Abstract
Severe deficiency of ADAMTS13, a plasma metalloprotease, leads to thrombotic thrombocytopenic purpura. ADAMTS13 contains 10 putative N-glycosylation sites in or near its metalloprotease sequence, spacer region, thrombospondin type 1 repeat no. 4 (TSR no. 4), and CUB domains. Tunicamycin treatment markedly decreased the secretion of ADAMTS13 into the culture medium of transfected cells. Nevertheless, the protease was efficiently secreted from N-acetylglucosaminyltransferase I–deficient Lec1 Chinese hamster ovary cells, indicating that N-glycosylation in the endoplasmic reticulum, but not the conversion of oligomannose to complex N-glycans in the Golgi complex, is important for secretion. However, ADAMTS13 with oligomannose N-glycans cleaved its substrate, von Willebrand factor (VWF) multimers, less effectively, with a higher Km but similar kcat value. In mutagenesis analysis, decreased secretion and VWF cleaving activity was observed with the N146Q and N828Q mutants, while decreased secretion only was observed with the N552Q mutant of ADAMTS13. Enzymatic removal of N-glycans from ADAMTS13 did not affect its VWF cleaving activity. Thus, N-glycosylation is necessary for efficient secretion of ADAMTS13, while conversion of the N-glycans from oligomannose to complex type in the Golgi complex enhances the proteolytic activity of the protease toward VWF multimers. After its secretion, ADAMTS13 does not require N-glycans for its VWF cleaving activity.
Introduction
ADAMTS13, a member of the ADAMTS (a disintegrin and metalloprotease with thrombospondin type 1 repeat) metalloprotease family, cleaves von Willebrand factor (VWF) when this platelet-aggregating protein becomes conformationally unfolded by shear stress in the circulation or in vitro by chaotropic agents such as guanidine chloride or urea.1 This proteolysis decreases the adhesive activity of VWF, thereby preventing unwarranted platelet aggregation in the microvasculature. Severe deficiency of ADAMTS13, due to genetic mutations or autoimmune inhibitory antibodies, leads to thrombotic thrombocytopenic purpura (TTP), a serious disorder with characteristic arteriolar and capillary thrombosis and extremely high risk of death if not properly treated with plasma therapy to replenish the protease
ADAMTS13 protein consists of a series of domains, including a signal peptide, a propeptide, a metalloprotease domain, a disintergrin domain, a thrombospondin type 1 repeat (TSR), a cysteine-rich region, a spacer domain, 7 additional TSRs (nos. 2-8), and 2 CUB (complement component Clr/Cls, Uegf, bone morphogenetic protein 1) domains.1 The mature ADAMTS13 protein consists of 1353 amino acid residues but has a molecular weight of 190 kDa as determined by either gel filtration or sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), substantially higher than the expected 145 kDa. This large difference suggests that ADAMTS13 is highly glycosylated. In glycoproteins, the glycans attached to the polypeptide backbone via covalent N- or O-linkage may regulate structure and biologic functions. Glycosylation may promote appropriate folding, assembly, or targeting of nascent proteins and/or their biologic activity.2-4 A recent study demonstrates that the N-glycans of VWF have a modulatory effect on its interaction with ADAMTS13.5 Separately, ADAMTS13 has been shown to be O-fucosylated in at least 6 TSR repeats.6 O-fucosylation is required for secretion of the protease. In this study, we have investigated the role of N-glycans in the secretion and VWF cleaving activity of ADAMTS13.
Methods
Materials
Alpha-modified Eagle medium, fetal bovine serum, Dulbecco modified Eagle medium (DMEM), and Lipofectamine 2000 were from Invitrogen (Carlsbad, CA); Centricon YM-30, from Millipore (Bedford, MA); recombinant peptide N-glycanase F (PNGase F), from Prozyme/Glyco (San Leandro, CA); endoglycosidase H, from Roche Applied Science (Indianapolis, IN); FRETS-VWF73, from Peptides International (Louisville, KY); Sephadex G-100 beads, from GE Bio-Sciences (Piscataway, NJ); monoclonal anti-V5 antibody, from AbD Serotec (Raleigh, NC); horseradish peroxidase (HRP)–conjugated goat anti–mouse IgG, HRP-conjugated goat anti–rabbit IgG and chemiluminescence reagents, from Pierce (Rockford, IL); polyclonal rabbit anti–human VWF, from DAKO North America (Carpinteria, CA); and tunicamycin, from Sigma-Aldrich (St Louis, MO).
Plasmid constructs and site-directed mutagenesis
pcDNA3.1/V5-His plasmids for expression of human ADAMTS13 or its truncated forms with C-terminal V5 and His6 tags were constructed as described previously.7 Mutations of specific amino acids were introduced into ADAMTS13 by polymerase chain reaction site-directed mutagenesis. The mutant constructs were confirmed by sequencing on both strands across the mutated site.
Cell cultures and transfections
The Pro-5 Chinese hamster ovary (CHO) parent cell line and the CHO Pro-5 Lec1.3C mutant, which lacks N-acetylglucosaminyltransferase I (GlcNAc-T-I) activity and is termed Lec1 in this paper, were isolated previously.8,9 Both lines were grown on 10-cm plates in alpha-modified Eagle medium with 10% (vol/vol) fetal bovine serum in 5% (vol/vol) CO2 at 37°C. HEK 293T cells were maintained in DMEM as previously described.7 Subconfluent cells were transiently transfected in plates with ADAMTS13 plasmids using Lipofectamine 2000.7 To suppress N-glycosylation, HEK 293T cells were treated with 2 μg/mL tunicamycin 24 hours before transfection was performed and maintained in serum-free culture medium containing the same concentration of tunicamycin. The conditioned serum-free media were collected at 72 hours and concentrated 30-fold on a Centricon YM-30 filter. The cells were collected in an equal volume of SDS-PAGE sample buffer and stored at −70°C.
Deglycosylation, SDS-PAGE, and immunoblotting
N-glycans were removed from proteins using recombinant PNGase F or endoglycosidase H. Digestion with PNGase F was performed overnight at 37°C in the presence of 0.05 mM EDTA (ethylenediaminetetraacetic acid), 0.1% SDS, and 50 mM β-mercaptoethanol according to the manufacturer's recommendation. In experiments involving analysis of VWF cleaving activity after the digestion step, the incubation was performed in the absence of EDTA, SDS, and β-mercaptoethanol. In some experiments, PNGase F was subsequently removed by chromatography over a Sephadex G-100 column. Digestion of ADAMTS13 by endoglycosidase H was performed overnight at 37°C in a 0.1 M phosphate buffer, pH 6.0, containing 2 mM EDTA, 0.1 g/mL SDS, and 50 mM β-mercaptoethanol. Recombinant ADAMTS13 proteins were separated by SDS-PAGE and visualized by immunoblotting with monoclonal anti-V5 antibody, horseradish peroxidase (HRP)–conjugated goat anti–mouse IgG, and chemiluminescence as previously described.7 The molar concentrations of recombinant ADAMTS13 proteins were estimated by comparing their respective optical densities against a reference curve constructed from serial dilutions of an AD5 protein (consisting of residues 1-745 of ADAMTS13; Figure 1), whose concentration had previously been calibrated.10 Percentage of protein secretion was defined as the concentration in the culture medium divided by the concentrations in the culture medium and cell lysate multiplied by 100%. To compare the efficiency of ADAMTS13 protein secretion, all the protein secretion percentages were normalized against 100% for the mean of the wild-type (WT) ADAMTS13.
Measurement of ADAMTS13 activity
Measurement of ADAMTS13 proteolytic activity toward VWF multimers was based on the generation of a dimeric 176-kDa fragment resulting from cleavage of VWF at the Y1605-M1606 bond as previously described.7 Briefly, VWF multimers isolated from normal human plasma and dissolved in 1.5 M guanidine chloride were incubated at a ratio of 1 to 9 (vol/vol) with protease samples at 37°C. The reaction was terminated at 60 minutes with 4 volumes of SDS-PAGE sample buffer containing 5 mM EDTA. The proteolytic fragments of VWF were analyzed in 5% SDS-PAGE under nonreducing conditions, followed by immunoblotting with polyclonal rabbit anti–human VWF, horseradish peroxidase–conjugated goat anti–rabbit IgG, and chemiluminescence. The activity level of an ADAMTS13 protease, represented by the optic density of the dimeric 176-kDa fragment intrapolated against a reference curve constructed from serial dilutions of normal human plasma (NHP, pooled from 10 healthy volunteer donors), was expressed in U/nmol protease, with the amount of activity in 1 mL NHP defined as 1 unit (U). For comparison among the recombinant proteases, the ADAMTS13 activity values were depicted as a percentage of the activity level in the WT ADAMTS13.
To measure the values of Michaelis-Menten constant (Km) and catalytic rate constant (kcat) of a protease, reaction of a protease with varying concentrations of VWF multimer substrate was stopped at designated time points. The level of the dimeric 176-kDa VWF fragment in the mixtures was determined by SDS-PAGE and immunoblotting as in the ADAMTS13 activity assay. The velocity of fragment increase was fitted to the Michaelis-Menten equation as previously described.10 Because the VWF fragment level was not measured in molar concentration, this analysis yielded equivalent Km and kcat values that are suitable only for intra-assay comparison. Consequently, the kinetic values were depicted as a percentage of the corresponding values obtained with the WT ADAMTS13 protein. Because the size of VWF multimers affected the proteolytic response, all comparisons were based on measurements using the same lot of VWF and analyzed on the same blots. The kinetic analysis was also performed as previously described against FRETS-VWF73 (Peptides International, Louisville, KY) as the substrate at varying concentrations.10 The reaction velocity was determined from the increase of fluorescent intensity measured at intervals of 5 minutes for up to 60 minutes.
Statistics
All results are expressed as mean plus or minus standard deviation (SD) unless otherwise indicated. The difference among experimental groups was analyzed by analysis of variance (ANOVA) and Dunnett posttest against the indicated controls. Statistical and curve fitting analyses were performed using Prism (GraphPad Software, La Jolla, CA). Difference between 2 proteases in Km or kcat was determined using extra sum-of-squares F test.
Results
ADAMTS13 contains complex N-glycans
Analysis using artificial neural networks (http://www.cbs.dtu.dk/services/NetNGlyc/) that examine the sequence context of the N-X-S/T sequons revealed that full-length ADAMTS13 contains 8 N-X-S/T sequons with N-glycosylation potential above the threshold of 0.5. Two of these sequons (N142 and N146) are in the metalloprotease domain, 5 (N552, N579, N614, N667, and N707) are in or near the spacer domain, and 1 (N828) is in the TSR no. 4 domain (Figure 1A). Two additional candidate residues (N1235 and N1354) present in the CUB domains have glycosylation potentials slightly less than the threshold of 0.5.
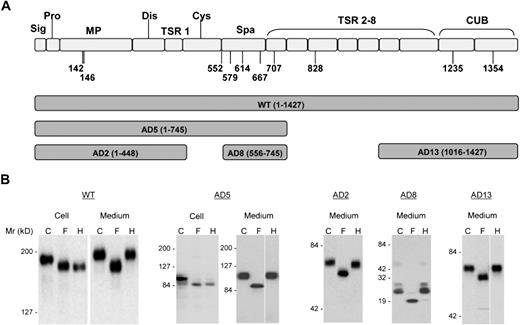
Expression of ADAMTS13 and its truncation variants in HEK 293T cells. (A) The predicted domain structure of ADAMTS13 (WT) and the range of amino acid sequences encompassed in the 5 truncated constructs (AD2, AD5, AD8, and AD13) used in this study. (B) Expressed ADAMTS13 and its truncation variants in either cell lysate (Cell) and/or concentrated culture medium (Medium) were digested with PNGase F (F) or endoglycosidase H (H) and analyzed by SDS-PAGE and immunoblotting with anti-V5. C indicates control proteins not digested with either enzyme. The numbers to the left of the gels indicate the position of molecular size markers in kilodaltons. The cell lysate data are not shown for AD2, AD8, and AD13. The space in AD5 and AD13 indicates repositioned gel lanes.
Expression of ADAMTS13 and its truncation variants in HEK 293T cells. (A) The predicted domain structure of ADAMTS13 (WT) and the range of amino acid sequences encompassed in the 5 truncated constructs (AD2, AD5, AD8, and AD13) used in this study. (B) Expressed ADAMTS13 and its truncation variants in either cell lysate (Cell) and/or concentrated culture medium (Medium) were digested with PNGase F (F) or endoglycosidase H (H) and analyzed by SDS-PAGE and immunoblotting with anti-V5. C indicates control proteins not digested with either enzyme. The numbers to the left of the gels indicate the position of molecular size markers in kilodaltons. The cell lysate data are not shown for AD2, AD8, and AD13. The space in AD5 and AD13 indicates repositioned gel lanes.
To determine whether ADAMTS13 contains N-glycans in these regions, we treated recombinant WT ADAMTS13 (residues 1-1427) and 4 truncated forms, AD2 (residues 1-448), AD5 (residues 1-745), AD8 (residues 556-745), and AD13 (residues 1016-1427) with PNGase F. SDS-PAGE analysis revealed that all forms in both cell lysates and medium concentrates were reduced in molecular weight by PNGase F (Figure 1B). Therefore, all forms of ADAMTS13 carried N-glycans of the complex or oligomannose type. To distinguish between these types of N-glycans, the ADAMTS13 proteins were treated with endoglycosidase H, which cleaves only oligomannose N-glycans. AD5 and WT ADAMTS13 in cell lysates were digested by endoglycosidase H to the same size generated by PNGase F digestion, indicating that intracellular AD5 and WT ADAMTS13 contained predominantly oligomannose N-glycans and were primarily located in the endoplasmic reticulum or cis Golgi compartment. Both forms of ADAMTS13 were converted to complex N-glycans before secretion, as shown by the resistance of the proteins in culture medium to digestion with endoglycosidase H (Figure 1B). The other forms of ADAMTS13 (AD2, AD8, and AD13) were also secreted with complex N-glycans sensitive to removal by PNGase F but not by endoglycosidase H (Figure 1B), as would be expected from the results with AD5 and WT. Taken together, these findings suggest that complex N-glycans are present in and near the metalloprotease, spacer, TSR no. 4, and CUB domains of the recombinant ADAMTS13. Analysis of 8 N-glycan site mutants (N142Q, N146Q, N552Q, N579Q, N614Q, N667Q, N707Q, and N828Q) in HEK 293T secretion by SDS-PAGE under reducing conditions showed that each N to Q mutation decreased the molecular weight of ADAMTS13 by approximately 5 kDa (Figure S1; available on the Blood website; see the Supplemental Materials link at the top of the online article), suggesting that each of the N residues was indeed glycosylated in the WT ADAMTS13.
