Abstract
The molecular mechanisms that underlie the development of primitive myeloid cells in vertebrate embryos are not well understood. Here we characterize the role of cebpa during primitive myeloid cell development in Xenopus. We show that cebpa is one of the first known hematopoietic genes expressed in the embryo. Loss- and gain-of-function studies show that it is both necessary and sufficient for the development of functional myeloid cells. In addition, we show that cebpa misexpression leads to the precocious induction of myeloid cell markers in pluripotent prospective ectodermal cells, without the cells transitioning through a general mesodermal state. Finally, we use live imaging to show that cebpa-expressing cells exhibit many attributes of terminally differentiated myeloid cells, such as highly active migratory behavior, the ability to quickly and efficiently migrate toward wounds and phagocytose bacteria, and the ability to enter the circulation. Thus, C/EPBα is the first known single factor capable of initiating an entire myelopoiesis pathway in pluripotent cells in the embryo.
Introduction
Hematopoiesis occurs in 2 distinct phases during development.1-4 The first wave, also known as primitive hematopoiesis, often occurs in extraembryonic hemogenic sites and provides the embryos with a transient population of blood cells. The second wave gives rise to blood progenitors, which persist into late development and adulthood in successive hemogenic sites, and is therefore called definitive hematopoiesis. Whereas much is known about the molecular and cellular pathways responsible for definitive hematopoiesis, relatively little is known about the pathways responsible for the specification of the primitive blood lineages.
In the past decade, aquatic vertebrate species have emerged as powerful model organisms for the investigation of primitive and definitive hematopoiesis.5-8 Favored aquatic model organisms for hematopoiesis include the teleost fish, zebrafish, and the amphibian, Xenopus laevis, and its diploid relative, X tropicalis.9,10 Fish and frog embryos can be produced in large numbers, and they develop externally, allowing the visualization of the development and behavior of blood lineages in vivo. Because the embryos are large, embryologic manipulations, such as tissue transplantations, can be performed with relative ease. In addition, the molecular and genetic basis of hematopoiesis in these organisms is largely conserved with those of mammals.5-8
Studies in zebrafish have shown that primitive hematopoiesis consists of 2 separate events: primitive myelopoiesis and primitive erythropoiesis, which take place in 2 distinct hematopoietic compartments.11,12 Similarly in Xenopus, the primitive myeloid-forming compartment is located in the anterior ventral blood islands (aVBIs), which are derived from dorsal/anterior gastrula mesoderm, while primitive erythropoiesis occurs primarily in the posterior ventral blood islands, which are derived from ventral/posterior gastrula mesoderm.1,13-15
We recently showed that primitive myeloid cells are the first blood cells to differentiate and become functional in the Xenopus embryo.15 We also showed that these early differentiating myeloid cells are quickly and efficiently recruited to embryonic wounds before a functional vasculature is established.15 We are particularly interested in investigating the role of these primitive myeloid cells during tissue formation and repair in the embryo. Before addressing these questions, however, we sought to learn more about how these cells are specified. Given that many of the pathways responsible for definitive hematopoiesis are also used during primitive hematopoiesis, we have endeavored to address the role of C/EBPα, a member of CCAAT/enhancer binding protein family, during primitive myelopoiesis.
C/EPBα has long been recognized as an important factor during definitive myelopoiesis, where it is a key factor in driving common myeloid progenitors to differentiate into granulocyte-monocyte progenitors.16-18 Subsequently, C/EBPα has a recurrent role in driving myeloid progenitor cells toward terminal differentiation.16,17 In addition, mutations in C/EBPα are commonly found in patients with acute myeloid leukemias.19,20 In zebrafish, overexpression of a dominant interfering cebpa construct promotes primitive erythropoiesis, without a discernable effect on granulopoiesis.21
Here we characterize the function of cebpa during primitive myeloid cell development in Xenopus. We found that cebpa is one of the first known hematopoietic genes expressed in Xenopus embryos. Furthermore, gain- and loss-of-function studies show that cebpa is both necessary and sufficient for the differentiation of functional myeloid cells in the embryo. Finally, we show that cebpa can induce naive embryonic cells along the myelopoiesis pathway, resulting in precocious differentiation of functional myeloid cells.
Methods
Isolation of cebpa and construct design
A full-length clone encoding X tropicalis cebpa (TEgg058d02) was identified in the Gurdon Institute X tropicalis full-length database (http://informatics.gurdon.cam.ac.uk/online/xt-fl-db.html).22 HA-cebpa and pCS2 cebpa were generated by polymerase chain reaction (PCR) from TEgg058d02, using the following primers: 5′ GGA TCC ATG GAT CAA GCC AAC TTC 3′ and 5′ GAA TTC TTA TGC ACA GTT GCC C 3′. After digestion with EcoRI and BamHI, the PCR product was subcloned into both pFTX11 and pCS2 vector, and verified by sequencing.
Whole-mount in situ hybridizations
Whole-mount in situ hybridizations were performed according to established protocols.23 Probe synthesis for cebpa, spib, spi1, scl, mpo, fli1, lcp, mmp7, and globin was done according to Costa et al.15 In addition, probe synthesis for csf1r (colony stimulating factor 1 receptor) was done using IMAGE clone:7015756 (EcoRV, T7) and for gsc using clone TNeu077f20 (EcoRI, T7). For X-gal staining, fixed embryos were incubated in a solution containing 4 mM potassium hexacyanoferrate (III), 4 mM potassium hexacyanoferrate (II), 2 mM MgCl2, and 0.4 mg/mL X-gal at 37°C for 1 hour. The extent of migration was scored according to Tomlinson et al.24 Image acquisition was performed on a Leica MZAPO stereomicroscope using Northern Eclipse software (Empix Imaging Inc).
RNA synthesis and embryo microinjections
Capped mRNA was synthesized using the mMessage Machine kit according to the manufacturer's directions (Ambion, cebpa: SP6; HA-cebpa: T7). Embryos were cultured and injected with RNAs according to established protocols.23 Developmental staging was done according to Nieuwkoop and Faber.25 The same quantity of eGFP mRNA was injected as a control. Frog embryo experiments are covered by a project license issued by the Home Office in the United Kingdom
Morpholino design and microinjections
cebpa is an intronless gene and can be knocked down by injecting translational blocking morpholino oligonucleotides overlapping the start coding of the gene.26 MOs were purchased from Gene Tools LLC. X tropicalis embryos were injected with 10 ng total MOs before first cleavage, and generally a fluorescent control MO (Li std; Gene Tools) was injected as control. MO sequences are listed as follows:
cebpaATG 5′-CTCGTAGAAGTTGGCTTGATCCATG-3′
standard control MO 5′-CCTCTTACCTCAGTTACAATTTATA-3′
Immunoblotting
Synthetic HA-cebpa or cebpa mRNA (300 pg) was injected into X laevis embryos at 1-cell-stage, with or without 10 ng of cebpaATG MO. Embryos were harvested at stage 12 and homogenized in 1× RIPA lysis buffer with the addition of Complete protease inhibitor cocktail (Roche). The equivalent of one embryo was loaded and run on a 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel. After electrophoresis, proteins were transferred to PDVF membrane (Millipore) and probed with 1:1000 goat anti-CEBPA polyclonal antibody (C/EBPα (C-18):sc-9314, Santa Cruz) or 1:1000 mouse anti-HA HRP (clone 3F10, 1667475, Roche).
RNA isolation and real-time PCR
Total RNA was extracted from pools of embryos or animal caps using TRIzol reagent according to manufacturer's instructions (Invitrogen). cDNA was synthesized using BioScript reverse transcriptase (Bioline), and PCR reactions were performed using Taq polymerase (Roche) according to established protocols. Primer sequences are shown in the supplementary table (available on the Blood website; see the Supplemental Materials link at the top of the online article). The data for each sample were normalized relative to the expression level of ribosomal protein L8 (rpl8) and calculated by the 2−ΔCt method. Real-time RT-PCR analysis was performed using a Chromo4 Real-Time PCR Detector (Bio-Rad).
Animal cap assay and mesodermal inhibition
X laevis embryos were injected into the animal pole at the 2-cell stage, and animal cap explants were excised at stage 8. Caps were then incubated and cultured in 75% Normal Amphibian Medium (NAM) containing 0.2% bovine serum albumin with or without 10 μg/mL cycloheximide (Sigma) until the required stage.23 To block mesoderm formation, both an fibroblast growth factor inhibitor, 20 μM SU5402, and a nodal signaling inhibitor, 200 μM SB505124, were added into the medium from 2-cell stage. The same volume of dimethyl sulfoxide was added as control.
Animal cap transplantation and live imaging
HA-cebpa mRNA (200 pg) was coinjected with eGFP mRNA (200 pg) into the animal pole of X laevis embryos at the one-cell stage. As a control, only eGFP mRNA was injected. Animal caps were harvested using an eyebrow knife at stage 8. Meanwhile, synchronized unlabeled host embryos were prepared by removing a similar size piece of animal cap. The labeled animal caps were placed on the host embryos, such that the margins between the transplant and host embryo matched. Transplants were allowed to heal in 0.4× Marc Modified Ringers (MMR) for 1 hour, and then the chimeric embryos were transferred to 0.1× MMR for long-term culture.23 Wounds were created using forceps. A suspension of mCherry-positive bacteria in 1× PBS was injected for the phagocytosis experiments. Time-lapse fluorescent image acquisition was done using a Leica MZFLIII stereoscope and Northern Eclipse software (Empix Imaging Inc). Confocal imaging was done using an Olympus IX81 FluoView FV1000 microscope (Olympus Inc).
Results
Characterization of cebpa expression in X tropicalis embryos
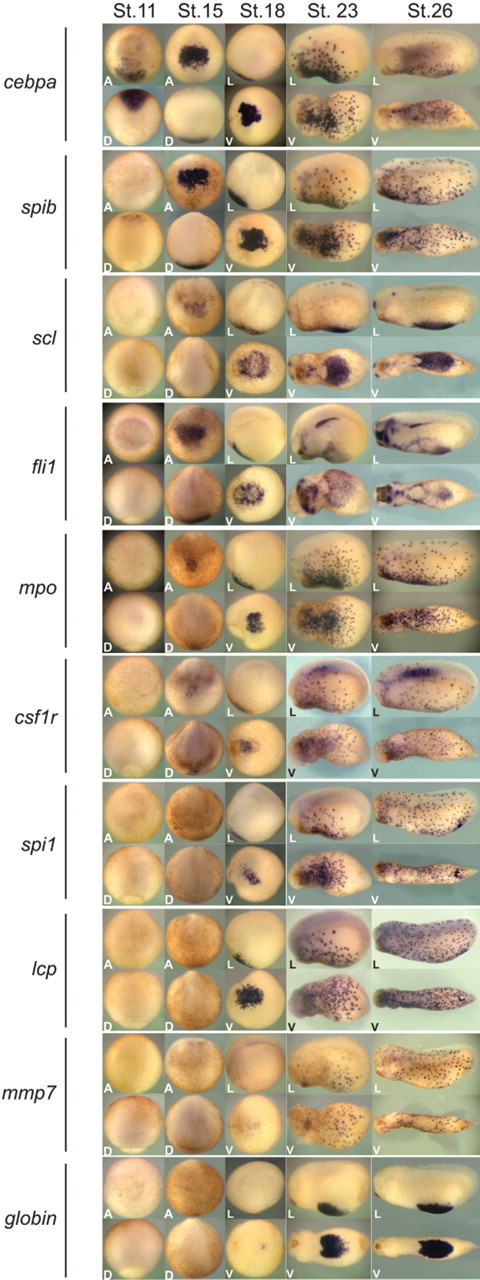
To better understand the molecular mechanisms of primitive hematopoiesis in X tropicalis, we identified a panel of hematopoietic genes from our X tropicalis expressed sequence tag database22 and analyzed their embryonic expression by whole-mount in situ hybridization (Figure 1). Whereas cebpa expression could be detected robustly at the mid-gastrula stage (Figure 1, stage 12; see supplemental Figure 1A-C, available on the Blood website; see the Supplemental Materials link at the top of the online article), the timing of expression of all the other early hematopoietic markers tested could not be seen until the neurula stages (stage 15; Figure 1). In the gastrula stage, the region that expressed cebpa corresponded to the anterior mesendoderm, marked by the expression of gsc (supplemental Figure 1D-F). This region of the gastrula is fated to give rise to the aVBI.1,13 During the neurula stages, cebpa expression was restricted to the aVBI (Figure 1, stages 15 and 18). At the early to mid-tailbud stages (Figure 1, stages 23 and 26, respectively), cebpa expression showed a punctate pattern, consistent with the initiation of migration of primitive myeloid cells throughout the embryo. The spatial expression pattern of cebpa during the neurula stage and tailbud stages resembled that seen by several primitive myeloid markers, including spib, mpo, mmp7, lcp (l-plastin), colony stimulating factor 1 receptor (csf1r), and spi1 (also known as PU.1), suggesting that cebpa is expressed in primitive myeloid cells (Figure 1). However, the whole-mount in situ hybridization data showed that cebpa was detectable considerably earlier than any other blood marker, including scl and fli1, early hemangioblast markers,27 or spib, an early marker of primitive myeloid cells15 (Figure 1). Consistent with this result, temporal RT-PCR analysis also showed that cebpa was expressed before all tested myeloid genes.15 These data suggested that cebpa may function very early in the hematopoietic gene regulatory cascade.
Expression pattern of hematopoietic genes in X tropicalis. Whole-mount in situ hybridization analysis of cebpa, spib, scl, fli1, mpo, csfr1, spi1, lcp, mmp7, and globin at stage 12, 15, 18, 23, and 26. Expression of cebpa was first detected at stage 12 in dorsal anterior mesoderm (and see supplemental Figure 1). From stage 15 until the end of neurulation, cebpa was expressed in the aVBI. Shortly after, cebpa-positive cells showed a punctate pattern throughout the embryo. A indicates anterior view; D, dorsal view; L, lateral view (anterior is to the left and dorsal to the top); and V, ventral view (anterior is to the left).
Expression pattern of hematopoietic genes in X tropicalis. Whole-mount in situ hybridization analysis of cebpa, spib, scl, fli1, mpo, csfr1, spi1, lcp, mmp7, and globin at stage 12, 15, 18, 23, and 26. Expression of cebpa was first detected at stage 12 in dorsal anterior mesoderm (and see supplemental Figure 1). From stage 15 until the end of neurulation, cebpa was expressed in the aVBI. Shortly after, cebpa-positive cells showed a punctate pattern throughout the embryo. A indicates anterior view; D, dorsal view; L, lateral view (anterior is to the left and dorsal to the top); and V, ventral view (anterior is to the left).
C/EBPα depletion down-regulates the expression of myeloid markers and inhibits terminal myeloid differentiation
The timing and pattern of expression of cebpa encouraged us to investigate its function during primitive hematopoiesis. We first determined the effect of knocking down cebpa function in embryos using antisense morpholino oligonucleotides (MOs). Given that the coding sequence of cebpa is not disrupted by introns, we designed a MO complementary to the start codon of cebpa, hereafter named cebpaATG. To test whether the cebpaATG MO could efficiently inhibit translation of cebpa, we coinjected in vitro transcribed cebpa mRNA (300 pg) with the cebpaATG MO (10 ng) into embryos and performed immunoblotting with a commercially available C/EBPα antibody (supplemental Figure 2). We found that coinjection of the cebpaATG MO completely inhibited the translation of cebpa. However, by adding an HA tag upstream of the start codon of cebpa, the cebpaATG MO was unable to inhibit translation. Thus we concluded that the cebpaATG MO was efficient in inhibiting the translation of wild-type cebpa but not the HA-cebpa construct.
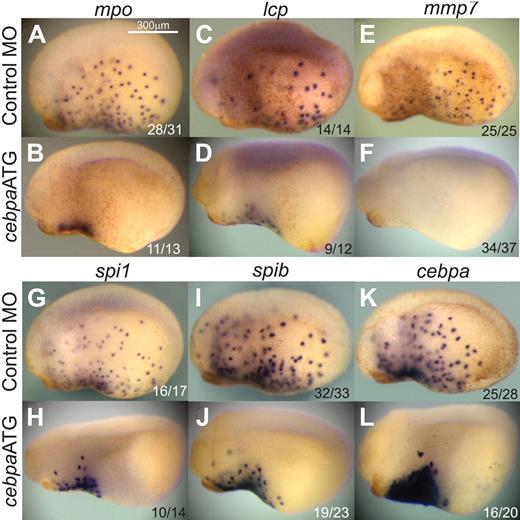
We then tested the effect of knocking down cebpa in X tropicalis embryos by assaying the expression of a large panel of primitive myeloid markers, including mpo, lcp, mmp7, spi1, spib, and cebpa (Figure 2). In control MO-injected mid-tailbud staged embryos (stage 23), cells expressing these markers have begun to migrate away from the aVBI and spread throughout the embryo (Figure 2A,C,E,G,I,K). However, the migration of these cells in cebpaATG MO-injected embryos was severely inhibited, suggesting that the cells were not differentiating properly into functional myeloid cells (Figure 2B,D,F,H,J,L). In addition, terminal differentiation markers, such as mpo, lcp, and mmp7 were significantly reduced or absent in the cebpaATG MO-injected embryos (Figure 2B,D,F). To quantify this effect, we performed real-time PCR analysis on mpo, lcp, mmp7, spi1, and spib (Figure 3C). With the exception of spib, all of these markers were significantly down-regulated (P < .05). However, hemangioblast markers, such as scl and fli, as well as the erythroid marker globin, were not affected by knocking down cebpa (supplemental Figure 3). Interestingly the expression level of endogenous cebpa was up-regulated in the cebpa morphants (Figure 2J), suggesting either that the endogenous mRNA is stabilized by the cebpaATG MO or that cebpa is involved in its own down-regulation.
cebpa loss-of-function phenotype. X tropicalis embryos injected with control MO (10 ng; A, C, E, G, I, K) or cebpaATG MO (10 ng; B, D, F, H, J, L), fixed at stage 23 and analyzed by whole mount in situ hybridization for the myeloid markers mpo (A, B), lcp (C, D), mmp7 (E, F), spi1 (G, H), spib (I, J), and cebpa (K, L). All images are lateral views, anterior to the left. The ratio of embryos showing the phenotype is shown in the bottom right corner of each image.
cebpa loss-of-function phenotype. X tropicalis embryos injected with control MO (10 ng; A, C, E, G, I, K) or cebpaATG MO (10 ng; B, D, F, H, J, L), fixed at stage 23 and analyzed by whole mount in situ hybridization for the myeloid markers mpo (A, B), lcp (C, D), mmp7 (E, F), spi1 (G, H), spib (I, J), and cebpa (K, L). All images are lateral views, anterior to the left. The ratio of embryos showing the phenotype is shown in the bottom right corner of each image.
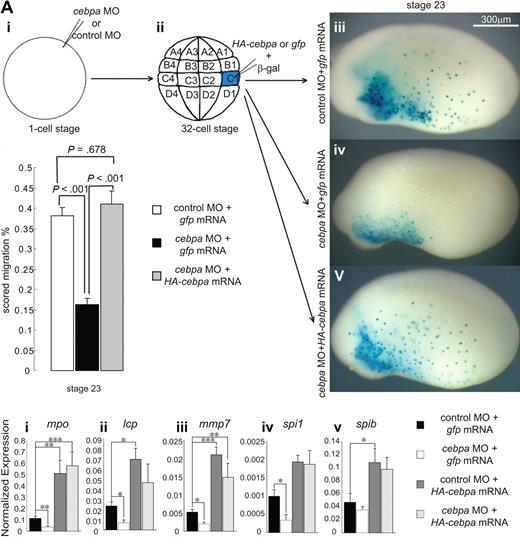
HA-cebpa mRNA rescued cebpa loss-of-function phenotype. (A) X tropicalis embryos injected with cebpaATG or control MO at one-cell stage (i) were allowed to develop to the 32-cell stage (ii), and then coinjected with HA-cebpa mRNA or eGFP mRNA and X-gal tracer mRNA into the 2 C1 blastomeres, which are fated to give rise to the aVBI. The embryos were fixed at stage 23 and stained for X-gal (iii-v) to assess the migration of the injected cells. (B) The extent of migration of X-gal+ cells was scored by placing a grid over the embryos and determining the percentage of the total area of each embryo that contained X-gal+ cells. Error bars represent SEM of 10 embryos. Statistical ANOVA was done using the SPSS software package. Compared with the control MO-injected embryos, the cebpaATG-injected morphants showed a significantly reduced extent of migration (P < .001), which was rescued by HA-cebpa mRNA to a level similar to that seen in the control embryos (P = .678). (C) Real-time PCR analysis on embryos at stage 23 injected with cebpaATG or control MO, coinjected with HA-cebpa mRNA or eGFP mRNA. Primitive myeloid markers mpo, lcp, mmp7, and spi1 were significantly reduced by cebpaATG MO (i-iv) and were rescued to normal or even higher levels by HA-cebpa. Expression levels were normalized relative to rpl8. Error bars represent SEM of 4 independent experiments. Statistical ANOVA was done using the SPSS software package. The experimental conditions that showed a significant difference in expression level relative to the control MO + gfp mRNA-injected embryos are labeled as *P < .05, **P < .01, or ***P < .001.
HA-cebpa mRNA rescued cebpa loss-of-function phenotype. (A) X tropicalis embryos injected with cebpaATG or control MO at one-cell stage (i) were allowed to develop to the 32-cell stage (ii), and then coinjected with HA-cebpa mRNA or eGFP mRNA and X-gal tracer mRNA into the 2 C1 blastomeres, which are fated to give rise to the aVBI. The embryos were fixed at stage 23 and stained for X-gal (iii-v) to assess the migration of the injected cells. (B) The extent of migration of X-gal+ cells was scored by placing a grid over the embryos and determining the percentage of the total area of each embryo that contained X-gal+ cells. Error bars represent SEM of 10 embryos. Statistical ANOVA was done using the SPSS software package. Compared with the control MO-injected embryos, the cebpaATG-injected morphants showed a significantly reduced extent of migration (P < .001), which was rescued by HA-cebpa mRNA to a level similar to that seen in the control embryos (P = .678). (C) Real-time PCR analysis on embryos at stage 23 injected with cebpaATG or control MO, coinjected with HA-cebpa mRNA or eGFP mRNA. Primitive myeloid markers mpo, lcp, mmp7, and spi1 were significantly reduced by cebpaATG MO (i-iv) and were rescued to normal or even higher levels by HA-cebpa. Expression levels were normalized relative to rpl8. Error bars represent SEM of 4 independent experiments. Statistical ANOVA was done using the SPSS software package. The experimental conditions that showed a significant difference in expression level relative to the control MO + gfp mRNA-injected embryos are labeled as *P < .05, **P < .01, or ***P < .001.
To address the specificity of the cebpaATG knock down, we asked whether the HA-cebpa construct, which we previously showed was not affected by the cebpaATG MO, could rescue the knock down effect of the MO, both in terms of migration and gene expression. To determine whether the HA-C/EBPα construct could rescue the migration defect, we injected either the control or experimental cebpaATG MOs at the one-cell stage (Figure 3Ai) and then we coinjected 20 pg of HA-cebpa or gfp mRNA with 50 pg of β-gal mRNA at the 32-cell stage, into the 2 C1 blastomeres, which give rise to aVBI (Figure 3Aii). When the embryos reached the mid-tailbud stage (stage 23), the embryos were fixed and stained with X-gal (Figure 3Aiii-v). Whereas control-injected embryos contained X-gal+ cells covering 40% of the embryo (Figure 3Aiii,B), the cebpaATG MO-injected embryos contained X-gal+ cells in less than 20% of the embryo (Figure 3Aiv,B), which is essentially the size of the aVBI. However, the cebpaATG MO-injected embryos rescued with the HA-cebpa construct contained X-gal stained cells in more than 40% of the embryo (Figure 3Av,B), suggesting that the HA-cebpa construct was able to fully restore the migratory behavior of the myeloid cells emanating from the aVBI.
We then determined whether the HA-C/EBPα could rescue the expression of myeloid markers in cebpaATG MO-injected embryos. As seen in Figure 3C, not only were all markers restored in the HA-cebpa–injected embryos, but all the myeloid markers were significantly up-regulated over their normal levels. Indeed, even spib, which was not significantly down-regulated by the cebpaATG MO, was up-regulated by the HA-cebpa mRNA (Figure 3Cv). These data suggested that the cebpa knock down phenotypes were specific and that overexpression of cebpa mRNA leads to an up-regulation of all myeloid marker genes tested.
Overexpression of cebpa induces ectopic expression of blood markers in X tropicalis embryos
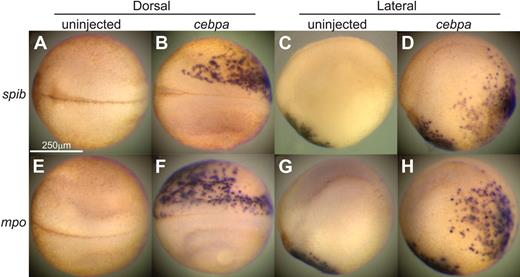
A potential gain-of-function phenotype was suggested by the morpholino rescue experiments, in which primitive myeloid markers were up-regulated by HA-cebpa overexpression. To further investigate the ability of C/EBPα to induce primitive hematopoiesis, we ectopically expressed cebpa in X tropicalis embryos by injecting cebpa mRNA randomly into a single cell at the 8-cell stage. Given that high levels of expression of cebpa can lead to embryonic death (data not shown), we injected a low amount of cebpa mRNA into embryos (10 pg per cell at the 8-cell stage), which resulted in an more than 95% survival rate. Under these conditions, C/EBPα caused extensive ectopic expression of several blood markers in a significant percentage of embryos (Figure 4 and supplemental Figure 4; spib: 54.2%, n = 35; mpo: 36.1%, n = 36; mmp7: 19.5%, n = 41; lcp: 31.2%, n = 32; fli1: 36.8%, n = 38; spi1: 18.9%, n = 37; and scl: 25.7%, n = 35). The normal expression of all these markers at stage 18 is localized to the aVBI (Figure 1). However, in embryos injected with cebpa mRNA, ectopic expression was detected in many regions of the embryo, including the neural folds, and lateral and posterior regions of the embryo. Thus, misexpression of cebpa can lead to a dramatic expansion in the expression of early hematopoietic and myeloid markers throughout the embryo. The expansion could be due to excessive proliferation and premature migration of existing pool of myeloid progenitors. Alternatively, this expansion of myeloid-expressing cells could be the result of an ectopic induction of myelopoiesis in the embryos.
cebpa gain-of-function phenotype. Overexpression of cebpa induced ectopic expression of blood markers on X tropicalis embryos. Embryos randomly injected with 10-pg cebpa mRNA into one cell at 8-cell stage were fixed at stage 18 for whole mount in situ hybridization for spib (A-D) and mpo (E-H). Panels A, B, E, and F are dorsal views (anterior to the left). Panels C, D, G, and H are lateral views (anterior to the left and dorsal to the top).
cebpa gain-of-function phenotype. Overexpression of cebpa induced ectopic expression of blood markers on X tropicalis embryos. Embryos randomly injected with 10-pg cebpa mRNA into one cell at 8-cell stage were fixed at stage 18 for whole mount in situ hybridization for spib (A-D) and mpo (E-H). Panels A, B, E, and F are dorsal views (anterior to the left). Panels C, D, G, and H are lateral views (anterior to the left and dorsal to the top).
cebpa induces the expression of blood markers in pluripotent cells
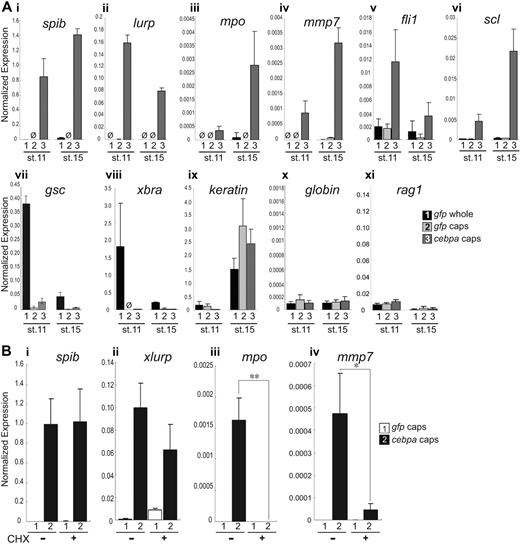
To address whether C/EBPα could induce ectopic expression of myeloid genes, we performed gain-of-function analyses in pluripotent embryonic cells isolated from early Xenopus embryos. Although normally fated to become epidermis, cells in the animal pole of blastula stage embryos (referred to as animal caps) are competent to differentiate into a broad range of tissue types, including neural, mesodermal or endodermal cell types, given the appropriate signals.28 We wondered whether C/EBPα could exert an effect on these pluripotent embryonic cells. When cebpa was misexpressed in animal caps, all blood markers tested, with the exception of globin and rag1, were significantly up-regulated (Figure 5A). Moreover, the induction of these hematopoietic genes occurred precociously, well before they would normally be expressed in the embryo. To further annotate these results, we divided all hematopoietic genes examined into 2 categories with respect to how quickly they responded to misexpression of cebpa. Early up-regulated genes included spib, lurp, and fli1; by the mid gastrula stage (stage 11), these genes had already been highly up-regulated in cebpa mRNA-injected animal caps (Figure 5Ai-ii,v). Late up-regulated genes included mpo, mmp7, and scl; these genes were dramatically up-regulated at the neurula stage (stage 15) but much less so at stage 11 (Figure 5Aiii-iv,vi). The erythroid marker, globin, and lymphoid marker, rag1,29 were not activated by cebpa misexpression (Figure 5Ax). Furthermore, neither the early liver marker, for1,30 nor the early lung marker, nkx2.1,31 were up-regulated precociously by cebpa (data not shown). Similarly, neither the early mesodermal markers, xbra and gsc, nor the ectodermal marker keratin were up-regulated in cebpa mRNA-injected animal caps (Figure 5Avii-ix), suggesting that cebpa specifically induced the early hematopoietic and myeloid genes.
cebpa induces blood markers in animal caps. (A) Injection of cebpa mRNA in the animal pole at one-cell stage induced precocious expression of all blood markers, with the exception of globin and rag1, in X laevis animal caps. eGFP mRNA was injected as control. Animal caps were excised at stage 8 and cultured in 0.2% bovine serum albumin, 75% NAM, until their sibling whole embryos reached stage 11 or 15. At stage 11, spib (i), lurp (ii), and fli1 (v) were strongly up-regulated, whereas mpo (iii), mmp7 (iv), and scl (vi) were strongly up-regulated at stage 15. In contrast, globin (x), rag1 (xi), and several nonhematopoietic markers, including gsc (vii), xbra (viii), and keratin (ix), were not induced. Ø: undetectable expression level. (B) The early induced genes, spib and lurp, were activated in the presence of CHX (i-ii), while the late induced genes mpo and mmp7 were not (iii-iv). Expression levels were normalized relative to rpl8. Error bars represent SEM of 3 independent experiments. Statistical ANOVA was done using the SPSS software package. Conditions that showed a significant difference in expression level comparing the cebpa-overexpressing animal caps with and without CHX are labeled *P < .01; **P < .001.
cebpa induces blood markers in animal caps. (A) Injection of cebpa mRNA in the animal pole at one-cell stage induced precocious expression of all blood markers, with the exception of globin and rag1, in X laevis animal caps. eGFP mRNA was injected as control. Animal caps were excised at stage 8 and cultured in 0.2% bovine serum albumin, 75% NAM, until their sibling whole embryos reached stage 11 or 15. At stage 11, spib (i), lurp (ii), and fli1 (v) were strongly up-regulated, whereas mpo (iii), mmp7 (iv), and scl (vi) were strongly up-regulated at stage 15. In contrast, globin (x), rag1 (xi), and several nonhematopoietic markers, including gsc (vii), xbra (viii), and keratin (ix), were not induced. Ø: undetectable expression level. (B) The early induced genes, spib and lurp, were activated in the presence of CHX (i-ii), while the late induced genes mpo and mmp7 were not (iii-iv). Expression levels were normalized relative to rpl8. Error bars represent SEM of 3 independent experiments. Statistical ANOVA was done using the SPSS software package. Conditions that showed a significant difference in expression level comparing the cebpa-overexpressing animal caps with and without CHX are labeled *P < .01; **P < .001.
To address whether some of these genes might be direct transcriptional targets of C/EBPα, we asked which genes could be activated in the absence of new protein synthesis. In Xenopus, zygotic transcription does not normally start until the mid-blastula transition, which occurs at stage 8.32 Therefore, we allowed translation of the cebpa mRNA after injection at the one-cell stage until stage 8. We then isolated the animal caps and tested for the activation of the marker with or without the protein synthesis inhibitor cycloheximide (CHX). From this experiment, we found that the early induced genes, spib and lurp, were activated in the presence of CHX, whereas the late-induced genes (mpo and mmp7) were not (Figure 5B). These results suggest that spib and lurp are direct targets of C/EPBα, whereas mpo and mmp7 are indirect targets of C/EPBα. We next scanned the promoter region of X tropicalis spib and lurp for putative C/EBP-binding sites. From this analysis, we found that the X tropicalis spib promoter contains a putative C/EBP-binding site at position −160 through −171 and the lurp promoter contains another at position −147 through −156 (data not shown). Interestingly, the murine Spib promoter also contains a putative C/EBP binding site in a conserved position.33
C/EBPα does not induce general mesodermal markers in animal caps
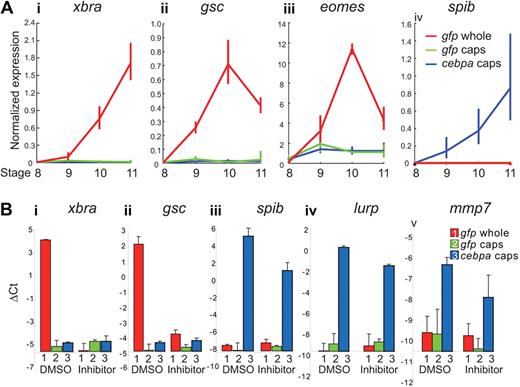
Our animal cap data suggest that cebpa overexpression is capable of driving pluripotent embryonic cells along the myelopoiesis pathway without the cells transitioning through an early general mesodermal state. However, given that cebpa induces myeloid markers precociously, we wondered whether the cells were passing through a very transient general mesodermal state, which had been missed in our experiments. To address this possibility, we assayed the expression of the early mesodermal markers xbra, gsc, and eomes by real-time PCR from the beginning of zygotic transcription (ie, stage 8; Figure 6A). We isolated animal caps from embryos at stage 8, and we analyzed the expression level of xbra, gsc and eomes at stages 8 through 11. The resulting real-time PCR data showed that cebpa did not induce the expression of the early mesodermal marker genes at any point in animal caps, although these genes could be detected robustly in whole embryos starting from stage 9 (Figure 6A). In contrast, an early myeloid marker, spib, was robustly induced in the cebpa overexpressing animal caps, starting from stage 9, even though spib expression was not detected in the whole embryos or the control animal caps at any stage between stages 8 through 11.
cebpa does not induce mesodermal markers in animal caps. (A) One-cell stage embryos were injected with either gfp or cebpa mRNA. Animal caps were excised, and RNA was collected when sibling embryos reached stage 8, 9, 10, and 11. Real-time PCR showed that xbra (i), gsc (ii), and eomes (iii) were highly up-regulated during gastrulation stage in the whole embryo (stage 8-11). However, these mesodermal marker genes were not induced in either the gfp or the cebpa mRNA-injected animal caps (i-iii). In contrast, the myeloid marker spib (iv) was rapidly up-regulated in cebpa-injected animal caps but remained undetectable in gfp-injected animal caps and whole embryos. (B) One-cell stage embryos were injected with either gfp or cebpa mRNA, and treated with dimethyl sulfoxide or a combination of 2 inhibitors (SU5402 and SB505124), which inhibit the FGF and nodal receptors, respectively, from the 2-cell stage onward. RNA was isolated from whole embryos and animal caps at stage 11. Real-time PCR analysis showed that cebpa induced the expression of the myeloid markers spib (iii), lurp (iv), and mmp7 (v), even when the combination of FGF and nodal inhibitors inhibited the expression of the early mesodermal genes xbra (i) and gsc (ii). Expression levels and ΔCt values were normalized relative to rpl8. Error bars represent SEM of 3 independent experiments.
cebpa does not induce mesodermal markers in animal caps. (A) One-cell stage embryos were injected with either gfp or cebpa mRNA. Animal caps were excised, and RNA was collected when sibling embryos reached stage 8, 9, 10, and 11. Real-time PCR showed that xbra (i), gsc (ii), and eomes (iii) were highly up-regulated during gastrulation stage in the whole embryo (stage 8-11). However, these mesodermal marker genes were not induced in either the gfp or the cebpa mRNA-injected animal caps (i-iii). In contrast, the myeloid marker spib (iv) was rapidly up-regulated in cebpa-injected animal caps but remained undetectable in gfp-injected animal caps and whole embryos. (B) One-cell stage embryos were injected with either gfp or cebpa mRNA, and treated with dimethyl sulfoxide or a combination of 2 inhibitors (SU5402 and SB505124), which inhibit the FGF and nodal receptors, respectively, from the 2-cell stage onward. RNA was isolated from whole embryos and animal caps at stage 11. Real-time PCR analysis showed that cebpa induced the expression of the myeloid markers spib (iii), lurp (iv), and mmp7 (v), even when the combination of FGF and nodal inhibitors inhibited the expression of the early mesodermal genes xbra (i) and gsc (ii). Expression levels and ΔCt values were normalized relative to rpl8. Error bars represent SEM of 3 independent experiments.
We next asked whether cebpa could induce myeloid markers under conditions where mesoderm formation is blocked using the fibroblast growth factor (FGF) receptor and nodal (ALK5) receptor inhibitors, SU5402 and SB505124, respectively.34,35 When we treated embryos with both inhibitors from the 2-cell stage, the embryos failed to express xbra and gsc at the gastrula stages (Figure 6Bi-ii), suggesting that these 2 inhibitors can completely block the formation of mesoderm. We then asked whether cebpa could induce myeloid markers in the presence of these 2 inhibitors. We found that the myeloid genes spib, lurp, and mmp7 were still induced in cebpa overexpressing animal caps at stage 11, even in the presence of both inhibitors (Figure 6Biii-v). Taken together, these data suggest that cebpa can induce primitive myeloid markers without the cells transitioning through an early mesodermal state.
Ectopic expression of C/EBPα induces an entire myelopoiesis program in pluripotent embryonic cells
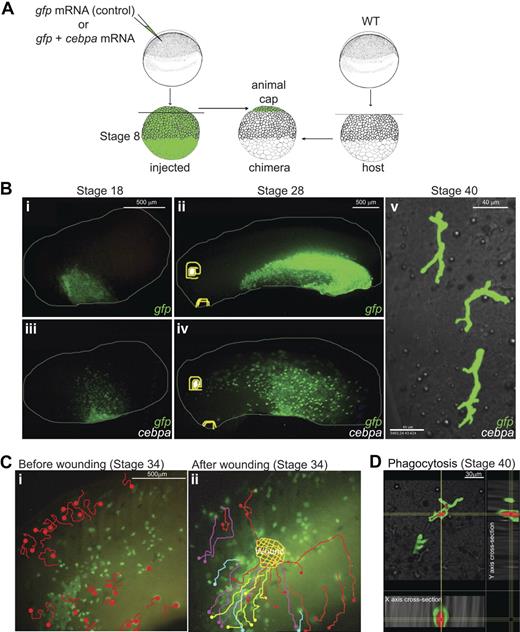
While the real-time PCR data were suggestive that C/EPBα might be initiating the hematopoietic regulatory cascade in the animal cap cells, we wanted to test whether the cells were becoming functional myeloid cells. To answer this question, we injected gfp mRNA into the animal pole of embryos with and without cebpa mRNA and performed animal cap transplantations into unlabeled host embryos (Figure 7A). By using live time-lapse fluorescence imaging, we were able to monitor the behavior of the graft-derived cells during later stages of development (Figure 7B). Whereas control eGFP labeled animal cap cells remained contiguous, underwent epiboly, and formed epidermis, eGFP-labeled animal cap cells expressing cebpa started to migrate away from the graft from the late gastrula stages (see supplemental Video 1). The cebpa expressing transplanted cells became migratory nearly a day before the endogenous primitive myeloid cells migrate away from the aVBI, consistent with the precocious up-regulation of blood markers seen in cebpa expressing animal cap explants (Figure 6). Whereas the boundaries between eGFP+ control transplants and the nonfluorescent host cells remained distinct throughout development (Figure 7Bi-ii), the cebpa expressing transplanted cells could be found in a spotty pattern throughout the embryo at the tailbud stages (Figure 7Biii-iv). We noted from these transplantation experiments that, whereas many C/EBPα/eGFP+ cells became migratory, some retained the morphology of epidermal cells, suggesting that not all C/EBPα/eGFP+ cells become myeloid cells. Later on, migratory cells showed ramified multipolar morphology (Figure 7Bv), characteristic of terminally differentiated macrophages.36 In addition, these cells displayed extremely active migratory behavior (see supplemental Video 2), characteristics of terminally differentiated primitive myeloid cells.15 Like terminally differentiated myeloid cells, a subset of these cells entered the circulation in the embryos (see supplemental Video 3).
Ectopic expression of C/EBPα induces myelopoiesis program in pluripotent embryonic cells. (A) Experimental setup: stage 8 animal caps were transplanted from cebpa and/or eGFP mRNA-injected embryos to unlabeled host embryos. Chimeras were recorded for live imaging (see supplemental Videos 1-9). (B) The boundaries between the eGFP+ transplants and nonfluorescent host embryos remain clear throughout embryonic development in the control eGFP transplants (i-ii). In contrast, blurred transplant margins and spotty pattern of eGFP+ cells were observed in the embryos containing cebpa expressing transplant cells (iii-iv). Confocal imaging at stage 40 showed that a subset of these scattered cells showed ramified morphology, reminiscent of macrophages (v). (C) Migration route of cebpa induced primitive myeloid cells before and after wounding. At stage 34, C/EBPα(+) graft derived migratory cells patrolled randomly before wounding (i, also see Video 4). Note that only a subset of GFP/CEBPα cells are migratory, whereas others retain the morphology of epidermal cells. After wounding, migratory GFP/CEBPα cells were immediately recruited to the wound site (ii; also see supplemental Video 5). Panels i and ii show the migratory paths of the cells before (i) and after (ii) wounding, as traced manually in supplemental Videos 4 and 5, respectively. Different colors were used to distinguish the migration routes of different cells. (D) Three-dimensional xyz confocal sections (20× objective, 1.0× zoom) of a macrophage (eGFP, green channel) that has phagocytosed bacteria (mCherry in red). Red bacteria are present in intracellular compartments surrounded by cytoplasm in the macrophages (green). For panels Bi-iv and C, images were obtained on a fluorescence stereoscope Leica MZ FLIII attached to a Sony CCD camera DXC-950 image capture system controlled by Northern Eclipse software 7.0 (Empix Image); 0.1× MMR was used as imaging medium. For panels B (v) and D, images were taken under Olympus IX81 FluoView FV1000 confocal microscope; 0.1× MMR, 2% methylcellulose (Sigma) 0.01% MS222 was used as imaging medium.
Ectopic expression of C/EBPα induces myelopoiesis program in pluripotent embryonic cells. (A) Experimental setup: stage 8 animal caps were transplanted from cebpa and/or eGFP mRNA-injected embryos to unlabeled host embryos. Chimeras were recorded for live imaging (see supplemental Videos 1-9). (B) The boundaries between the eGFP+ transplants and nonfluorescent host embryos remain clear throughout embryonic development in the control eGFP transplants (i-ii). In contrast, blurred transplant margins and spotty pattern of eGFP+ cells were observed in the embryos containing cebpa expressing transplant cells (iii-iv). Confocal imaging at stage 40 showed that a subset of these scattered cells showed ramified morphology, reminiscent of macrophages (v). (C) Migration route of cebpa induced primitive myeloid cells before and after wounding. At stage 34, C/EBPα(+) graft derived migratory cells patrolled randomly before wounding (i, also see Video 4). Note that only a subset of GFP/CEBPα cells are migratory, whereas others retain the morphology of epidermal cells. After wounding, migratory GFP/CEBPα cells were immediately recruited to the wound site (ii; also see supplemental Video 5). Panels i and ii show the migratory paths of the cells before (i) and after (ii) wounding, as traced manually in supplemental Videos 4 and 5, respectively. Different colors were used to distinguish the migration routes of different cells. (D) Three-dimensional xyz confocal sections (20× objective, 1.0× zoom) of a macrophage (eGFP, green channel) that has phagocytosed bacteria (mCherry in red). Red bacteria are present in intracellular compartments surrounded by cytoplasm in the macrophages (green). For panels Bi-iv and C, images were obtained on a fluorescence stereoscope Leica MZ FLIII attached to a Sony CCD camera DXC-950 image capture system controlled by Northern Eclipse software 7.0 (Empix Image); 0.1× MMR was used as imaging medium. For panels B (v) and D, images were taken under Olympus IX81 FluoView FV1000 confocal microscope; 0.1× MMR, 2% methylcellulose (Sigma) 0.01% MS222 was used as imaging medium.
We have previously shown that terminally differentiated primitive myeloid cells can be quickly and efficiently recruited to embryonic wounds.15 Therefore, we tested whether these animal cap-derived cells expressing C/EBPα and eGFP in chimeric embryos could respond to embryonic wounds. Before wounding, a subset of C/EBPα/eGFP+ cells patrolled the embryo randomly (see supplemental Video 4 and Figure 7C), but after wounding, these cells immediately responded by migrating toward the wound site (see supplemental Video 5 and Figure 7C). In contrast, control eGFP transplanted cells failed to respond to wounds (see supplemental Video 6).
We next asked whether cebpa expressing animal cap cells were capable of phagocytosis. We transplanted animal cap cells expressing either C/EBPα and eGFP or eGFP alone into unlabeled host embryos and allowed these chimeric embryos to develop into the tailbud and tadpole stages. We then injected mCherry expressing E coli into these chimeric embryos. As shown in supplementary supplemental Video 7, the transplanted C/EBPα/eGFP+ cells quickly migrated to the infection site and engulfed the red bacteria. Whereas the C/EBPα/eGFP+ cells can be visualized initially in the green channel only, after engulfing the bacteria, migratory myeloid cells became both green and red. Twenty-four hours after infection, the C/EBPα/eGFP+ cells, which have phagocytosed the red fluorescent bacteria, could be seen migrating throughout the embryo. We even detected these green/red myeloid cells reentering the circulation.
To obtain a better resolution of the morphology of the cells, we used time-lapse confocal microscopy. Using this method, we were able to follow the morphology and behavior of the C/EBPα/eGFP+ cells that had engulfed the red fluorescent bacteria (green/yellow cells in supplemental Video 8), C/EBPα/eGFP+ cells that had not engulfed bacteria (green cells in supplemental Video 8), and endogenous myeloid cells that had phagocytosed the fluorescent bacteria (red cells in supplemental Video 8). The ramified morphology of the C/EBPα/eGFP+ cells is reminiscent of macrophages grown on epithelial substrates.36 Analysis of the confocal images in 3 dimensions shows that the engulfed bacteria were present in intracellular compartments within the cytoplasm of the C/EBPα/eGFP+ macrophages (Figure 7D). Further analysis of the confocal videos, using Imaris software, allowed us to follow the morphology and behavior of the cells in 3 dimensions over time (see supplemental Video 9). This level of analysis not only allowed us to assess the changes in morphology of the cells over time but also the movement of the internalized bacteria within the intracellular compartments as the cells moved (see supplemental Video 9). In summary, these data show that C/EBPα is sufficient to precociously trigger an entire myelopoiesis pathway in naive pluripotent prospective ectodermal cells, resulting in terminally differentiated and fully functional primitive myeloid cells.
Discussion
Although much is known about the molecular and cellular pathways that underlie definitive myelopoiesis, relatively little is known about the mechanisms responsible for primitive myelopoiesis. In the mouse, the first hematopoietic cells arise from the blood islands in the yolk sac, soon after the completion of gastrulation.37 Whereas the main cell type that arises from the yolk sac is primitive erythroblasts, the yolk sac also contains a small population of mature macrophages, arising from the mother and from the yolk sac proper.38 Whereas the ontogeny of the yolk sac derived primitive myeloid cells has been investigated in the mammalian embryo, very little is known about the molecular events that lead their specification.39 Given the relative difficulty in studying this small population of cells in mammalian embryos, we have begun to study the molecular events that lead to the specification of primitive myeloid lineages in amphibian embryos.15
We have previously shown that primitive myeloid cells are the first blood cells to differentiate and become functional in the Xenopus embryo.15 Here we show that cebpa is one of the earliest genes to be activated in the hematopoietic pathway. Furthermore, we show that cebpa function is necessary for proper primitive myeloid cell differentiation. However, strikingly, we found that overexpression of C/EBPα was sufficient to initiate primitive myeloid development in pluripotent embryonic cells. Interestingly, C/EBPα initiated hematopoiesis without the cells transitioning through a general early mesodermal state, suggesting that C/EBPα can bypass the general mesodermal state and initiate myelopoiesis directly from pluripotent embryonic cells. Importantly, we found that C/EBPα was not only sufficient to induce a precocious expression of several hemotopoietic and myeloid genes, but C/EBPα was singularly capable of driving an entire primitive myelopoiesis pathway resulting in functional myeloid cells. To date, no other single factor has been described which is able to drive an entire myelopoiesis pathway in naive pluripotent cells.
Whereas overexpression of C/EBPα can induce precocious primitive myeloid development in pluripotent embryonic cells in Xenopus, knocking down cebpa in embryos did not completely prevent the initial specification of endogenous primitive myeloid progenitors. For example, spib, a gene known to be important for the specification of primitive myeloid cells,15 was not affected by knocking down cebpa, although overexpression of cebpa can potently induce this gene. Similarly, hemangioblast markers, scl and fli1, were not affected in the cebpa morphants but were induced by cebpa in animal caps and in embryos. These data suggest that, although cebpa is dispensable for the early specification of primitive myeloid progenitors, it can singularly initiate primitive myelopoiesis. There are several possible reasons for this apparent discrepancy. One possible reason is redundancy; experiments in mammals have suggested that members of the C/EBP family can functionally compensate for each other.40,41 For example, replacing C/EBPβ into the C/EBPα locus can completely restore hematopoiesis.41 Indeed, cebpb and other members of the C/EBP family appear to be expressed in early X tropicalis embryos, as suggested by the presence of expressed sequence tags for these genes in early embryonic cDNA libraries (http://informatics.gurdon.cam.ac.uk/online/xt-fl-db.html). Thus, cebpb or other C/EPBs could be acting in a partially redundant fashion with cebpa. Second, we have noted that, although cebpa is expressed in the anterior mesendoderm in gastrula embryos, this gene is also expressed maternally. Therefore, there could be some compensation in the knock down embryos from maternally provided protein. Finally, although cebpa can singularly initiate primitive myelopoiesis in the animal cap, in the embryo cebpa may normally act in combination with other factors to promote hematopoiesis. It will be important in the future to determine which other factors function in combination with C/EBPα to drive primitive myelopoiesis in the embryo.
Currently, a large amount of effort has been placed toward identifying factors that can drive pluripotent embryonic stem cells toward specific developmental fates. Recently C/EBPα has been shown to be a potent factor, which can induce either transdifferentiation or dedifferentiation, depending on the experimental regime. For example, forced expression of C/EBPα in lymphoid cells can induce transdifferentiation into myeloid cells in a PU.1-dependent manner.42,43 Furthermore, C/EBPα, in combination with PU.1, is able to convert fibroblasts into macrophage-like cells.44 Finally, C/EBPα was recently shown to help reprogram terminally differentiated B cells toward a pluripotent stem cell state in combination with Oct4, Sox2, Klf4, and c-Myc.45 Here we extend these findings by showing that C/EBPα alone can initiate and drive an entire primitive myelopoiesis program in pluripotent embryonic cells in Xenopus. In future studies, it will be important to determine whether C/EBPα can induce myelopoiesis in human or mouse embryonic stem cells. After all, when designing strategies to generate defined cell types for therapeutic purposes, single factors, such as C/EBPα, which can initiate an entire developmental program from pluripotent cells is of particular interest.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Karel Dorey and Kimberly Mace for comments on the manuscript.
This work was supported by 2 project grants (E.A.) and a studentship (Y.C.) from The Healing Foundation.
Authorship
Contribution: Y.C. performed and analyzed most of the experiments, prepared the figures, and cowrote the manuscript; R.M.B.C. and M.R. cloned and performed the initial expression analysis on cebpa; R.M.B.C also assisted in the animal cap transplants, the phagocytosis experiments, and confocal imaging; N.L. and X.S. assisted in the real-time PCR analyses; R.P. identified the putative C/EBP binding sites in the spib and lurp promoters; and E.A. assisted in the animal cap transplantations, guided the project, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Enrique Amaya, The Healing Foundation Centre, Michael Smith Bldg, Faculty of Life Sciences, University of Manchester, Oxford Rd, Manchester, M13 9PT, United Kingdom; e-mail: enrique.amaya@manchester.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal