using the appropriate morphologic classification, DLBCL versus BCLU, with features intermediate between BL and DLBCL, with an indication of the dominant oncogenes involved, including the MYC translocation partner and the BCL2 protein expression.
BCL2+/MYC+ lymphomas may be underrecognized because this diagnosis would be missed if genetic testing by karyotype and/or FISH analysis were not performed. These lymphomas represented 4% of selected lymphoma cases subjected to karyotype and FISH analysis in British Columbia. A selection bias in our study may have led to a modest overestimation of the incidence, given that cytogenetic analysis was selectively performed on lymphoma samples that had high-grade histology or aggressive clinical features. However, the incidence of MYC translocations may also have been underestimated because some non-IG/MYC rearrangements could have been missed by karyotype analysis alone. Furthermore, FISH analysis could miss uncommon MYC breakpoints that are centromeric or telomeric to the commercial break-apart probes used in most clinical laboratories.34,35 Indeed, one of our cases had a t(8;14) by karyotype and a far centromeric MYC breakpoint that was not detected using the commercial LSY MYC dual-color break-apart probe.34 Therefore, more sensitive MYC FISH probes may be needed to eliminate the false-negative results with the current commercial probes. With those limitations in mind, it is interesting that our estimate is comparable with that seen in a comprehensive review of 2175 lymphoma karyotypes.36 A 5% incidence of MYC translocations was reported among 355 cases of t(14;18)-positive lymphomas: 2% had t(8;14), 2% had t(8;22), and 1% had t(8;9).36 Furthermore, the incidence of dual MYC and BCL2 translocations in de novo DLBCL was found to be 3% to 4% in 2 independent studies.21,37
The IPI and tumor morphology appear to be the most powerful, independent predictors of outcome in this disease, the latter being highly associated with the MYC translocation partner. MYC is a crucial regulator of all aspects of cellular growth and proliferation (reviewed by Wierstra and Alves38 ). In the case of the t(8;14) and related IG/MYC translocations, MYC is under the regulation of a strong IG enhancer resulting in constitutive MYC expression that commonly manifests as a higher tumor grade. The gene expression profiling study by Hummel et al showed a strong correlation between the MYC partner, MYC expression, and resultant morphology.39 In our study, non-IG/MYC translocations were common and the “classic” t(8;14) translocation was present in only 15% of our samples. The chromosomal band 9p13 was the most common non-IG MYC translocation partner and the breakpoints flanked several candidate genes, including ZCCHC7, and were approximately 200 kb 5′ of PAX5.33 These genes are transcription factors that are very important in B-cell development and could potentially lead to MYC deregulation when translocated near the 8q24 locus.40,41 Within both the DLBCL and BCLU categories, there was a trend toward a more favorable outcome for patients having non-IG/MYC translocation partners in their biopsies; thus, FISH analysis using only a MYC break-apart probe may not be sufficient for accurate prognostication. Future prospective studies to investigate the clinical impact of MYC translocation partners in larger cohorts of DLBCL and BCLU are required.
Mutations in the BCL2 gene corresponding to the flexible loop domain may be clinically important and may explain the observed discrepancies between the results with the different BCL2 antibodies (clone 124 and E17). Such discrepancies have been observed in FLs and are thought to be a consequence of ongoing somatic hypermutation given the proximity of BCL2 to the IGH locus.30,42 Such mutations may interfere with BCL2 function sufficiently to affect the caspase cleavage site at D34, the P53 binding site at amino acids 32 to 68, or the phosphorylation of S70, which is required for BCL2's full and potent antiapoptotic function.43,44 Thus, it is conceivable that the BCL2 “pseudo-negative” cases in our study may have a dysfunctional BCL2 protein that is associated with improved chemo-sensitivity to R-CHOP. Rituximab has been shown to partially overcome the chemotherapy resistance associated with BCL2 protein expression (positive by clone 124) in DLBCL.45 Stolz et al have recently shown that rituximab can induce apoptosis through the “mitochondrial” pathway that is regulated by the BCL2 protein family and the addition of BH3 mimetics that target BCL2 function can restore chemo-sensitivity in primary rituximab-resistant cell lines.46 Indeed, the addition of the BH3 mimetic ABT-737 to cyclophosphamide was synergistic at inducing long-term remissions in 78% (14 of 18) of mice with BCL2+/MYC+ lymphomas.47 Thus, pharmacologic modulation of BCL2 in humans with BH3 mimetics with or without rituximab may render the aggressive BCL2+/MYC+ lymphomas chemo-sensitive and should be studied further in the context of clinical trials.
In conclusion, a comprehensive cytogenetic analysis of BCL2 and MYC status is currently feasible in most reference clinical laboratories and should be performed on all aggressive lymphomas as this may not only rule out BL as a potential diagnosis, but it may identify more homogeneous populations of lymphomas within the current BCLU category.32 Concurrent BCL2 and variant (non-IG) MYC rearrangements may be more common than previously appreciated in DLBCL. Prospective analysis of their incidence and prognostic significance, including BCL2 protein expression, in patients with DLBCL treated with R-CHOP may lead to the identification of very-high-risk patients who might derive the most benefit from rituximab and BCL2-targeted therapy. Furthermore, it may pave the way to a better understanding of the complex synergism that arises when these 2 dominant oncogenes are deregulated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the cytogenetic technologists of the Cancer Genetic Laboratory, Department of Pathology, British Columbia Cancer Agency for their the hard work, and the dedication and the generous support of the Center for Translational and Applied Genomics laboratory for infrastructure support.
N.A.J. is a research fellow of the Terry Fox Foundation through an award from the National Cancer Institute of Canada (019005) and the Michael Smith Foundation for Health Research (ST-PDF-01793); she was previously supported by a training award from the Canadian Institute of Health Research (no. STP-53912) during which part of this work was performed. R.S. was supported by the Deutsche Kresbhilfe and the KinderKrebsInitiative Buchholz, Holm-Seppensen. J.M.C., R.D.G., and D.E.H. were supported by the National Cancer Institute of Canada, Terry Fox Foundation Program Project (grant 019001). This work was also supported in part by the National Institutes of Health (grant UO1-CA84967-1, Vancouver). R.D.G. and J.M.C. were supported by Roche Canada.
National Institutes of Health
Authorship
Contribution: N.A.J. designed and performed the research, analyzed the data, and wrote the manuscript; O.L., S.B.-N., and C.S. performed FISH experiments; K.J.S., M.J.S.D., R.S., J.K., J.M.C., and R.K. contributed to data collection; R.W. performed the statistical analysis; R.D.G. performed the pathologic review; and D.E.H. designed the research. All coauthors read and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Douglas E. Horsman, Cytogenetic and Molecular Genetic Laboratories, British Columbia Cancer Agency, 600 West 10th Ave, Vancouver, BC, V5Z 4E5, Canada; e-mail: dhorsman@bccancer.bc.ca.