N-glycosylation is necessary for efficient secretion of ADAMTS13
To determine whether N-glycosylation is essential for the secretion of ADAMTS13, the recombinant glycoproteins were expressed in the presence of tunicamycin, which blocks N-glycosylation by inhibiting the activity of dolichol-P:N-aceytlglucosamine phosphotransferase. SDS-PAGE gel analysis shows that in the presence of tunicamycin, WT ADAMTS13, AD5, or AD2 were synthesized at lower levels and had lower molecular weights (Figure 2A). Furthermore, no secreted forms were detected in the medium of tunicamycin-treated cells, indicating that tunicamycin profoundly decreased the secretion of these proteins. The suppressive effect of tunicamycin on the secretion of AD2 suggests that one or both of the N-glycans in the metalloprotease domain may be critical for effective ADAMTS13 secretion.
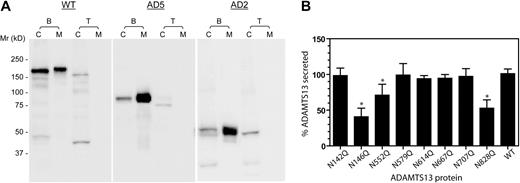
N-glycosylation affects the secretion of ADAMTS13. (A) Tunicamycin suppresses the secretion of ADAMTS13. HEK 293T cells were transfected in the absence (B) or presence (T) of tunicamycin. Cell lysates (C) and concentrated culture media (M) were collected 72 hours after transfection and analyzed at equal volumes by SDS-PAGE and immunoblotting with anti-V5. The numbers to the left of the gels indicate the position of molecular weight markers in kilodaltons. (B) The effect of N to Q substitution on the secretion of ADAMTS13. Transfection of wild-type ADAMTS13 (WT) cDNA or N-glycosylation site mutants was performed in the absence of tunicamycin. Each percent ADAMTS13 secretion was determined from the concentration of ADAMTS13 in the culture medium, divided by the sum of protease concentration in the culture medium and cell lysate. The values were normalized against 100% for the mean of the WT ADAMTS13 secretion. Mean and SD values shown on the y-axis were based on at least 5 batches of transfections. *P < .01 by ANOVA with Dunnett postanalysis against the WT ADAMTS13.
N-glycosylation affects the secretion of ADAMTS13. (A) Tunicamycin suppresses the secretion of ADAMTS13. HEK 293T cells were transfected in the absence (B) or presence (T) of tunicamycin. Cell lysates (C) and concentrated culture media (M) were collected 72 hours after transfection and analyzed at equal volumes by SDS-PAGE and immunoblotting with anti-V5. The numbers to the left of the gels indicate the position of molecular weight markers in kilodaltons. (B) The effect of N to Q substitution on the secretion of ADAMTS13. Transfection of wild-type ADAMTS13 (WT) cDNA or N-glycosylation site mutants was performed in the absence of tunicamycin. Each percent ADAMTS13 secretion was determined from the concentration of ADAMTS13 in the culture medium, divided by the sum of protease concentration in the culture medium and cell lysate. The values were normalized against 100% for the mean of the WT ADAMTS13 secretion. Mean and SD values shown on the y-axis were based on at least 5 batches of transfections. *P < .01 by ANOVA with Dunnett postanalysis against the WT ADAMTS13.
To determine whether any of the asparagine (N) residues or its glycosylation plays a role in the effective secretion of ADAMTS13, we substituted each of the 8 N residues in or near the metalloprotease or spacer domains with glutamine (Q), and compared the secretion of the mutants with that of the WT ADAMTS13. This analysis revealed that, compared with the WT ADAMTS13, 3 (N146Q, N552Q, and N828Q) mutants were secreted somewhat less efficiently (Figure 2B; P < .01). Nevertheless, individual N residue substitution alone was not sufficient to reproduce the profound decrease of secretion observed with tunicamycin, suggesting that more than 1 N-glycosylation contributes to the efficient secretion of ADAMTS13.
ADAMTS13 with oligomannose N-glycans are efficiently secreted but are less active
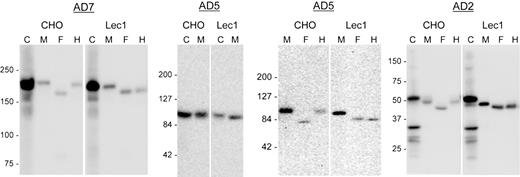
To determine whether conversion of N-glycans from the oligomannose to complex type affects the secretion or biologic function of ADAMTS13, we expressed full-length ADAMTS13 (WT) and its truncated variants (AD2 and AD5) in N-acetylglucosaminyltransferase I–deficient cells (Lec1 CHO mutant). Lec1 cells do not convert oligomannose N-glycans to complex N-glycans due to an inactivating mutation in the Mgat1 gene.11 The efficiency of secretion of AD2, AD5, and WT ADAMTS13 from Lec1 cells was compared with that from parent CHO cells. As illustrated in Figure 3, there was no apparent difference in the efficiency of secretion from CHO versus Lec1 cells. As expected, the proteins secreted from Lec1 cells were equally sensitive to digestion with both PNGase F and endoglycosidase H, and gave forms of the same molecular weight, confirming the presence of solely oligomannose N-glycans on the proteins secreted from Lec1 cells.
Lec1 cells secreted ADAMTS13 proteins with oligomannose N-glycans. ADAMTS13 (WT) and its truncated variants, AD5 and AD2, were expressed in CHO or the Lec1 mutant cells (Lec1). Cell lysates (C) and serum-free culture media (M) were collected for SDS-PAGE and immunoblotting for V5 tag 72 hours after transfection. Each secreted protein was also digested with PNGase F (F) or endoglycosidase H (H) before analysis. The numbers to the left of each gel indicate the position of the molecular weight markers in kilodaltons. Smaller molecular weight bands in the cell lysates of WT and AD2 might have resulted from proteolysis of expressed protein.
Lec1 cells secreted ADAMTS13 proteins with oligomannose N-glycans. ADAMTS13 (WT) and its truncated variants, AD5 and AD2, were expressed in CHO or the Lec1 mutant cells (Lec1). Cell lysates (C) and serum-free culture media (M) were collected for SDS-PAGE and immunoblotting for V5 tag 72 hours after transfection. Each secreted protein was also digested with PNGase F (F) or endoglycosidase H (H) before analysis. The numbers to the left of each gel indicate the position of the molecular weight markers in kilodaltons. Smaller molecular weight bands in the cell lysates of WT and AD2 might have resulted from proteolysis of expressed protein.
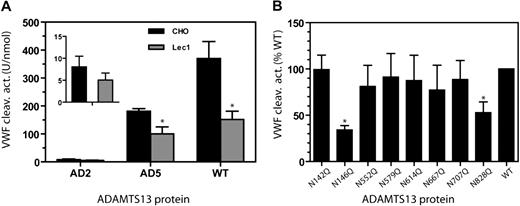
The enzymatic activity of AD2, AD5, and WT ADAMTS13 was analyzed using VWF multimers as the substrate. As shown in Figure 4A, WT and AD5 in secretions from Lec1 cells were less effective at cleaving VWF multimers, exhibiting 30% to 50% of the WT activity. This lower activity was consistently observed whether the assay incubation reaction was terminated at 3, 10, or 20 minutes (data not shown) instead of the usual 60 minutes. AD2 expressed in Lec1 cells also exhibited a trend toward lower proteolytic activity. Nevertheless, because of the low activity of AD2, this difference did not reach statistical significance.
N-glycans affect the VWF cleaving activity of ADAMTS13. (A) The mean VWF cleaving activity (± SD) from 4 measurements is depicted for WT ADAMTS13, AD5, and AD2 in CHO or Lec1 cell secretion. The insert is a magnification of the AD2 results. *P < .05 by Student t test. (B) VWF cleaving activity of WT ADAMTS13 and 8 N-residue mutants. ADAMTS13 proteins were from secretion of HEK 293T cells. For each mutant, the VWF cleaving activity level per nanomole protein is expressed as a percentage of WT ADAMTS13. *P < .01 by ANOVA with Dunnett posttest against the WT ADAMTS13 (N = 4-6).
N-glycans affect the VWF cleaving activity of ADAMTS13. (A) The mean VWF cleaving activity (± SD) from 4 measurements is depicted for WT ADAMTS13, AD5, and AD2 in CHO or Lec1 cell secretion. The insert is a magnification of the AD2 results. *P < .05 by Student t test. (B) VWF cleaving activity of WT ADAMTS13 and 8 N-residue mutants. ADAMTS13 proteins were from secretion of HEK 293T cells. For each mutant, the VWF cleaving activity level per nanomole protein is expressed as a percentage of WT ADAMTS13. *P < .01 by ANOVA with Dunnett posttest against the WT ADAMTS13 (N = 4-6).
The lower VWF cleaving activity observed with both WT/Lec1 and AD5/Lec1 suggested that the N-glycans in the metalloprotease-spacer-TSR no. 4 region contribute to the proteolytic activity of ADAMTS13. To further explore this possibility, we analyzed the VWF cleaving activity of ADAMTS13 with substitution at 1 of the 8 N residues in this region (Figure 4B). The VWF multimer cleaving activity, in U/nmol protein for each protease derived from HEK 293T cell secretion, was normalized to 100% for the WT ADAMTS13. The results revealed that the N146Q and N828Q mutants were 30% to 50% as active as wild-type ADAMTS13 in cleaving VWF multimers (P < .05). None of the other mutants showed a significantly diminished VWF cleaving activity.
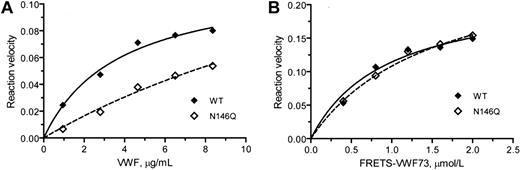
Michaelis-Menten analysis of the VWF cleaving reaction using a previously described procedure10 showed that the lower proteolytic activity of the N146Q mutant was primarily due to a higher Km (641% of the WT, P < .05) while the kcat was not significantly different (86% of WT; Figure 5A). No such difference was observed between WT and N146Q mutant when FRETS-VWF73 was used as the substrate (Figure 5B). Similar results were obtained with the N828Q mutant (data not shown).
N146Q mutant is less effective than WT ADAMTS13 in cleaving VWF multimers but not in cleaving FRETS-VWF73. (A) Kinetic analysis of VWF multimer cleavage by WT ADAMTS13 (concentration 0.87 nmol/L) or N146Q mutant (concentration 1.08 nmol/L). (B) Kinetic analysis of FRETS-VWF cleavage by WT ADAMTS13 (0.2 nmol/L) or N146Q mutant (0.18 nmol/L). The reaction velocity (WT:♦; N146Q: ◇), based on the increase of the dimeric 176-kDa fragment level on immunoblots against a reference curve constructed from serial dilutions of normal human plasma for panel A and on the increase of fluorescence intensity for panel B, was curve-fitted to Michaelis-Menten equation (solid and dashed lines) to obtain the equivalent values of Km and kcat. Because neither the reaction velocity nor the unit of VWF multimers was measured in molar concentrations, this analysis yielded kinetic value equivalents that were suitable only for intra-assay comparison. The difference between N146Q and WT ADAMTS13 against VWF multimers was significant (<.05) for Km but not for kcat. No significant difference was detected in either Km or kcat against FRETS-VWF73. Similar results were obtained in a repeated analysis.
N146Q mutant is less effective than WT ADAMTS13 in cleaving VWF multimers but not in cleaving FRETS-VWF73. (A) Kinetic analysis of VWF multimer cleavage by WT ADAMTS13 (concentration 0.87 nmol/L) or N146Q mutant (concentration 1.08 nmol/L). (B) Kinetic analysis of FRETS-VWF cleavage by WT ADAMTS13 (0.2 nmol/L) or N146Q mutant (0.18 nmol/L). The reaction velocity (WT:♦; N146Q: ◇), based on the increase of the dimeric 176-kDa fragment level on immunoblots against a reference curve constructed from serial dilutions of normal human plasma for panel A and on the increase of fluorescence intensity for panel B, was curve-fitted to Michaelis-Menten equation (solid and dashed lines) to obtain the equivalent values of Km and kcat. Because neither the reaction velocity nor the unit of VWF multimers was measured in molar concentrations, this analysis yielded kinetic value equivalents that were suitable only for intra-assay comparison. The difference between N146Q and WT ADAMTS13 against VWF multimers was significant (<.05) for Km but not for kcat. No significant difference was detected in either Km or kcat against FRETS-VWF73. Similar results were obtained in a repeated analysis.
Similar kinetic analysis revealed that the Km of the full-length ADAMTS13 in Lec1 cell secretion was 525% compared with the protease in CHO cell secretion, while the kcat was not significantly different (83% of WT). The oligomannose variant of ADAMTS13 from Lec1 cell secretion exhibited no difference from the complex type ADAMTS13 from WT CHO cell secretion in either Km (116% of the WT) or kcat (122% of the WT) when FRETS-VWF73 was used as the substrate.
Presence of N-glycans is not necessary for the VWF cleaving activity of ADAMTS13
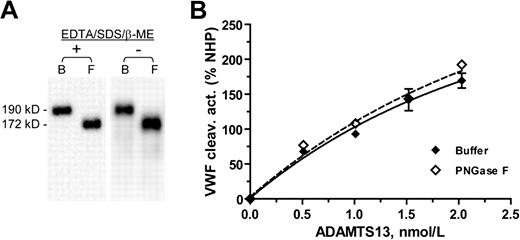
To determine whether the N-glycans are necessary for the VWF cleaving activity of ADAMTS13, we treated WT ADAMTS13 with PNGase F in the absence of EDTA, SDS, and β-mercaptoethanol. SDS-PAGE analysis of the digested ADAMTS13 showed that this modified incubation led to same removal of the N-glycans as the original incubation (Figure 6A). The VWF cleaving activity analysis shows that N-deglycosylation by PNGase F does not affect the VWF cleaving activity of ADAMTS13 (Figure 6B). In the VWF cleaving assay, the VWF fragments generated from cleavage by PNGase F treated ADAMTS13 or its buffer control were not different in molecular size, indicating that PNGase F in the reaction mixtures did not cause N-deglycosylation of the VWF substrate during the assay (data not shown).
Deglycosylation of ADAMTS13 by PNGase F does not affect its VWF cleaving activity. (A) The molecular weight of the WT ADAMTS13 in HEK 293T cell–secretion decreased from 190 kDa in the buffer control (B) to 172 kDa after overnight incubation at 37°C with PNGase F in the presence (+) or absence (−) of EDTA, SDS, and β-mercaptoethanol (β-ME). (B) VWF cleaving activity, expressed in percent of NHP, of the ADAMTS13 protein assayed at varying concentrations after incubation with buffer (♦) or PNGase F (◇) in the absence of EDTA/SDS/β-ME. NHP: normal human plasma.
Deglycosylation of ADAMTS13 by PNGase F does not affect its VWF cleaving activity. (A) The molecular weight of the WT ADAMTS13 in HEK 293T cell–secretion decreased from 190 kDa in the buffer control (B) to 172 kDa after overnight incubation at 37°C with PNGase F in the presence (+) or absence (−) of EDTA, SDS, and β-mercaptoethanol (β-ME). (B) VWF cleaving activity, expressed in percent of NHP, of the ADAMTS13 protein assayed at varying concentrations after incubation with buffer (♦) or PNGase F (◇) in the absence of EDTA/SDS/β-ME. NHP: normal human plasma.
Discussion
ADAMTS13 is a highly glycosylated plasma protease, with N- and O-glycans accounting for approximately 20% of its molecular mass. Recent work by Ricketts et al6 demonstrated that O-fucosylation of the TSR domains is important for secretion of ADAMTS13. In this study, we found that N-glycosylation is also critical for secretion of the protease, because no ADAMTS13 proteases were detected in culture media of cells treated with tunicamycin. On the other hand, the data from N-glycan site mutants of ADAMTS13 suggest that individually, each N-glycan has minimal or moderate impact on the efficiency of secretion. Thus, the dramatic impact of tunicamycin on ADAMTS13 secretion reflects the abolition of more than one N-glycans. A similar phenomenon was observed with O-fucosylation of the TSP repeats.6 Both N-glycan addition and O-fucosylation occur cotranslationally. Early glycosylation may promote ADAMTS13 secretion by ensuring correct protein folding, disulfide bonding, or other processing steps in the endoplasmic reticulum.
In the expression experiments, the intracellular pool of ADAMTS13 consisted essentially of oligomannose N-glycans in the endoplasmic reticulum or cis Golgi. Thus, the endoplasmic reticulum (ER) folding and/or Golgi translocation and early trafficking are likely to be major determinants of ADAMTS13 secretion. In contrast, ADAMTS13 proteases were secreted efficiently from N-acetylglucosaminyltransferase I–deficient Lec1 cells, suggesting that ADAMTS13 exits from the endoplasmic reticulum in a secretion-competent form, and that further processing of oligomannose N-glycans to the complex type in the Golgi is not necessary for secretion.
Previous studies of hereditary TTP have shown that the most common defect of ADAMTS13 mutations is impaired secretion, with retention of mutant proteases in the intracellular compartments.12-15 Although none of the known patient mutations affect the N-glycosylation sites, we speculate that exit block from the ER may involve a common defect in the signal for secretion. Thus, further investigation of the factors regulating the exit of ADAMTS13 from the endoplasmic reticulum may have important implications for understanding how ADAMTS13 mutations lead to secretion defects.
The predicted N-glycosylation sites of ADAMTS13 are clustered in the central part of the protease, with 5 of them (N552, N579, N614, N667, and N707) located within a 156-residue region of the spacer-TSR no. 2 domain. One additional site is located slightly downstream in the TSR no. 4 domain. Interestingly, previous studies using chemically or shear stress–unfolded VWF have shown that while truncation of the protease distal to the spacer domain decreases the VWF cleaving activity by approximately 20% to 60%, the residual protease activity is decreased to less than 2% (but not nil) when the truncation is upstream of the spacer domain.7,16 Thus, the spacer domain and, to a lesser extent, the downstream TSR-CUB domains are involved in the expression of VWF cleaving activity. The clustering of N-glycan sites in the spacer-TSR no. 2 and TSR no. 4 regions suggested that the N-glycans might affect the proteolytic activity of ADAMTS13. Our analysis of the proteases secreted by Lec1 cells shows that ADAMTS13 with oligomannose side chains are only 30% to 50% as active as the WT, confirming that N-glycans are functionally important for expression of VWF cleaving activity of the protease. Kinetic analysis suggests that the decrease in proteolytic activity of the oligomannose ADAMTS13 results primarily from higher Km, suggesting that conversion of N-glycans from oligomannose to complex type promotes the binding between the protease and VWF. Separately, in solid-phase binding analysis we found that in the presence of EDTA, the Lec1 variant of ADAMTS13 was not different from the WT protease in binding VWF (data not shown). This discrepancy indicates that ADAMTS13-VWF binding data obtained under noncatalytic conditions may not be applied to interpret functional ADAMTS13-VWF interaction.
Surprisingly, our analysis using the N-glycan site mutants reveals that the N residues located in the spacer-TSR no. 2 domains are not important for expression of proteolytic activity. Instead, N146 in the metalloprotease domain and N828 in the TSR no. 4 are important for VWF cleaving activity. Notably, both mutants, as well as N552Q, also exhibited secretion defect. Kinetic analysis shows that the lower VWF cleaving activity of the N146Q or N828Q ADAMTS13 mutant results primarily from a higher Km. Interestingly, the N142 residue located 4 residues upstream from N146 does not affect the secretion or proteolytic activity of ADAMTS13. It should be noted that studies using N-glycan site mutants do not distinguish between the effect resulting from loss of N-glycosylation and the effect directly from amino acid substitution.
Further activity analysis of the N-deglycosylated ADAMTS13 shows that removal of N-glycans by PNGase F did not affect the VWF cleaving activity of ADAMTS13. Thus, glycosylation at the N-residues most likely contributes to the ADAMTS13 activity by promoting correct protein folding or disulfide bonding for optimal binding with its VWF substrate. The N-glycans are not needed for VWF cleaving activity once the protease is correctly folded and secreted. In contrast, removal of N-glycans from VWF makes the substrate susceptible to cleavage by ADAMTS13.5
The lower proteolytic activity of the N146Q and N828Q mutants and the oligomannose variant ADAMTS13 in Lec1 cell secretion was not reproduced when FRETS-VWF73 was used as the substrate. In a previous report, we observed a similar phenomenon of differential response to the substrates: compared with full-length ADAMTS13, truncated ADAMTS13 variants (ie, AD5 and AD2) exhibited lower binding affinity (ie, higher Km) for VWF multimers.10 This difference in Km between full-length ADAMTS13 and its truncated variants markedly decreased when the proteases were assayed against FRETS-VWF73. Taken together, the data of these 2 studies suggest that N-glycans and the domains downstream of the catalytic site are important for protease-VWF multimer interaction but are less critical when the substrate is an abbreviated VWF peptide. We speculate that to interact effectively with VWF multimers, ADAMTS13 requires a stringent conformation. N-glycosylation and the presence of downstream domains of ADAMTS13 contribute to correct protein folding that is necessary for ADAMTS13 to assume this active conformation. For interaction with short peptide substrate, correct folding of ADAMTS13 is less important.
Analysis of the truncated variants of ADAMTS13 suggests that the recombinant ADAMTS13 is N-glycosylated at the metalloprotease, spacer, TSR no. 4 and CUB domains. Interpretation of the truncation variant data may be hampered by uncertainty over the impact of truncation mutation on the efficiency of N-glycosylation. Nevertheless, SDS-PAGE analysis demonstrates that the each N glycosylation site mutation in these domains decreases the molecular weight of the full-length ADAMTS13 by approximately 5 kDa. Together, these observations suggest that all 8 N residues are glycosylated in the recombinant ADAMTS13. The 2 potential N-glycosylation sites in the CUB domain will be investigated in future studies.
It is well recognized that the efficiency of N-glycosylation and the structures of the glycans may be affected by the type of cells synthesizing the protein. Consequently, the ADAMTS13 in plasma, which is believed to derive primarily from hepatic stellate cells17,18 and perhaps other types of cells,19-21 may be different from recombinant ADAMTS13 proteins in the fine structure of N-glycans. Nevertheless, we find plasma ADAMTS13 and recombinant ADAMTS13 have the same size by SDS-PAGE, and both are similarly decreased in size after N-deglycosylation (data not shown). Because the SDS-PAGE analysis has the resolution power to detect molecular weight difference resulting from the loss of a single N-glycosylation residue (Figure S1), our data suggest that plasma and recombinant ADAMTS13 most likely have the same number of N-glycosylated residues.
In conclusion, N-glycosylation in the ER is a critical step for efficient secretion of ADAMTS13. In contrast, the lower proteolytic activity observed with the oligomannose-type ADAMTS13 in Lec1 cell secretion suggests that conversion of oligomannose N-glycans to the complex type promotes the transformation of ADAMTS13 to a proteolytically more active conformation. N-glycosylation may contribute to VWF cleaving activity of ADAMTS13 by promoting correct protein folding or disulfide bonding, creating a conformation of ADAMTS13 optimal for interaction with VWF multimers. Nevertheless, once secreted, continued presence of N-glycans is not essential for ADAMTS13 interaction with VWF. N-glycosylation is less important for cleavage of FRETS-VWF73, a short VWF peptide substrate, perhaps because the interaction between ADAMTS13 and the short substrate requires less stringent conformation of the protease. Future studies are needed to determine the precise profile and structure of N-glycans in the plasma ADAMTS13 and the role of N-glycans in the conformational maturation of ADAMTS13 during its biosynthesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Pamela Stanley of the Albert Einstein College of Medicine (NY) for kindly providing the CHO Lec1 mutant cells used in this study and for her in-depth review and helpful comments on the manuscript, and Dr Robert Haltiwanger of the State University of New York at Stony Brook for his insightful comments.
This study was supported in part by a grant (R01HL62136) to H.-M.T. from the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH).
National Institutes of Health
Authorship
Contribution: W.Z. and H.-M.T. contributed to conception, design, execution, data interpretation, and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Han-Mou Tsai, MD, Penn State Milton S. Hershey Medical Center, Section of Hemostasis and Thrombosis, Division of Hematology/Oncology, 500 University Drive, Hershey, PA 17033; e-mail: htsai@hmc.psu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal