Abstract
Inhibitory killer cell immunoglobulin-like receptors (KIRs) preserve tolerance to self and shape the functional response of human natural killer (NK) cells. Here, we have evaluated the influence of selection processes in the formation of inhibitory KIR repertoires in a cohort of 44 donors homozygous for the group A KIR haplotype. Coexpression of multiple KIRs was more frequent than expected by the product rule that describes random association of independent events. In line with this observation, the probability of KIR acquisition increased with the cellular expression of KIRs. Three types of KIR repertoires were distinguished that differed in frequencies of KIR- and NKG2A-positive cells but showed no dependency on the number of self-HLA class I ligands. Furthermore, the distribution of self- and nonself-KIRs at the cell surface reflected a random combination of receptors rather than a selection process conferred by cognate HLA class I molecules. Finally, NKG2A was found to buffer overall functional responses in KIR repertoires characterized by low-KIR expression frequencies. The results provide new insights into the formation of inhibitory KIR repertoires on human NK cells and support a model in which variegated KIR repertoires are generated through sequential and random acquisition of KIRs in the absence of selection.
Introduction
Natural killer (NK) cell function is fine tuned by the integrated signaling through multiple activating and inhibitory receptors.1,2 Murine and human NK cells express inhibitory Ly49 and killer cell Ig-like receptors (KIRs), respectively, that interact with major histocompatibility complex (MHC) class I molecules.3,4 In addition, NK cells express inhibitory CD94/NKG2A heterodimers that recognize complexes of HLA-E (human) and Qa-1 (mice) bound to peptides derived from the leader sequences of other MHC class I molecules.5,6 The human KIR gene cluster is located within the leukocyte receptor complex on chromosome 19.7 KIR haplotypes can be divided into group A and B haplotypes that differ in gene contents.8 Four major inhibitory KIRs have been defined (KIR2DL2/3, KIR2DL1, KIR3DL1, and KIR3DL2). These receptors recognize HLA-C allotypes with asparagine 80 (C1), HLA-C allotypes with lysine 80 (C2), HLA-A and HLA-B allotypes with Bw4 motifs at positions 77-83, and HLA-A3/11, respectively.9
Since the discovery of inhibitory receptors for MHC class I molecules, several investigations have examined how the expression of these receptors is regulated. NK cells display a variegated expression pattern of inhibitory receptors that confers a broad specificity to sense the presence or absence of single MHC class I alleles.10,11 Furthermore, it has been observed that the frequency of NK cells that coexpress 2 inhibitory Ly49 or KIRs equals the product of their individual expression frequencies in the NK cell repertoire.10,12 This formula is referred to as the product rule and has led to the general perception that KIR acquisition is a random event that is independent of expression of other KIRs. In humans, most NK cells express 1 or 2 KIRs and most, albeit not all, NK cells express at least one inhibitory KIR specific for self-HLA class I molecules.10,13 However, the expression of 2 or more self-specific receptors is relatively uncommon and it has been proposed that this situation may be disfavored because it could decrease the ability to detect MHC alterations occurring during tumor transformation or viral infection.12
The fact that human and mouse NK cell receptor repertoires are characterized by expression of one but not many self-specific receptors has been taken as an argument for the existence of an active selection process conferred by interactions with cognate MHC class I molecules. However, early studies found no role for HLA in shaping the variegated KIR repertoire of human NK cells.14,15 Subsequent studies have corroborated the view that the KIR genotype is the major determinant for KIR repertoire formation but with a subtle modulating effect of the HLA genotype.16,17 The small effect of HLA class I, noted in KIR-identical but HLA-disparate siblings, was independent of self-KIR epitopes.17 Importantly, and unlike the process of T-cell selection, no NK cell clones are deleted following interactions with MHC class I.18 Recent investigations have shown that frequencies of NK cells with KIR2DL1 and KIR3DL1 were higher in individuals carrying the cognate KIR epitope.16 Although the picture emerging from these studies is that there is a minor effect of HLA on KIR expression frequencies, it is unclear to what extent this leads to a bias for expression of self-KIRs in the total repertoire.
Here, we have analyzed KIR repertoires and functional responses in a cohort of 44 individuals homozygous for the group A KIR haplotype. By limiting the study to donors with this haplotype, we avoided confounding effects of cross-reactive mAbs binding to activating KIRs. Moreover, this enabled a full assessment of all expressed inhibitory KIRs, since KIR2DL5 is not expressed in donors with this haplotype.19 We sought to investigate whether acquisition of KIRs is independent of previously expressed receptors and how the presence of cognate HLA class I molecules influences KIR repertoire formation on human NK cells. We then evaluated how the results fit with selection processes conferred by cognate HLA class I molecules.
Methods
Human subjects, blood samples, and cell processing
This study was approved by the regional ethics committee (Stockholm, Sweden, approval number 2006/229-31/3), as well as the Karolinska Institutet review board, and donor informed consent was obtained in accordance with the Declaration of Helsinki. Buffy coats were prepared from peripheral blood of healthy human donors and separated by density gradient centrifugation (Ficoll-Hypaque; GE Healthcare Bio-Sciences AB). When necessary, NK cells were isolated using the NK cell selection kit (magnetic-activated cell sorting [MACS]; Miltenyi Biotec) followed by sorting on a FACSAria Flow Cytometer (BD Biosciences).
KIR and HLA genotyping
Genomic DNA was isolated from 100 μL buffy coats using the DNeasy Blood & Tissue Kit (QIAGEN). KIR genotyping was performed as described previously using polymerase chain reaction using sequence-specific primers (PCR-SSP) technology and a KIR typing kit (Olerup-SSP).20 KIR ligands were determined using the KIR HLA ligand kit (Olerup-SSP) for detecting the HLA-Bw4, -C1, and -C2 motifs. For analysis of HLA-A3/A11, and specific HLA-B alleles, complementary HLA genotyping was performed with the HLA-A and -B low-resolution kit (Olerup-SSP).
Antibodies and flow cytometry
The antibody panel used in the present study was recently described in detail by Fauriat et al.21 The specific conjugations used are depicted in Figure 1. Data were acquired on a CyAn ADP LX 9-color flow cytometer (Dako) as previously described.22 Acquired data were compensated with the software-based compensation platform in FlowJo 8.6 (TreeStar) and analyzed with FlowJo 8.6.
CD107a assay
Peripheral blood mononuclear cells (PBMCs; 1.5 × 106cells/mL) were mixed with K562 cells at a ratio of 10:1 in a final volume of 200 μL in V-bottom 96-well plates and incubated for 6 hours at 37°C and 5% CO2. Cells were stained with NK cell markers and CD107a on ice for 20 minutes in cold fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 2% FBS and 2 mM EDTA). Cells were washed 3 times and analyzed by flow cytometry.
Calculation of expected values according to the product rule
The product rule is a special case of Bayes rule and valid for independent events. The expected probability for NK cells to express all 4 KIRs is given by PAll KIR = PKIR2DL1PKIR2DL3PKIR3DL1PKIR3DL2, where P is the total probability of expressing a given receptor including NK cells coexpressing other KIRs.
Combinatorial analysis
The number of combinations l of self- and nonself-KIRs on NK cells may be described as follows. Among N KIRs, m's are self-KIR and N-m's are nonself-KIRs. Further, the number of ways to combine n KIRs is the product of the possible combinations of k self-KIRs and n-k nonself-KIRs among m and N-m receptors, respectively.
Division by the number of possible ways to combine n receptors of N gives the probability of expressing k self- and n-k nonself-KIRs for NK cells with n KIRs.
This is the hypergeometric probability distribution that describes the number of successes in a sequence of n draws from a finite population without replacement.
Calculation of probability distributions
For the binomial probability distribution of KIR expression, it is assumed that each cell may acquire up to 4 KIRs and that success or failure to acquire a given KIR is independent of other receptors and has a constant probability p and q, (q = 1 − p), respectively. The binomial distribution is given by:
where P(n) is the probability of expressing n KIRs of N.
To account for individual KIR acquisition probabilities, the total frequencies of KIR expression were normalized and the resulting probabilities for each KIR were introduced into the binomial distribution.
To study the dependency of expression between different KIRs, probabilities were modeled using a correlated binomial model, the beta-binomial distribution.23,24
where n is the number of KIRs among N, and B(α, β) is the beta function with shape parameters α and β. The parameter values were obtained by fitting the beta-binomial model to the data. The shaping parameters that minimized the chi-square goodness of fit resulted in a correlation coefficient ρ = 0.14 where α = 1.80 and β = 4.26.
Calculation of conditional probabilities
Conditional probabilities corresponding to the beta-binomial distribution were given by
where ρ is the correlation coefficient and p is the conditional probability of expressing another KIR given that i KIRs have already been expressed and j KIRs have not been expressed.25 Conditional probabilities could also be determined from Bayes rule:
where P (A|B) indicate the probability of acquiring KIR A, given that KIR B is expressed. Bayes rule may be used to derive the relation between k-values and probabilities of acquiring additional KIRs. The probability of acquiring the nth KIR was given by
Statistical analysis
In Figures 2 through 7 differences were evaluated using Wilcoxon signed-rank test. Matched-pairs tests and tests against one predicted value were used when relevant. In Figure 7B (left panel), differences were evaluated using a 2-tailed Mann-Whitney test. In Figure 7B (right panel) and Figure 7C-D, a one-way ANOVA followed by Kruskal-Wallis test was used. (*P < .05; **P < .01; ***P < .001.)
Results
Coexpression of multiple KIRs is more frequent than expected by the product rule
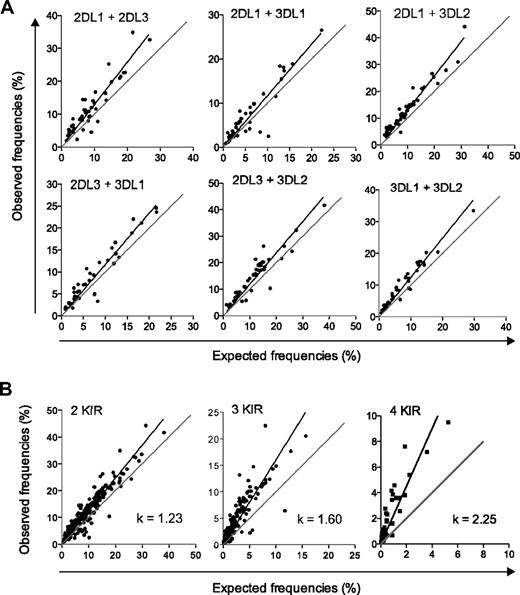
To address whether KIR acquisition occurs in a random and independent fashion, we tested the validity of the product rule for NK cells expressing 2, 3, and 4 KIRs in a cohort of 44 donors homozygous for the group A KIR haplotype. A Boolean gating strategy was used to identify subsets of NK cells expressing various combinations of KIRs (Figure 1). The linear regression of the observed versus expected frequencies of NK cells expressing combinations of 2 individual KIRs was plotted (Figure 2A). For all possible combinations of 2 KIRs, there was a small (on average 23%) but statistically significant deviation from the product rule (Figure 2B left panel). The deviation became more evident when examining the frequencies of NK cells coexpressing more than 2 KIRs (Figure 2B middle and right panels). Thus, coexpression of 3 and 4 KIRs was significantly more frequent (60% and 125%, respectively) than expected from a random and independent association of KIRs.
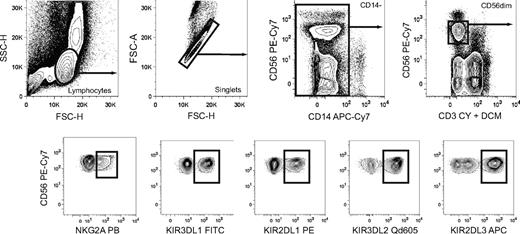
Boolean gating of NK cell subsets expressing discrete combinations of inhibitory KIRs and NKG2A. Gating scheme to identify subsets of CD56dim NK cells expressing single inhibitory KIR and combinations thereof with or without coexpression of NKG2A. Doublet cells and CD14+ cells were excluded based on an FSC-area versus FSC–height gate and CD14 expression, respectively. Gates were set on live CD3− cells as determined by staining with a dead cell marker (DCM) and anti-CD3.
Boolean gating of NK cell subsets expressing discrete combinations of inhibitory KIRs and NKG2A. Gating scheme to identify subsets of CD56dim NK cells expressing single inhibitory KIR and combinations thereof with or without coexpression of NKG2A. Doublet cells and CD14+ cells were excluded based on an FSC-area versus FSC–height gate and CD14 expression, respectively. Gates were set on live CD3− cells as determined by staining with a dead cell marker (DCM) and anti-CD3.
Coexpression of multiple KIRs is more frequent than expected by the product rule. (A) The observed frequencies of NK cells coexpressing the indicated KIRs in 44 haplotype A donors are plotted against those given by the product of the expression frequencies of the respective KIRs. Combinations including KIR3DL1 were based on 40 individuals, since 4 donors had KIR3DL1 alleles, which were not expressed at the cell surface.26 A perfect fit to the product rule is shown as a gray line, representing a 1:1 relationship between observed and expected frequencies. (B) The observed frequencies of NK cells coexpressing 2, 3, and 4 inhibitory KIRs are plotted against frequencies expected by the product rule. Pooled data from distinct combinations of 2 (left panel), 3 (middle panel), and 4 (right panel) KIRs are shown.
Coexpression of multiple KIRs is more frequent than expected by the product rule. (A) The observed frequencies of NK cells coexpressing the indicated KIRs in 44 haplotype A donors are plotted against those given by the product of the expression frequencies of the respective KIRs. Combinations including KIR3DL1 were based on 40 individuals, since 4 donors had KIR3DL1 alleles, which were not expressed at the cell surface.26 A perfect fit to the product rule is shown as a gray line, representing a 1:1 relationship between observed and expected frequencies. (B) The observed frequencies of NK cells coexpressing 2, 3, and 4 inhibitory KIRs are plotted against frequencies expected by the product rule. Pooled data from distinct combinations of 2 (left panel), 3 (middle panel), and 4 (right panel) KIRs are shown.
To exclude the possibility that the observed deviations were a consequence of difficulties in compensating for spectral overlaps from multiple conjugates, NK cells coexpressing all 4 KIRs were sorted out and reassessed for expression of the 4 KIRs using one anti-KIR antibody per staining (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cellular expression of KIRs increases the probability of acquiring additional KIRs
The observation that the frequency of NK cells expressing multiple KIRs was higher than expected by the product rule suggested that KIR acquisition is either nonrandom or dependent on previously expressed KIRs or both. To examine these possibilities, we plotted the frequencies of NK cells expressing zero to 4 KIRs and compared the results with theoretic values generated by different statistical distributions describing random independent or dependent KIR acquisition patterns (Figure 3).
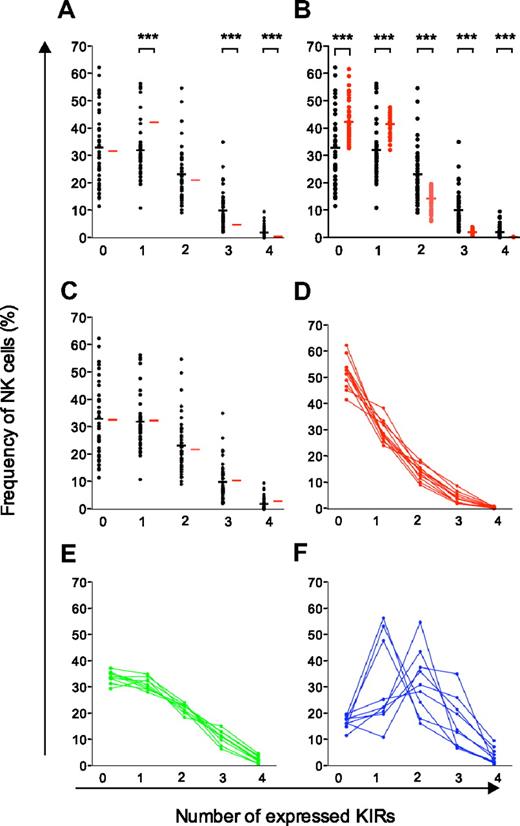
Comparison of experimental data with different models of KIR acquisition and identification of 3 distinct KIR repertoires. (A-C) Observed frequencies of NK cells expressing zero to 4 KIRs were compared with those predicted by sequential acquisition of KIRs under different conditions (red bars). (A) Binomial distribution with equal and constant probabilities (P = .25) of KIR acquisition. (B) Binomial distribution with individual probabilities for acquisition of different KIRs in each individual. (C) Beta-binomial distribution describing the best possible fit to the observed data and corresponding to increasing KIR acquisition probabilities depending on the number of KIRs expressed at the cell surface. (D-F) Identification of 3 distinct KIR repertoires characterized by (D) low-KIR, n = 9; (E) intermediate-KIR, n = 9; and (F) high-KIR, n = 11, expression. Low-KIR repertoires were defined as more than 40% KIR-negative NK cells and high-KIR repertoires were defined as less than 20% KIR-negative NK cells.
Comparison of experimental data with different models of KIR acquisition and identification of 3 distinct KIR repertoires. (A-C) Observed frequencies of NK cells expressing zero to 4 KIRs were compared with those predicted by sequential acquisition of KIRs under different conditions (red bars). (A) Binomial distribution with equal and constant probabilities (P = .25) of KIR acquisition. (B) Binomial distribution with individual probabilities for acquisition of different KIRs in each individual. (C) Beta-binomial distribution describing the best possible fit to the observed data and corresponding to increasing KIR acquisition probabilities depending on the number of KIRs expressed at the cell surface. (D-F) Identification of 3 distinct KIR repertoires characterized by (D) low-KIR, n = 9; (E) intermediate-KIR, n = 9; and (F) high-KIR, n = 11, expression. Low-KIR repertoires were defined as more than 40% KIR-negative NK cells and high-KIR repertoires were defined as less than 20% KIR-negative NK cells.
The binomial distribution describes how the KIR repertoire would appear if each KIR had an equal expression probability (P = .25; ie, 1 of 4 KIRs) regardless of whether the cell expressed other KIRs. The resulting calculated frequencies of NK cells expressing zero to 4 KIRs differed significantly from the observed expression frequencies (Figure 3A). In line with the observed deviation from the product rule for NK cells expressing multiple KIRs, this analysis revealed that the frequency of NK cells expressing only one inhibitory receptor was significantly lower than that expected from a binomial distribution, whereas those expressing 3 and 4 KIRs were more frequent (Figure 3A).
The deviation from a random and independent KIR acquisition process as modeled by the binomial distribution could be due to either of 2 assumptions made by this model. First, the 4 different KIRs are not likely to have the same intrinsic expression probabilities. Second, the probability of acquiring a KIR may not be constant during acquisition of additional receptors. To address the first possibility, we assumed that the probability of KIR acquisition was reflected by the frequency with which any given KIR was expressed in the individual. However, taking the individual expression probabilities into consideration did not result in an improved fit with the observed data (Figure 3B).
To address the possibility of a dynamic expression probability during KIR acquisition, we examined whether it was possible to describe how the probability of KIR acquisition changes as cells acquire additional receptors (see supplemental Figure 2 for schematic overview). Using the beta-binomial distribution, it was possible to get an exact fit with the observed data (Figure 3C). The obtained distribution described a positive correlation corresponding to increasing conditional probabilities for KIR acquisition as additional KIRs are expressed. The resulting conditional probabilities were 0.24, 0.35, 0.45, and 0.53 for acquisition of the first, second, third, and fourth KIR, respectively (Table 1; see “Calculation of conditional probabilities” for details).
Conditional probabilities of KIR acquisition
| KIR . | Type of distribution . | Values calculated from slope of deviation . | |||
|---|---|---|---|---|---|
| Binomial . | Beta-binomial . | Low . | Intermediate . | High . | |
| 1 | 0.25 | 0.24 | 0.19 | 0.29 | 0.36 |
| 2 | 0.25 | 0.35 | 0.22 | 0.40 | 0.50 |
| 3 | 0.25 | 0.45 | 0.24 | 0.48 | 0.64 |
| 4 | 0.25 | 0.53 | 0.26 | 0.53 | 0.65 |
| KIR . | Type of distribution . | Values calculated from slope of deviation . | |||
|---|---|---|---|---|---|
| Binomial . | Beta-binomial . | Low . | Intermediate . | High . | |
| 1 | 0.25 | 0.24 | 0.19 | 0.29 | 0.36 |
| 2 | 0.25 | 0.35 | 0.22 | 0.40 | 0.50 |
| 3 | 0.25 | 0.45 | 0.24 | 0.48 | 0.64 |
| 4 | 0.25 | 0.53 | 0.26 | 0.53 | 0.65 |
Conditional probabilities obtained from the degree of deviation (k-values) from the product rule for the 3 types of repertoires. The probabilities of acquiring the first KIR (values in italics) were estimated to equal the average frequency of KIR expression (2DL3+2DL1+3DL1+3DL2)/4.
Collectively, these results demonstrated that the probability of acquiring additional KIRs was higher in cells that already expressed KIRs at the cell surface. These results are in agreement with a previous investigation of NK cells expressing up to 3 inhibitory KIRs,27 and indicate that KIR acquisition is sequential and dependent on previously expressed receptors. As a consequence, the product rule that is valid for independent events cannot be used to accurately determine frequencies of NK cells coexpressing multiple KIRs.
Identification of 3 types of KIR repertoires with different probabilities of KIR acquisition
The beta-binomial distribution provided a good description of the average proportion of NK cells with zero to 4 KIRs in the cohort of 44 donors. However, it was still unclear how well the proposed model of a sequential and dependent KIR acquisition would describe the formation of the variegated KIR repertoires on an individual basis. To address this issue, 3 types of KIR repertoires were distinguished with high-, intermediate-, and low-KIR expression frequencies (Figure 3D-F and supplemental Figure 3). Interestingly, intermediate- and low-KIR repertoires deviated more from the product rule than high-KIR repertoires (Figure 4A).
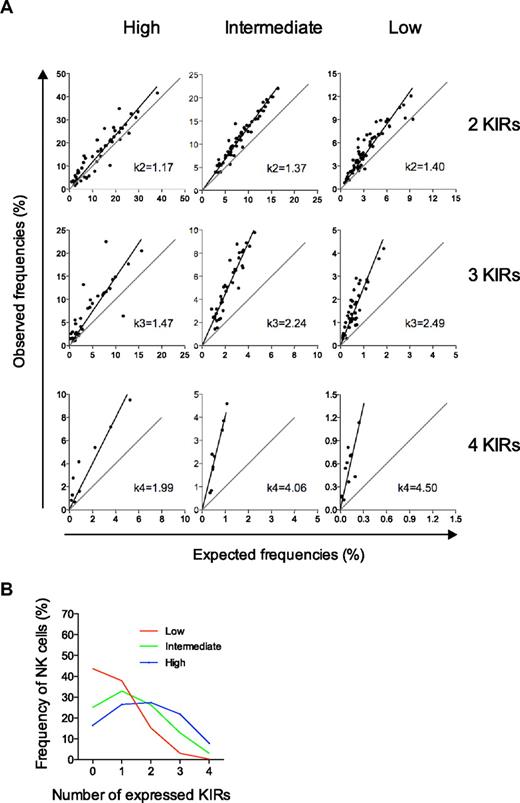
Deviation from the product rule differs for the 3 types of repertoires and defines their conditional probabilities for KIR acquisition. (A) Deviations from the product rule for NK cells expressing 2, 3, and 4 KIRs are shown for the 3 distinct repertoires defined by their high-, intermediate-, and low-KIR expression frequencies. k-values were derived from the slope of the linear regression of observed data relative to a perfect fit to the product rule. (B) The conditional probabilities (shown in Table 1) for acquiring the nth KIR were given by Bayes rule (see “Calculation of conditional probabilities” for details). The conditional probabilities were then run in an algorithm for random and dependent KIR acquisition and the corresponding repertoires are shown.
Deviation from the product rule differs for the 3 types of repertoires and defines their conditional probabilities for KIR acquisition. (A) Deviations from the product rule for NK cells expressing 2, 3, and 4 KIRs are shown for the 3 distinct repertoires defined by their high-, intermediate-, and low-KIR expression frequencies. k-values were derived from the slope of the linear regression of observed data relative to a perfect fit to the product rule. (B) The conditional probabilities (shown in Table 1) for acquiring the nth KIR were given by Bayes rule (see “Calculation of conditional probabilities” for details). The conditional probabilities were then run in an algorithm for random and dependent KIR acquisition and the corresponding repertoires are shown.
Next, we examined if and how we could link the degree of deviation from the product rule to the conditional probabilities that determined the likelihood for KIR acquisition given that NK cells already expressed one or more KIRs. The slopes obtained from the linear regression of the observed data for NK cells expressing 2, 3, and 4 KIRs were denoted k2, k3, and k4, respectively (Figure 4A). According to Bayes rule, these k-values may be used to approximate the conditional probabilities for KIR acquisition (see “Calculation of conditional probabilities” for details). However, an unknown factor was the probability of acquiring the first KIR. Since we could not determine in which order KIRs had been acquired on each NK cell, we assumed that the probability of KIR acquisition could be estimated from the average KIR expression frequency. These average KIR expression frequencies were 19%, 29%, and 36% for the low-, intermediate-, and high-KIR repertoires, respectively (supplemental Figure 3; Table 1).
Based on the estimated probabilities for acquisition of the first KIR and the k-values of the deviation from the product rule, we could determine the conditional probabilities of KIR acquisition for the 3 KIR repertoires (Table 1). High- and low-KIR repertoires were characterized by high- and low-KIR acquisition probabilities, respectively. Importantly, and validating this approach, the probabilities of the intermediate-KIR repertoires were similar to those obtained from the beta-binomial distribution that provided mean values of conditional probabilities for the whole cohort. Furthermore, the conditional probabilities for the 3 types of repertoires corresponded to their respective expression patterns of zero to 4 KIRs (Figure 4B) when introduced into the sequential binomial algorithm (supplemental Figure 2).
Together, these results demonstrate that variegated KIR repertoires can be generated through sequential and dependent KIR acquisition with gradually increasing conditional probabilities for acquiring new KIRs. The data also indicate that there are individual set points for KIR acquisition that determine the overall probability of acquiring KIRs and that translate into variegated KIR repertoires.
The presence of cognate HLA class I molecules does not influence KIR repertoire formation
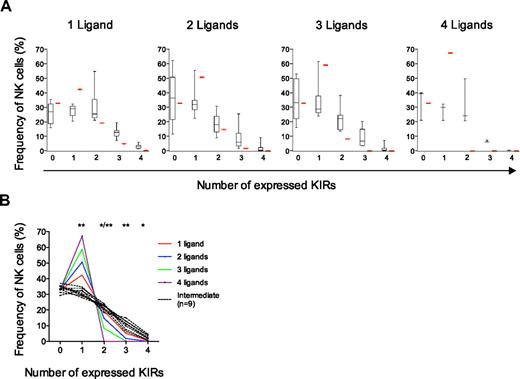
To address whether high- and low-KIR repertoires were the result of selection processes conferred by few and several KIR ligands, respectively, donors were genotyped for the presence of the 4 major KIR ligands, HLA-C1, -C2, -Bw4, and -A3/A11 (supplemental Table 1). We plotted the frequencies of NK cells expressing zero to 4 KIRs in individuals with 1 to 4 KIR ligands (Figure 5A). Interestingly, donors with 3 ligands had similar frequencies of NK cells that expressed multiple KIRs as those with only one ligand (Figure 5A). Moreover, although only 3 donors had all 4 ligands, the data did not reveal a profoundly skewed KIR repertoire in these donors. The results were similar when weak ligands, including HLA-A23, -A24, -A3, and -A11 were excluded (supplemental Figure 4).18,21
KIR acquisition is independent of cognate HLA ligands. The observed frequencies of NK cells were compared with those predicted by the sequential selection model. (A) Frequencies of NK cells expressing zero to 4 KIRs in individuals with 1 (n = 9), 2 (n = 20), 3 (n = 9), and 4 (n = 3) KIR ligands are shown. Importantly, nonfunctional KIR ligands including HLA-B13 and HLA-A25 were considered nonself-ligands.18,28 Boxes represent the median and the 5th and 95th percentile. Red bars indicate values obtained by modeling KIR expression frequencies selected by 1 to 4 self-KIR ligands. (B) Frequencies of NK cells expressing zero to 4 KIRs in individuals with intermediate-KIR repertoires (n = 9) and variable number of strong KIR ligands were compared with those generated under selection by 1, 2, 3, and 4 ligands, respectively.
KIR acquisition is independent of cognate HLA ligands. The observed frequencies of NK cells were compared with those predicted by the sequential selection model. (A) Frequencies of NK cells expressing zero to 4 KIRs in individuals with 1 (n = 9), 2 (n = 20), 3 (n = 9), and 4 (n = 3) KIR ligands are shown. Importantly, nonfunctional KIR ligands including HLA-B13 and HLA-A25 were considered nonself-ligands.18,28 Boxes represent the median and the 5th and 95th percentile. Red bars indicate values obtained by modeling KIR expression frequencies selected by 1 to 4 self-KIR ligands. (B) Frequencies of NK cells expressing zero to 4 KIRs in individuals with intermediate-KIR repertoires (n = 9) and variable number of strong KIR ligands were compared with those generated under selection by 1, 2, 3, and 4 ligands, respectively.
To test whether the analysis performed here was robust enough to detect the presence of selection, we modeled how KIR repertoires would appear given that KIR acquisition ceases upon interaction with a self-ligand as postulated by the sequential acquisition model for NK cell selection (Figure 5A-B and supplemental Figure 5A-B). The theoretic calculation of KIR expression frequencies under the influence of selection, depicted as red bars in Figure 5A and Figure S4, revealed a gradual increase of NK cells with only one KIR depending on the number of selecting ligands. This pattern differed significantly from the experimental data showing constant frequencies of NK cells with one KIR regardless of the number of KIR ligands (Figure 5A). The fact that KIR expression was similar within each of the 3 repertoires also allowed a more strict comparison of intermediate-KIR repertoires with theoretic repertoires formed under selection on 1 to 4 self-ligands (Figure 5B). These results support the notion that KIR repertoires are formed in a manner that is largely independent of cognate HLA class I ligands in the host.
Expression of self- and nonself-KIR is randomly regulated and not a consequence of selection by cognate HLA class I molecules
To explore whether gradually increasing KIR acquisition probabilities resulted in accumulation of self-specific receptors, we examined how the presence of cognate HLA class I molecules influenced the expression of self- and nonself-KIRs on NK cells. The frequency of NK cells expressing one self-KIR was nearly identical to the frequency of NK cells with one nonself-KIR and the data matched those obtained in the absence of selection. In contrast, experimental data differed significantly from theoretic KIR frequencies predicted by the sequential selection model (Figure 6A and supplemental Figure 4A-B).
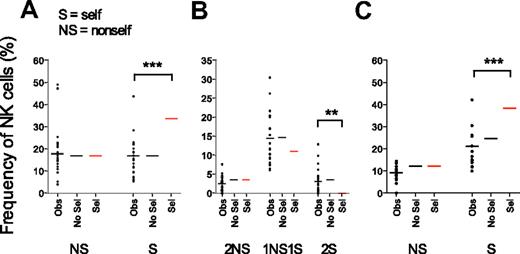
Expression of self- versus nonself-KIRs reflects a random combination of receptors. Observed frequencies of NK cells expressing nonself- (NS) and self- (S) KIRs were compared with theoretic values obtained by sequential acquisition of KIRs in the presence (Sel) or absence (No Sel) of selection by cognate ligands. (A) Frequencies of cells expressing one NS or one S KIR as their only KIR. To exclude any unintentional bias for expression of S- or NS-KIR, the analysis was restricted to individuals with 2 KIR ligands ensuring equal probabilities for both events. (B) Frequencies of NK cells expressing 2 NS, 1 NS and 1 S, or 2 S KIRs in individuals with 2 strong KIR ligands. (C) Frequencies of NK cells expressing 1 NS or 1 S KIR in individuals with 2 ligands when the KIR3DL2–HLA-A3/A11 interaction was excluded.
Expression of self- versus nonself-KIRs reflects a random combination of receptors. Observed frequencies of NK cells expressing nonself- (NS) and self- (S) KIRs were compared with theoretic values obtained by sequential acquisition of KIRs in the presence (Sel) or absence (No Sel) of selection by cognate ligands. (A) Frequencies of cells expressing one NS or one S KIR as their only KIR. To exclude any unintentional bias for expression of S- or NS-KIR, the analysis was restricted to individuals with 2 KIR ligands ensuring equal probabilities for both events. (B) Frequencies of NK cells expressing 2 NS, 1 NS and 1 S, or 2 S KIRs in individuals with 2 strong KIR ligands. (C) Frequencies of NK cells expressing 1 NS or 1 S KIR in individuals with 2 ligands when the KIR3DL2–HLA-A3/A11 interaction was excluded.
Similar comparisons for NK cells expressing 2 KIRs revealed that the frequency of NK cells coexpressing 1 self-KIR and 1 nonself-KIR was higher than those expressing either 2 self-KIRs or 2 nonself-KIRs (Figure 6B). However, the distribution of self- and nonself-KIRs was almost identical to that obtained when modeling KIR acquisition in the absence of selection (Figure 6B). The algorithm used for modeling KIR acquisition tested all possible combinations of self- and nonself-receptors revealing that the frequency of NK cells with 1 self- and 1 nonself-KIR was 4 times higher than the frequency of NK cells with 2 self- or 2 nonself-KIRs (Figure 6B). Indeed, there are 4 times as many possible ways to combine 1 self- and 1 nonself-KIR compared with 2 self- or 2 nonself-KIRs (see “Combinatorial analysis” for details). In contrast, repertoires formed under the sequential selection model differed from the observed values (Figure 6B).
Importantly, the results were similar when excluding KIR3DL2-HLA-A3/A11 as a functional inhibitory receptor-ligand pair (Figure 6C). Again, the observed values were in the range of those predicted for sequential KIR acquisition in the absence of selection but differed significantly from those predicted by the selection model. Thus, despite the observation of gradually increasing KIR acquisition probabilities, self- and nonself-KIRs were randomly distributed and there was no accumulation of self-specific receptors.
NKG2A expression is independent of the number of self-KIR ligands and buffers the functional response in repertoires characterized by low-KIR expression
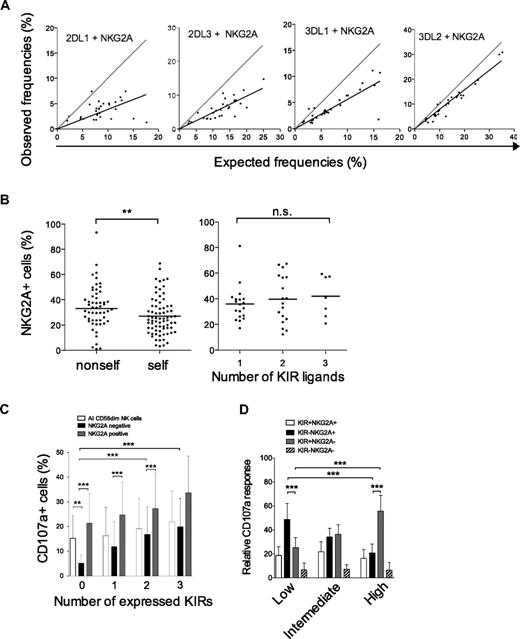
We then examined whether NKG2A expression was influenced by the presence of cognate ligands on individual NK cells and also in the total repertoire. In agreement with previous studies, we observed that coexpression of either KIR2DL1, KIR3DL1, or KIR2DL3 with NKG2A was less frequent than expected from the product rule (Figure 7A).10,18 Surprisingly, KIR3DL2 was coexpressed with NKG2A in a fashion that could nearly be calculated by the product rule. We recently showed that KIR3DL2 single-positive NK cells are hyporesponsive in HLA-A3/A11–positive individuals and that coexpression with NKG2A is associated with full functional competence.21 Therefore, we speculated that NKG2A expression is increased on hypofunctional, noneducated NK cells (ie, NK cells expressing nonself-KIRs). Indeed, we observed a small increase in the frequency of NKG2A expression on NK cells expressing a nonself-KIR (Figure 7B). However, the presence of strong KIR ligands had no effect on NKG2A expression in the total NK cell repertoire, since individuals with several ligands had no general decrease in NKG2A expression (Figure 7B).
NKG2A buffers NK cell function on individual NK cells and in KIR repertoires characterized by low-KIR expression. (A) The observed frequencies of NK cells coexpressing the indicated KIR with NKG2A are plotted against frequencies obtained by multiplying their individual expression frequency according to the product rule. A perfect fit to the product rule is shown as a gray line, representing a 1:1 relationship between the observed and expected frequencies. (B) (Left panel) The frequency of NKG2A expression on NK cells expressing one self- or nonself-KIR (KIR3DL2 excluded). Importantly, this analysis was restricted to NK cells expressing one single inhibitory KIR since NKG2A expression is less frequent on NK cells expressing multiple KIRs21 and those have a higher chance of expressing a self-KIR. (Right panel) Frequencies of NKG2A expression in individuals with 1 to 3 strong KIR ligands. (C) CD107a expression was monitored on NK cells expressing zero to 4 KIRs with and without NKG2A upon interaction with HLA class I–negative K562 cells. (D) Gates were set on CD107a+ cells to monitor the contribution of different subsets of cells to the total NK cell response in individuals with the 3 repertoires. The relative contribution of the indicated subsets to the overall NK cell response in the 3 types of KIR repertoires is shown.
NKG2A buffers NK cell function on individual NK cells and in KIR repertoires characterized by low-KIR expression. (A) The observed frequencies of NK cells coexpressing the indicated KIR with NKG2A are plotted against frequencies obtained by multiplying their individual expression frequency according to the product rule. A perfect fit to the product rule is shown as a gray line, representing a 1:1 relationship between the observed and expected frequencies. (B) (Left panel) The frequency of NKG2A expression on NK cells expressing one self- or nonself-KIR (KIR3DL2 excluded). Importantly, this analysis was restricted to NK cells expressing one single inhibitory KIR since NKG2A expression is less frequent on NK cells expressing multiple KIRs21 and those have a higher chance of expressing a self-KIR. (Right panel) Frequencies of NKG2A expression in individuals with 1 to 3 strong KIR ligands. (C) CD107a expression was monitored on NK cells expressing zero to 4 KIRs with and without NKG2A upon interaction with HLA class I–negative K562 cells. (D) Gates were set on CD107a+ cells to monitor the contribution of different subsets of cells to the total NK cell response in individuals with the 3 repertoires. The relative contribution of the indicated subsets to the overall NK cell response in the 3 types of KIR repertoires is shown.
Finally, we addressed how NKG2A modulated functional responses at the single-cell and at the population level. NK cells lacking KIRs were more dependent on NKG2A for their functional response than those positive for KIR (Figure 7C). Overall functional responses were independent of the number of strong KIR ligands and similar in the 3 identified KIR repertoires defined by high-, intermediate-, and low-KIR expression frequencies (data not shown). However, NK cell repertoires dominated by KIR-negative NK cells and with higher frequencies of NKG2A-positive cells (supplemental Figure 3) were dependent on KIR−NKG2A+ cells for their overall functional responsiveness (Figure 7D). This pattern gradually changed toward a KIR+NKG2A− dominated response for high-KIR repertoires.
Overall, these results illustrate how NKG2A may buffer functional responses in the individual cell, as well as on a population level in different KIR repertoires, in a manner that is largely independent of self-HLA class I ligands.
Discussion
The inhibitory KIR repertoire on human NK cells is highly variegated. We have analyzed this diversity in a cohort of 44 individuals homozygous for the group A KIR haplotype. Our results suggests that the variegated repertoires are generated through sequential and random acquisition of KIRs in a manner that is independent of the presence or absence of cognate HLA class I but, likely, determined by genetically hardwired KIR acquisition probabilities.
We examined to what extent self-HLA class I molecules select the human inhibitory KIR repertoire. At least 2 models describe how inhibitory NK cell receptor repertoires are shaped by interactions with cognate MHC class I molecules.12,29 The 2-step selection model postulates a random acquisition of KIRs followed by positive selection of NK cells expressing at least one inhibitory receptor to self-HLA class I and negative selection of cells that express too many such receptors. The sequential acquisition model postulates that NK cells sequentially express KIRs until a threshold for inhibition is reached, leading to the fixation of the KIR repertoire for that particular cell.30-32 In our analysis, 3 lines of evidence argue against these 2 models. First, KIR acquisition probabilities increased with the cellular expression of other KIRs as indicated by a significant deviation from the product rule in NK cells expressing multiple KIRs. This result supports a sequential acquisition of KIRs and is not compatible with an independent and simultaneous appearance of multiple KIRs as postulated by the 2-step selection model. Second, the frequency of NK cells expressing multiple inhibitory KIRs was independent of the number of cognate HLA class I ligands. These data disagree with predictions made by both former models of NK cell selection, that is, that donors with 3 and 4 KIR ligands should have fewer cells with multiple KIRs, since KIR acquisition should either have stopped (sequential model) or because such cells would be deleted (2-step selection model). Indeed, KIR repertoires modeled under the influence of selection by 1 to 4 cognate ligands differed from the observed repertoires and contained gradually increasing frequencies of NK cells with 1 KIR. Third, the observed frequencies of NK cells expressing self- and nonself-KIRs were near those predicted by a random sequential acquisition of KIRs but differed significantly from those predicted by the sequential selection model.
Interestingly, many of the desired properties of an NK cell repertoire, thought to require active selection, were fulfilled despite such random KIR acquisition. Since there are more ways to combine a self-KIR with a nonself-KIR than to combine multiple self- or nonself-receptors, the random combinatorial process results in NK cell repertoires that are dominated by NK cells expressing only one self-specific inhibitory KIR. This enables NK cells to sense the loss of single HLA alleles or their expression. Furthermore, the random distribution of self- and nonself-KIR on NK cells indicates that the increasing probability of KIR acquisition does not depend on other types of selection processes that control which type of receptor (self or nonself) that is expressed.
Our results are in agreement with a previous investigation showing that self-HLA ligands do not influence the overall KIR repertoire,17 but do not exclude the possibility that cognate HLA class I has a minor impact on the inhibitory KIR repertoire as described for KIR3DL1 and KIR2DL1.16 Addressing this question in a reliable way and excluding confounding selection events imposed by complementary KIR-HLA pairs requires larger numbers of studied individuals. Furthermore, such studies should be based on allele-level KIR genotyping since KIR polymorphism affects the level and frequency of KIR expression.16,18 However, as shown here, selection is not required to generate KIR repertoires that are dominated by NK cells expressing self-KIRs.
The increasing probability of KIR acquisition depending on the cellular expression of other receptors was described by Bayes rule for conditional probabilities. Key questions that still need to be resolved are how to determine the probability of acquiring the first KIR and whether the same KIR always appears first. Some insights have been provided by NK cell differentiation experiments performed in vitro indicating that KIR expression is genetically hardwired and that KIR2DL3 appears early, sequentially followed by KIR2DL1 or KIR3DL1 regardless of the HLA background.33,34 However, since most individuals have NK cells expressing any of the 4 major KIRs as their only KIR, it is most likely that KIR acquisition may occur in any order although some are more probable.
Here we assumed that the probability of acquiring the first KIR could be estimated by the average frequencies of KIR expression. Based on this assumption and Bayes rule, we were able to calculate conditional probabilities that reflected 3 types of KIR repertoires. Thus, formation of variegated KIR repertoires could be described by a model in which acquisition of individual KIRs is based on genetically predisposed expression probabilities, and in which KIR coexpression is randomly regulated, but characterized by gradually increasing probabilities of expression.
The molecular basis for the sequential increase in probabilities and individual set points for KIR acquisition remains unknown. KIR genes are epigenetically regulated with histone acetylation of the KIR locus followed by demethylation of the KIR promotors.35,36 Recently, models have been proposed to account for the regulation of KIR and Ly49 expression at the genetic level.37 These models imply that intergenic RNAs produced via probabilistic mechanisms control the selective activation or silencing of KIR alleles.37,38 Moreover, polymorphism in transcription factor binding sites was recently shown to affect the functional activity of bidirectional KIR promoters that are associated with distinct frequencies of receptor expression.39
Future experiments in which KIR promoter polymorphism is studied in donors with the 3 distinct KIR repertoires may provide a molecular explanation for the set points of KIR acquisition probabilities. Also relevant would be a detailed exploration on how monoallelic and biallelic expression of the same KIR fit into the proposed sequential acquisition of inhibitory KIR in a random but dependent manner. In one study on NK cell clones, biallelic expression of 2 alleles of 3DL1 was greater than expected from the product rule, possibly indicating that initial transcript at one allele increases the probability of expression at the second allele.40 Others have found that KIR genes have a largely monoallelic hypomethylation pattern indicating that most NK cells express only one allele of a given KIR gene.41
In this study, we have distinguished 3 types of KIR acquisition patterns with different overall KIR expression frequencies (high, intermediate, and low). The 3 defined KIR repertoires show partial overlap with those previously described by Yawata et al.18 The high-KIR repertoire corresponds roughly to the KIR-dominant type-I and type-III repertoires and the low-KIR repertoire resembles the NKG2A-dominant type-V repertoire.18 However, the inclusion of KIR3DL2 in our analysis precluded an exact comparison of the data sets. A potentially important difference between the 2 studies that requires further investigation was the observation by Yawata et al, that the number of strong KIR ligands sets the balance between KIR and NKG2A dominance.18 This outcome differs from our results, since NKG2A expression was found to be independent of the number of strong ligands in the host and high-KIR (KIR-dominant) and low-KIR (NKG2A-dominant) repertoires developed independently of cognate HLA class I ligands. Differences between the 2 studies may partly depend on different ethnicities and on the fact that the present study was restricted to individuals with the group A KIR haplotype.
There is accumulating evidence for functional modulation of NK cells through the interaction with MHC class I molecules.42 Data suggest that NK cell responses are quantitatively determined by the strength of inhibitory signals during NK cell education.18,43 NKG2A expression, correlating inversely with expression of KIRs,17,21 may endow NK cells with functional competence.18,21,44 It was recently proposed that NKG2A may buffer the overall responsiveness in distinct NK cell repertoires.18 In line with this, we have shown that NKG2A significantly contributes to functional responses on an individual cell level but also in repertoires characterized by low-KIR acquisition probabilities.
Our results support a model in which KIRs are sequentially acquired in a random but dependent manner. This study provides new perspectives for understanding how variegated KIR repertoires dominated by self-KIRs form in the absence of a selection process. Instead of shaping KIR and NKG2A expression patterns, the primary role of MHC class I is to balance the responsiveness of any given receptor repertoire.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all blood donors and the staff of the blood bank at Karolinska University Hospital, Stockholm.
This work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, the Swedish Cancer Society, the Swedish Children's Cancer Foundation, the Cancer Society of Stockholm, the Royal Swedish Academy of Sciences, the Tobias Foundation, the Söderberg foundation, The Belvén Foundation, the Åke Wiberg Foundation, and the Karolinska Institutet (all in Stockholm, Sweden).
Authorship
Contribution: S.A. and C.F. designed and performed experiments, and analyzed and interpreted data; J.-A.M. analyzed and interpreted data, designed and performed mathematical modeling, performed statistical analysis, and contributed to writing the paper; H.-G.L. interpreted data and contributed to writing the paper; and K.-J.M. designed the study, analyzed and interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karl-Johan Malmberg, Center for Infectious Medicine (CIM), F59, Department of Medicine, Karolinska Institutet, Karolinska University Hospital, 14186 Stockholm, Sweden; e-mail: kalle.malmberg@ki.se.
References
Author notes
*S.A., C.F., and J.-A.M. contributed equally to this work and are listed in alphabetic order.

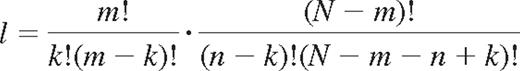
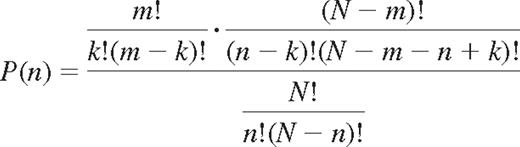
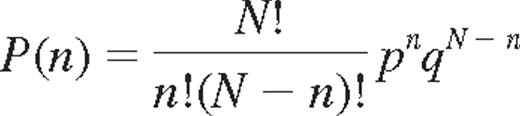
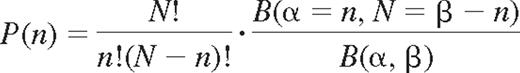
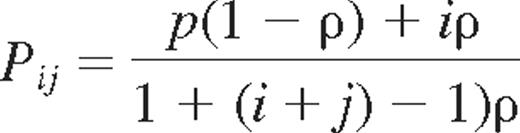
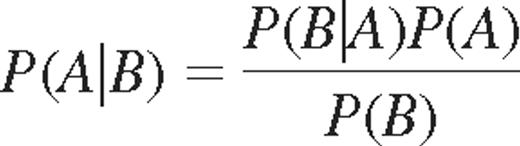
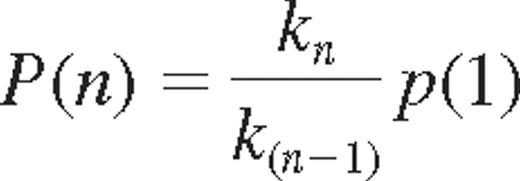
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal