Abstract
Mice with total and constitutive iron regulatory protein 2 (IRP2) deficiency exhibit microcytosis and altered body iron distribution with duodenal and hepatic iron loading and decreased iron levels in splenic macrophages. To explore cell-autonomous and systemic context-dependent functions of IRP2 and to assess the systemic consequences of local IRP2 deficiency, we applied Cre/Lox technology to specifically ablate IRP2 in enterocytes, hepatocytes, or macrophages, respectively. This study reveals that the hepatic and duodenal manifestations of systemic IRP2 deficiency are largely explained by cell-autonomous functions of IRP2. By contrast, IRP2-deficient macrophages from otherwise IRP2-sufficient mice do not display the abnormalities of macrophages from systemically IRP2-deficient animals, suggesting that these result from IRP2 disruption in other cell type(s). Mice with enterocyte-, hepatocyte-, or macrophage-specific IRP2 deficiency display normal red blood cell and plasma iron parameters, supporting the notion that the microcytosis in IRP2-deficient mice likely reflects an intrinsic defect in hematopoiesis. This work defines the respective roles of IRP2 in the determination of critical body iron parameters such as organ iron loading and erythropoiesis.
Introduction
As both lack and excess of iron compromise survival, homeostatic regulatory mechanisms have evolved to maintain systemic and cellular iron balance and ensure sufficient iron supply while preventing toxic iron accumulation. At the systemic level, adequate blood iron levels are maintained by coordinating dietary iron absorption in the duodenum, iron recycling from effete erythrocytes by resident macrophages (mainly in the spleen), iron utilization by erythroid precursors, and iron storage by hepatocytes.1,2 The hepatic “iron hormone” hepcidin (HAMP) negatively controls blood iron levels by triggering the degradation of the iron exporter ferroportin (FPN1)/solute carrier family 40 member A1 (SLC40A1)3 that releases iron from absorptive duodenal enterocytes and iron-recycling macrophages.4 Importantly, hepatic HAMP synthesis is stimulated by iron overload and inflammation, and suppressed by enhanced erythropoiesis.5
Cellular iron levels are balanced by iron regulatory proteins 1 (IRP1) and IRP2 (also named ACO1 and IREB2, respectively) that coordinate the posttranscriptional expression of proteins involved in iron uptake, utilization, storage, and export by binding to cis-regulatory iron-responsive elements (IREs) in the respective mRNAs.2,6,7 Either of the 2 IRPs inhibits translation by binding to the IRE located within the 5′ untranslated region (UTR) of the ferritin H- (FTH1) and L- (FTL) chain, the mitochondrial aconitase (ACO2), the erythroid 5′-aminolevulinic acid synthase (eALAS), or the hypoxia inducible factor 2α (HIF2α/EPAS1) mRNAs, whereas the IRPs increase mRNA stability when bound to the IREs present in the 3′ UTR of the transferrin receptor 1 (TFR1/TFRC) mRNA. The combined IRE-binding activity of both IRPs is increased in iron deficiency and conversely decreased in iron-loaded cells.2,6,7 Although the role of the IRP/IRE regulatory network has been extensively investigated in cultured cells, the understanding of its systemic in vivo functions remained elusive until animal models of IRP deficiency were created.8-12
We generated conditional alleles of the murine Aco1 and Ireb2 genes11,12 to study the functions of IRP1 and IRP2 in the context of the entire organism and to determine how these proteins modulate iron fluxes through cells and tissues to maintain adequate body iron levels and distribution. Although mice lacking either of the 2 IRPs are viable and fertile, lack of both proteins results in embryonic lethality,13,14 showing that the IRP/IRE system is essential for development, and that IRP1 and IRP2 can largely substitute for each other. Interestingly, IRP1-deficient mice display no overt phenotype under standard laboratory conditions,9 whereas mice with total and constitutive IRP2 deficiency suffer from microcytosis15,16 and altered body iron distribution with iron loading of the duodenal mucosa and the liver associated with FTH1 and FTL up-regulation,9,10,16 and a relative iron deficiency in splenic macrophages associated with down-regulation of FTH1, FTL, and FPN1.16 These studies uncovered a role of IRP2 in securing physiologic body iron balance and in normal erythropoiesis. To delineate tissue-specific and systemic context-dependent functions of IRP2, we selectively ablated IRP2 in intestinal enterocytes, hepatocytes, or macrophages, respectively, and determined the consequences for local and systemic iron metabolism. The results define the respective cell type–specific functions of IRP2 in the control of critical body iron parameters.
Methods
Mice
The generation of mouse lines carrying truncated (Ireb2Δ/Δ) or floxed (Ireb2flox/flox) Ireb2 alleles has been described.12 Ireb2Δ/Δ and Ireb2+/+ littermates were obtained from Ireb2+/Δ intercrosses. For tissue-specific removal of the floxed Ireb2 allele, Ireb2flox/flox animals were first bred to Cre-deletor strains; Cre-positive, Ireb2+/flox mice of the progeny were then crossed with Ireb2flox/flox animals to produce Ireb2flox/flox mice with or without Cre. Mice expressing Cre under the control of the murine Villin promoter (B6SJL-Tg(Vil-Cre)997Gum)17 were obtained from The Jackson Laboratory (Bar Harbor, ME). Mouse lines expressing Cre under the control of the Albumin/Alpha fetoprotein regulatory elements18 and of the LysozymeM promoter,19 respectively, were kindly provided by G. Schütz (Deutsches Krebsforschungszentrum, Heidelberg, Germany) and by I. Förster (Institut für Umweltmedizinische Forschung, Düsseldorf, Germany). Animals were kept under a constant light/dark cycle and had access to water and food ad libitum. They were killed at 8 to 10 weeks of age by CO2 inhalation and heparinized blood was collected by cardiac puncture. Tissues were flash-frozen into liquid nitrogen or embedded. To ensure optimal comparability of the results, all mouse lines were raised in parallel and analyzed using the same reagent lots and equipment settings. Animal handling was in accordance with guidelines approved by the European Molecular Biology Laboratory (EMBL).
Hematology and tissue iron determination
Red blood cell profiles and hemoglobin content were determined using the ABC Vet apparatus (ABX Diagnostics, Montpellier, France). Tissue nonheme iron content was measured using the bathophenanthroline method as described.16
Preparation of peritoneal and bone marrow–derived macrophages
For the isolation of peritoneal macrophages, mice were injected intraperitoneally with 1 mL 2.98% water-solubilized thiogylcolate medium (Sigma-Aldrich, Taufkirchen, Germany). Cells were harvested 4 days later by peritoneal lavage using ice-cold PBS and were pelleted at 300g for 10 minutes at 4°C. They were resuspended in RPMI1640 medium (Invitrogen, Karlsruhe, Germany) supplemented with 10% heat-inactivated fetal calf serum (FCS; Hyclone, Thermo Fisher Scientific, South Logan, UT), and seeded at a density of approximately 2 × 105 cells/cm2 onto collagen-coated plates (Falcon, BD Biosciences, Heidelberg, Germany). Cells were allowed to adhere for 5 hours at 37°C in a 5% CO2 atmosphere. Nonadherent cells were washed away with PBS and adherent cells were collected.
Bone marrow–derived macrophages (BMDMs) were flushed out from the femur with ice-cold Hanks-balanced salt solution (HBSS). The cell suspension was filtered through a 80-μm cell strainer (Falcon, BD Biosciences) and cells were seeded at a density of approximately 5 × 104 cells/cm2 in RPMI1640 + Glutamax (Invitrogen) supplemented with 20% of heat-inactivated FCS (Hyclone, Thermo Fisher Scientific), 1% penicillin + streptomycin (Invitrogen), and 100 ng/mL mouse colony-stimulating factor (Sigma-Aldrich). Cells were grown at 37°C in a 5% CO2 atmosphere. After 4 days, nonadherent cells were removed by washing with HBSS and the medium was subsequently replaced daily until cells were harvested (typically 6-7 days after seeding).
The proportion of macrophages in the peritoneal and bone marrow–derived cell preparations was estimated to exceed 90% by labeling with an Alexa Fluor 488 coupled-rat monoclonal antibody against the F4/80 macrophage-specific marker (Serotec, Düsseldorf, Germany).
DNA analyses
Mice were genotyped by polymerase chain reaction (PCR) using the A51/A52 primer pairs to distinguish the floxed and wild-type Ireb2 alleles as described.12 The Cre transgene was detected using the Cre-5/Cre-6 primer pair as described.13 Cre-mediated recombination of the floxed Ireb2 allele was analyzed by Southern blotting of EcoRI-treated genomic DNA using a PCR probe generated with the Ireb2-S/Ireb2-R primers as described.11 The intact floxed and the truncated Ireb2 alleles give rise to, respectively, approximately 4-kb and 3-kb fragments.
RNA analyses
Total RNA was extracted using the Trizol reagent (Invitrogen). HAMP1 and β-actin mRNA levels were determined by real-time quantitative PCR (qPCR) as described.13 For each sample, qPCR analysis was performed in triplicate, and the average of the triplicate was used for subsequent calculations.
Protein analyses
Protein extracts were prepared and subjected to Western blot analysis as described.11,16 Ferritin H- and L-subunits, IRP1, IRP2, FPN1, and β-actin were detected as described.16 Cre was detected using a rabbit polyclonal anti-Cre antibody (Novagen, Merck Chemicals, Nottingham, United Kingdom). The goat polyclonal anti-SLC27A4 antibody (sc-5835) was from Santa Cruz Biotechnology (Santa Cruz, CA).
Statistical analyses
Differences between the mean values were evaluated using 2-tailed Student t test. A P value less than .05 was considered significant.
Results
Efficient and tissue-specific ablation of IRP2
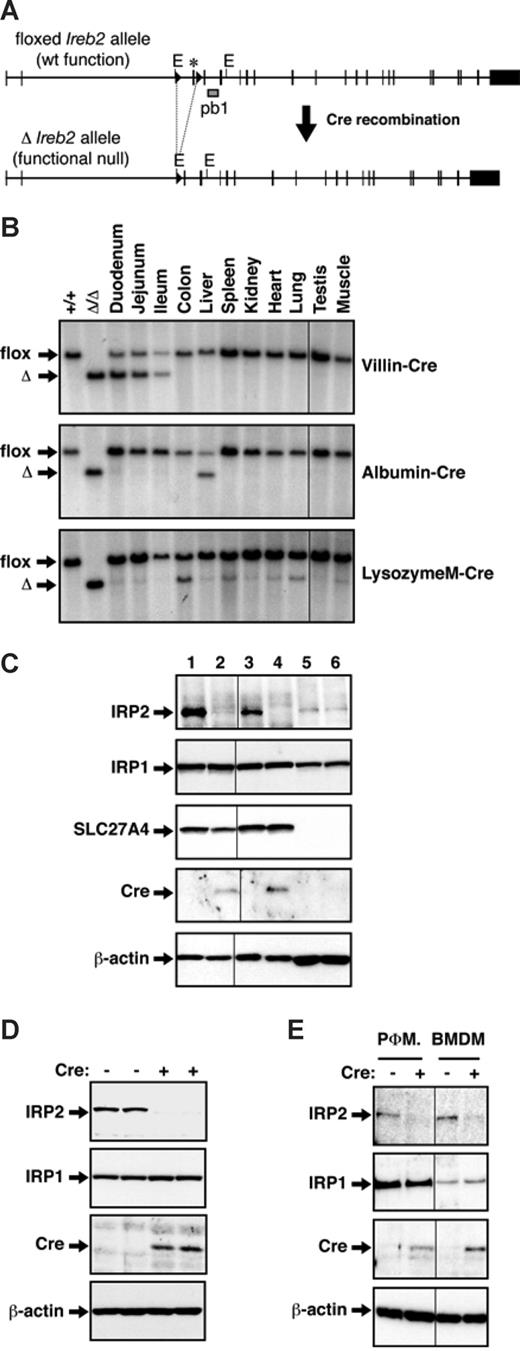
Tissue-specific ablation of IRP2 was achieved using Cre/Lox technology.20,21 Mice carrying floxed Ireb2 alleles (Ireb2flox/flox) have been described12 ; in this line, Cre-mediated removal of the third exon induces a frameshift and generates a functional null allele (Figure 1A). A mouse line with total and constitutive IRP2 deficiency (designated Ireb2Δ/Δ) was derived from Ireb2flox/flox mice12 using the PGK-Cre deletor strain that expresses the Cre recombinase ubiquitously including the germ line.22 To selectively ablate IRP2 in intestinal epithelial cells (IECs) or in hepatocytes, respectively, Ireb2flox/flox animals were bred to transgenic strains expressing the Cre recombinase under the control of the Villin promoter17 or under the control of both mouse Albumin regulatory elements and the Alpha fetoprotein enhancers18 ; the obtained lines are designated, respectively, Ireb2VilCre and Ireb2AlfpCre. Elimination of IRP2 expression in monocytes/macrophages was obtained by breeding to a knock-in strain with an insertion of the Cre cDNA into the LysozymeM locus19 ; the obtained line is designated Ireb2LysMCre. The presence (+) or the absence (−) of Cre is indicated. Ireb2VilCre(+), Ireb2AlfpCre(+), and Ireb2LysMCre(+) mice were born in Mendelian ratios and presented with no overt abnormalities (data not shown).
Specific and efficient ablation of IRP2 in intestinal epithelial cells (IECs), hepatocytes, or macrophages. (A) Schematic representation of the floxed (flox) and the truncated (Δ) Ireb2 alleles. The exon flanked by loxP sites (triangles) is marked with an asterisk. Excision of this exon upon Cre-mediated recombination generates a functional null allele.12 (B) The tissue specificity of Cre-mediated truncation of the floxed Ireb2 allele was determined by Southern blot analysis of various tissues from Ireb2flox/flox mice expressing the Cre recombinase under the control, respectively, of the Villin (Ireb2VilCre), the Albumin/Alpha fetoprotein (Ireb2AlfpCre), or the LysozymeM (Ireb2LysMCre) promoter, as indicated. The EcoRI restriction sites (indicated E) and the probe used (pb1) are shown in panel A. Tail DNA from mice with intact floxed Ireb2 alleles (Ireb2flox/flox) or from mice with total and constitutive truncation of the Ireb2 locus (Ireb2Δ/Δ) was used as a control. The floxed (flox) and truncated (Δ) alleles are indicated by arrows. The efficiency of IRP2 ablation in the intestine, the liver, and macrophages, respectively, of Ireb2VilCre (C), Ireb2AlfpCre (D), and Ireb2LysMCre (E) mice expressing (+) or not expressing (−) Cre was assessed by Western blotting. (C) IRP2 expression in whole duodenal tissue samples (lanes 1- 2), in mucosal scrapings (lanes 3-4), and in the submucosal, nonepithelial tissue (lanes 5-6) from Ireb2VilCre(+) mice (lanes 2, 4, and 6) versus Ireb2VilCre(−) littermates (lanes 1, 3, and 5). SLC27A4 was used as a mucosal marker. (D) IRP2 levels were assayed in whole liver samples from Ireb2AlfpCre(+) mice versus Ireb2AlfpCre(−) littermates, as indicated. (E) IRP2 expression was examined in peritoneal (PΦM) and bone marrow–derived macrophages (BMDMs) from Ireb2LysMCre(+) mice versus Ireb2LysMCre(−) littermates, as indicated. IRP1 levels and Cre expression were verified in every sample analyzed. β-Actin was used as a loading control. Representative results of at least 3 experiments are presented. Vertical lines have been inserted to indicate repositioned gel lanes.
Specific and efficient ablation of IRP2 in intestinal epithelial cells (IECs), hepatocytes, or macrophages. (A) Schematic representation of the floxed (flox) and the truncated (Δ) Ireb2 alleles. The exon flanked by loxP sites (triangles) is marked with an asterisk. Excision of this exon upon Cre-mediated recombination generates a functional null allele.12 (B) The tissue specificity of Cre-mediated truncation of the floxed Ireb2 allele was determined by Southern blot analysis of various tissues from Ireb2flox/flox mice expressing the Cre recombinase under the control, respectively, of the Villin (Ireb2VilCre), the Albumin/Alpha fetoprotein (Ireb2AlfpCre), or the LysozymeM (Ireb2LysMCre) promoter, as indicated. The EcoRI restriction sites (indicated E) and the probe used (pb1) are shown in panel A. Tail DNA from mice with intact floxed Ireb2 alleles (Ireb2flox/flox) or from mice with total and constitutive truncation of the Ireb2 locus (Ireb2Δ/Δ) was used as a control. The floxed (flox) and truncated (Δ) alleles are indicated by arrows. The efficiency of IRP2 ablation in the intestine, the liver, and macrophages, respectively, of Ireb2VilCre (C), Ireb2AlfpCre (D), and Ireb2LysMCre (E) mice expressing (+) or not expressing (−) Cre was assessed by Western blotting. (C) IRP2 expression in whole duodenal tissue samples (lanes 1- 2), in mucosal scrapings (lanes 3-4), and in the submucosal, nonepithelial tissue (lanes 5-6) from Ireb2VilCre(+) mice (lanes 2, 4, and 6) versus Ireb2VilCre(−) littermates (lanes 1, 3, and 5). SLC27A4 was used as a mucosal marker. (D) IRP2 levels were assayed in whole liver samples from Ireb2AlfpCre(+) mice versus Ireb2AlfpCre(−) littermates, as indicated. (E) IRP2 expression was examined in peritoneal (PΦM) and bone marrow–derived macrophages (BMDMs) from Ireb2LysMCre(+) mice versus Ireb2LysMCre(−) littermates, as indicated. IRP1 levels and Cre expression were verified in every sample analyzed. β-Actin was used as a loading control. Representative results of at least 3 experiments are presented. Vertical lines have been inserted to indicate repositioned gel lanes.
Tissue specificity of Cre-mediated truncation of the floxed Ireb2 allele was tested by Southern blotting. The truncated Ireb2 allele was detected exclusively in the small intestine of Ireb2VilCre(+) mice, including the proximal part of the duodenum that is the main site of iron absorption (Figure 1B top). In Ireb2AlfpCre(+) animals, recombination of the floxed Ireb2 gene was adequately restricted to the liver (Figure 1B middle). Truncation of the Ireb2 allele was expectedly detected in the spleen of Ireb2LysMCre(+) mice (Figure 1B bottom), and additionally in other tissues such as the lung and the liver, likely due to the presence of resident macrophages.
A faint signal corresponding to the full-length floxed Ireb2 allele was still detectable in the liver of Ireb2AlfpCre(+) animals (Figure 1B). Because the Southern blot analysis was performed on organ tissue rather than hepatocytes, the remaining signal corresponding to the intact allele could reflect the contribution of nonparenchymal cells not expressing the Cre recombinase. Such cells can represent up to one third of liver cells.23 In any case, Western blotting of liver extracts reveals that IRP2 expression is undetectable in samples from Ireb2AlfpCre(+) mice (Figure 1D).
The residual full-length floxed Ireb2 allele in the intestine of Ireb2VilCre(+) animals also appeared to be relatively abundant, and even prevailed in all organs from Ireb2LysMCre(+) mice (Figure 1B). To test whether the remaining intact allele reflects the contribution of connective tissue cells and others that do not express the Cre recombinase or is due to inefficient Cre-mediated recombination in the targeted cells, we determined the efficiency of IRP2 ablation specifically in Cre-expressing cells (Figure 1C,E). Western blot analysis of duodenal tissue samples shows a profound, yet not totally complete, reduction of IRP2 expression in Ireb2VilCre(+) mice compared with control Ireb2VilCre(−) littermates (Figure 1C compare lane 2 with lane 1). In mucosal scrapings that are highly enriched in enterocytes, as assessed by increased amounts of the SLC27A4 (solute carrier family 27, member 4) mucosal marker and of the Cre recombinase (Figure 1C compare lane 4 with lane 2), IRP2 is undetectable in Ireb2VilCre(+) samples versus control (Figure 1C compare lane 4 with lane 3). As expected, IRP2 levels are unchanged in the underlying, SLC27A4-negative tissue that does not express the Cre recombinase (Figure 1C lanes 5 and 6). Notably, the decrease in IRP2 protein levels in the whole duodenum (Figure 1C) is not associated with a commensurate disappearance of the intact floxed Ireb2 allele (Figure 1B); this suggests that the IRP2 signal in the Western blot analysis is accounted for mainly by IECs. In Ireb2LysMCre(+) mice, IRP2 levels are strongly, although not completely, reduced in peritoneal and bone marrow–derived macrophages (Figure 1E), consistent with the reported efficiency of the LysM::Cre deletor strain19 and with RNase protection analysis of the IRP2 mRNA in peritoneal macrophages (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Altogether, these data reveal efficient and specific ablation of IRP2 in IECs, hepatocytes, and macrophages of Ireb2VilCre(+), Ireb2AlfpCre(+), and Ireb2LysMCre(+) mice, respectively. Importantly, the Western blot analyses indicate lack of compensatory up-regulation of IRP1 protein levels (Figure 1C-E) and IRE-binding activity (Figure S2) in IECs, hepatocytes, or macrophages deficient in IRP2 expression.
Red blood cell parameters in mice with total versus tissue-specific IRP2 deficiency
Having established mouse lines with selective ablation of IRP2 in IECs, hepatocytes, or macrophages, we determined which of the phenotypic abnormalities detected in Ireb2Δ/Δ mice lacking IRP2 expression in all tissues was also present in the Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mouse lines, respectively. First we looked at red blood cell parameters. In agreement with previous reports,12,15,16 both male and female Ireb2Δ/Δ mice with total and constitutive IRP2 deficiency exhibit microcytosis associated with reduced serum hemoglobin and hematocrit values at 8 to 10 weeks of age (Table 1). By contrast, neither Ireb2VilCre(+), Ireb2AlfpCre(+), nor Ireb2LysMCre(+) mice, respectively, display microcytosis and have normal serum hemoglobin and hematocrit values compared with their respective control littermates lacking Cre expression (Table 1); Ireb2AlfpCre(+) animals even exhibit a rather modest, yet statistically significant, macrocytosis (Table 1). These data demonstrate that the microcytosis and the decreased hematocrit and serum hemoglobin values in Ireb2Δ/Δ mice are independent of IRP2 expression in IECs, hepatocytes, or macrophages.
Hematologic parameters in mice with total IRP2 deficiency versus mice lacking IRP2 in enterocytes, hepatocytes, or macrophages
| Genotype . | n . | RBC, 1012/L . | Hematocrit, L/L . | Hemoglobin, g/L . | MCV, fL . |
|---|---|---|---|---|---|
| Males | |||||
| Ireb2+/+ | 8 | 10.9 ± 0.1 | 0.58 ± 0.01 | 161 ± 1 | 52.8 ± 0.2 |
| Ireb2Δ/Δ | 8 | 10.2 ± 0.3 | 0.49 ± 0.01‡ | 138 ± 1‡ | 48.0 ± 0.3‡ |
| Ireb2VilCre(−) | 10 | 11.1 ± 0.2 | 0.58 ± 0.01 | 161 ± 2 | 52.7 ± 0.4 |
| Ireb2VilCre(+) | 9 | 11.3 ± 0.2 | 0.59 ± 0.01 | 169 ± 6 | 53.0 ± 0.3 |
| Ireb2AlfpCre(−) | 8 | 10.9 ± 0.3 | 0.57 ± 0.01 | 157 ± 3 | 52.8 ± 0.3 |
| Ireb2AlfpCre(+) | 8 | 10.8 ± 0.2 | 0.58 ± 0.01 | 161 ± 2 | 54.1 ± 0.3* |
| Ireb2LysMCre(−) | 6 | 10.7 ± 0.2 | 0.53 ± 0.01 | 169 ± 1 | 52.2 ± 0.3 |
| Ireb2LysMCre(+) | 9 | 10.7 ± 0.2 | 0.55 ± 0.01 | 168 ± 1 | 51.9 ± 0.3 |
| Females | |||||
| Ireb2+/+ | 8 | 10.3 ± 0.2 | 0.55 ± 0.01 | 160 ± 1 | 53.5 ± 0.4 |
| Ireb2Δ/Δ | 8 | 9.8 ± 0.1* | 0.48 ± 0.01‡ | 138 ± 1‡ | 49.8 ± 0.2‡ |
| Ireb2VilCre(−) | 10 | 10.9 ± 0.4 | 0.59 ± 0.01 | 166 ± 4 | 54.2 ± 0.8 |
| Ireb2VilCre(+) | 10 | 10.7 ± 0.2 | 0.58 ± 0.01 | 163 ± 3 | 54.3 ± 0.3 |
| Ireb2AlfpCre(−) | 8 | 10.7 ± 0.2 | 0.57 ± 0.01 | 161 ± 1 | 53.3 ± 0.2 |
| Ireb2AlfpCre(+) | 8 | 10.4 ± 0.1 | 0.57 ± 0.01 | 160 ± 1 | 54.6 ± 0.3† |
| Ireb2LysMCre(−) | 7 | 9.8 ± 0.4 | 0.52 ± 0.01 | 172 ± 3 | 52.7 ± 0.8 |
| Ireb2LysMCre(+) | 9 | 10.7 ± 0.3 | 0.56 ± 0.02 | 176 ± 2 | 52.6 ± 0.3 |
| Genotype . | n . | RBC, 1012/L . | Hematocrit, L/L . | Hemoglobin, g/L . | MCV, fL . |
|---|---|---|---|---|---|
| Males | |||||
| Ireb2+/+ | 8 | 10.9 ± 0.1 | 0.58 ± 0.01 | 161 ± 1 | 52.8 ± 0.2 |
| Ireb2Δ/Δ | 8 | 10.2 ± 0.3 | 0.49 ± 0.01‡ | 138 ± 1‡ | 48.0 ± 0.3‡ |
| Ireb2VilCre(−) | 10 | 11.1 ± 0.2 | 0.58 ± 0.01 | 161 ± 2 | 52.7 ± 0.4 |
| Ireb2VilCre(+) | 9 | 11.3 ± 0.2 | 0.59 ± 0.01 | 169 ± 6 | 53.0 ± 0.3 |
| Ireb2AlfpCre(−) | 8 | 10.9 ± 0.3 | 0.57 ± 0.01 | 157 ± 3 | 52.8 ± 0.3 |
| Ireb2AlfpCre(+) | 8 | 10.8 ± 0.2 | 0.58 ± 0.01 | 161 ± 2 | 54.1 ± 0.3* |
| Ireb2LysMCre(−) | 6 | 10.7 ± 0.2 | 0.53 ± 0.01 | 169 ± 1 | 52.2 ± 0.3 |
| Ireb2LysMCre(+) | 9 | 10.7 ± 0.2 | 0.55 ± 0.01 | 168 ± 1 | 51.9 ± 0.3 |
| Females | |||||
| Ireb2+/+ | 8 | 10.3 ± 0.2 | 0.55 ± 0.01 | 160 ± 1 | 53.5 ± 0.4 |
| Ireb2Δ/Δ | 8 | 9.8 ± 0.1* | 0.48 ± 0.01‡ | 138 ± 1‡ | 49.8 ± 0.2‡ |
| Ireb2VilCre(−) | 10 | 10.9 ± 0.4 | 0.59 ± 0.01 | 166 ± 4 | 54.2 ± 0.8 |
| Ireb2VilCre(+) | 10 | 10.7 ± 0.2 | 0.58 ± 0.01 | 163 ± 3 | 54.3 ± 0.3 |
| Ireb2AlfpCre(−) | 8 | 10.7 ± 0.2 | 0.57 ± 0.01 | 161 ± 1 | 53.3 ± 0.2 |
| Ireb2AlfpCre(+) | 8 | 10.4 ± 0.1 | 0.57 ± 0.01 | 160 ± 1 | 54.6 ± 0.3† |
| Ireb2LysMCre(−) | 7 | 9.8 ± 0.4 | 0.52 ± 0.01 | 172 ± 3 | 52.7 ± 0.8 |
| Ireb2LysMCre(+) | 9 | 10.7 ± 0.3 | 0.56 ± 0.02 | 176 ± 2 | 52.6 ± 0.3 |
Hematologic parameters were determined at 10 weeks of age in Ireb2+/+ versus Ireb2Δ/Δ mice with total and constitutive IRP2 deficiency and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with (+) or without (−) selective IRP2 ablation in intestinal epithelial cells, in hepatocytes, or in macrophages, respectively. Data are given as average plus or minus SEM. The sample size (n) is indicated.
RBC indicates red blood cell; MCV, mean corpuscular volume; and WBC, white blood cell.
Data were compared between control and IRP2-deficient animals within each group:
P < .05;
P < .01;
P < .001, Student t test.
Cell-autonomous functions of IRP2 in duodenal enterocytes
Ireb2Δ/Δ mice with total and constitutive IRP2 deficiency exhibit duodenal iron loading (Figure 2A) associated with increased expression of the ferritin H- and L-subunits (Figure 2B), as reported before.12,16 Selective removal of IRP2 in IECs causes comparable iron accumulation in the duodenum (Figure 2A) and concomitant up-regulation of the ferritin H- and L-chains (Figure 2B). By contrast, ablation of IRP2 in hepatocytes or in macrophages has no detectable impact on these parameters (Figure 2A,B). To check whether iron mismanagement in the intestine of Ireb2VilCre(+) mice could result from Cre expression rather than from IRP2 ablation,24 we measured the duodenal iron content of mice with wild-type Ireb2 alleles with (Cre) or without (noCre) expression of the Villin::Cre transgene. As expected, expression of Cre recombinase alone does not affect duodenal iron content (481 ± 73 μg iron per milligram of dry tissue in Cre males [n = 8] versus 521 ± 127 in noCre [n = 8] and 611 ± 120 μg iron per milligram of dry tissue in Cre females [n = 8] versus 602 ± 128 in noCre [n = 8]). These results demonstrate that iron deposition and ferritin up-regulation in the duodenum are caused by intrinsic, cell-autonomous iron mismanagement in IECs.
Analysis of duodenal iron content and ferritin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Relative total nonheme iron levels and (B) Western blot analysis of ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression in the duodenum of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), in hepatocytes (hepat.), or in macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B) A representative Western blot experiment is presented. β-Actin was used as a loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Analysis of duodenal iron content and ferritin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Relative total nonheme iron levels and (B) Western blot analysis of ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression in the duodenum of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), in hepatocytes (hepat.), or in macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B) A representative Western blot experiment is presented. β-Actin was used as a loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Cell-autonomous functions of IRP2 in hepatocytes
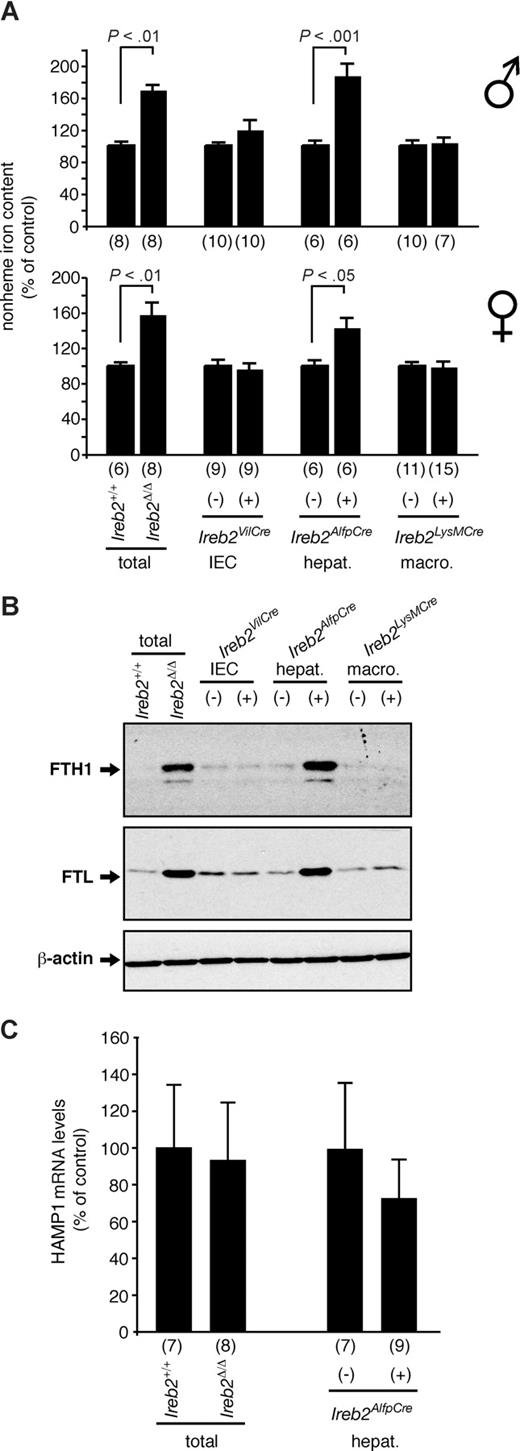
Similar to the duodenum, the liver of Ireb2Δ/Δ mice is iron loaded, associated with an up-regulation of the ferritin H- and L-chains (Figure 3). Interestingly, Ireb2VilCre(+) mice display normal iron and ferritin levels in the liver (Figure 3A-B), showing that iron loading and ferritin up-regulation in the duodenum (Figure 2) have no detectable impact on steady-state hepatic iron metabolism. Analogous to Ireb2VilCre(+) mice, Ireb2LysMCre(+) animals exhibit unchanged iron parameters in the liver (Figure 3A-B). By contrast, selective ablation of IRP2 in the hepatocytes of Ireb2AlfpCre(+) mice completely phenocopies the hepatic iron loading and the concomitant up-regulation of the ferritin H- and L-subunits (Figure 3A-B). Similar to Ireb2VilCre(+) mice, iron mismanagement in the liver of Ireb2AlfpCre(+) animals is dependent on IRP2 ablation since Cre expression alone does not alter hepatic iron metabolism.25 These data show that iron metabolism changes in the liver of IRP2-deficient mice result from intrinsic, cell-autonomous iron mismanagement in hepatocytes.
Analysis of hepatic iron content and ferritin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Relative total nonheme iron levels and (B) Western blot analysis of ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression in the liver of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), hepatocytes (hepat.), or macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B) A representative Western blot experiment is shown. The faint band below the ferritin H-chain signal likely corresponds to a truncated product. β-Actin was used as a loading control. (C) Hepatic levels of HAMP1 mRNA were determined by qPCR in Ireb2Δ/Δ mice versus Ireb2+/+ littermates, and in Ireb2AlfpCre mice carrying (+) or not carrying (−) the Cre transgene. HAMP1 mRNA levels were standardized to β-actin mRNA expression. The number of mice per group (n) is indicated. Data are presented as mean plus or minus SD. Within each group, HAMP1 expression in control mice was set to 100%. In Ireb2Δ/Δ mice, the antagonistic effects of liver iron loading (A) and microcytic anemia (Table 1) on Hamp1 expression may neutralize each other. However, a Student t test revealed no statistically significant change in HAMP1 mRNA levels in Ireb2AlfpCre(+) mice with an iron-loaded liver (A) and without microcytosis (Table 1). The histogram shows data obtained from female mice; similar results were obtained from males.
Analysis of hepatic iron content and ferritin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Relative total nonheme iron levels and (B) Western blot analysis of ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression in the liver of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), hepatocytes (hepat.), or macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B) A representative Western blot experiment is shown. The faint band below the ferritin H-chain signal likely corresponds to a truncated product. β-Actin was used as a loading control. (C) Hepatic levels of HAMP1 mRNA were determined by qPCR in Ireb2Δ/Δ mice versus Ireb2+/+ littermates, and in Ireb2AlfpCre mice carrying (+) or not carrying (−) the Cre transgene. HAMP1 mRNA levels were standardized to β-actin mRNA expression. The number of mice per group (n) is indicated. Data are presented as mean plus or minus SD. Within each group, HAMP1 expression in control mice was set to 100%. In Ireb2Δ/Δ mice, the antagonistic effects of liver iron loading (A) and microcytic anemia (Table 1) on Hamp1 expression may neutralize each other. However, a Student t test revealed no statistically significant change in HAMP1 mRNA levels in Ireb2AlfpCre(+) mice with an iron-loaded liver (A) and without microcytosis (Table 1). The histogram shows data obtained from female mice; similar results were obtained from males.
We also reported that mice with IRP2 deficiency following insertion of a gene-trap cassette into the second intron of the Ireb2 gene11 exhibit unchanged HAMP mRNA levels despite microcytic anemia and increased hepatic iron levels.16 Since IRP2 deficiency appears not to disrupt physiologic Hamp regulation,16 we hypothesized that the antagonistic effects of microcytosis and liver iron loading on Hamp expression could neutralize each other or that liver iron loading had not reached the threshold required for Hamp to respond. Ireb2AlfpCre(+) animals should allow for the discrimination between these 2 alternatives as they display increased hepatic iron levels comparable with mice with total and constitutive IRP2 deficiency (Figure 3A) but no microcytosis (Table 1). We assayed hepatic HAMP1 mRNA levels by qPCR (Figure 3C) (HAMP2 expression was not assessed because only HAMP1 is relevant for iron metabolism in the mouse26 ). Similar to IRP2-deficient mice carrying a gene-trap cassette within the Ireb2 locus,16 Ireb2Δ/Δ mice expectedly show normal Hamp1 expression in the liver compared with wild type (Figure 3C). Strikingly, Ireb2AlfpCre(+) animals display normal HAMP1 mRNA levels despite liver iron loading. These data indicate that liver iron accumulation due to hepatocytic IRP2 deficiency is not sufficient to modulate Hamp1 expression, or that IRP2 activity is required to mediate increased HAMP1 synthesis.
Different splenic phenotypes of mice with total versus macrophage-specific IRP2 deficiency
In contrast to the iron-loaded duodenum and liver, the spleen of Ireb2Δ/Δ mice exhibits relative iron deficiency (Figure 4A) associated with reduced expression of the ferritin H- and L-subunits (Figure 4B) and FPN1 down-regulation (Figure 4C). By contrast, total iron levels in the spleen of Ireb2LysMCre(+) mice remain unchanged (Figure 4A). Because we measured iron levels in the whole spleen and macrophages represent only a limited fraction of cells in the spleen,27 we analyzed splenic sections by Perl staining. This analysis reveals iron loading of the macrophages of the red pulp, and confirms that the iron content of splenic macrophages is normal in Ireb2LysMCre(+) mice (Figure S3). Similar to iron levels, splenic expression of the ferritin H- and L-subunits and FPN1 remains unchanged in Ireb2LysMCre(+) mice (Figure 4B,C). Thus, contrasting with the effects of IRP2 ablation in IECs or hepatocytes on duodenal and hepatic iron metabolism, respectively, selective removal of IRP2 in macrophages does not recapitulate the splenic phenotype of animals with total and constitutive IRP2 deficiency. These observations suggest that iron mismanagement in macrophages of Ireb2Δ/Δ mice is not cell autonomous, and requires IRP2 deficiency in one or several other cell types. However, neither duodenal iron accumulation in Ireb2VilCre(+) mice nor liver iron loading in Ireb2AlfpCre(+) animals affects the measured spleen iron parameters (Figure 4). Hence, iron mismanagement in macrophages of Ireb2Δ/Δ mice cannot be explained, respectively, by duodenal or hepatocytic IRP2 deficiency.
Analysis of splenic iron content, and ferritin and ferroportin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Total nonheme iron levels and Western blot analysis of (B) ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression and of (C) ferroportin (indicated FPN) in the spleen of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), hepatocytes (hepat.), or macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B,C) Representative Western blots are shown. The slight variation in FTH1, FTL, and FPN levels in Ireb2AlfpCre(−) versus Ireb2AlfpCre(+) mice reflects interindividual variability but not a difference between groups. β-Actin was used as a loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Analysis of splenic iron content, and ferritin and ferroportin expression of mice with tissue-specific versus total IRP2 deficiency. (A) Total nonheme iron levels and Western blot analysis of (B) ferritin H- (indicated FTH1) and L- (indicated FTL) chain expression and of (C) ferroportin (indicated FPN) in the spleen of Ireb2Δ/Δ mice lacking IRP2 expression in all tissues (total) versus Ireb2+/+ littermates, and in Ireb2VilCre, Ireb2AlfpCre, and Ireb2LysMCre mice with selective IRP2 ablation in intestinal epithelial cells (IECs), hepatocytes (hepat.), or macrophages (macro.), respectively, versus their corresponding control littermates; the presence (+) or absence (−) of the Cre transgene is indicated. (A) Data are presented as average plus or minus SEM. Within each group, the iron level of control mice was set to 100%. Top histogram represents male mice; bottom, female mice. The number of animals per group (n) is indicated. P: Student t test. (B,C) Representative Western blots are shown. The slight variation in FTH1, FTL, and FPN levels in Ireb2AlfpCre(−) versus Ireb2AlfpCre(+) mice reflects interindividual variability but not a difference between groups. β-Actin was used as a loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Discussion
Since its discovery 2 decades ago, the IRP/IRE regulatory network has emerged as the central system for the control of cellular iron homeostasis. The generation of different mouse models with IRP deficiency offered the possibility to explore the functions of the IRP/IRE network in systemic iron metabolism.9-12 These animal models uncovered the importance of IRP2 for normal erythropoiesis and for securing physiologic iron distribution between major sites of systemic iron balance, the duodenum, the liver, and the spleen, linking a classic regulator of cellular iron metabolism to systemic iron homeostasis.15,16 Here we used Cre-Lox technology to unveil the tissue-specific functions of IRP2 in enterocytes, hepatocytes, or macrophages for organ and organismic iron metabolism.
Role of IRP2 in erythropoiesis
Ireb2Δ/Δ mice display microcytosis despite normal serum iron parameters.12,16 Therefore, we hypothesized that the microcytosis was likely independent of altered iron distribution as a consequence of the observed abnormalities in the duodenum, the liver, and the spleen.16 Our findings reported here that Ireb2VilCre(+), Ireb2AlfpCre(+), and Ireb2LysMCre(+) mice, respectively, with selective IRP2 ablation display normal erythropoietic and plasma iron parameters directly support the hypothesis that microcytosis in Ireb2Δ/Δ mice is a cell-autonomous phenotype of erythroid cells that is explained mainly by impaired iron acquisition as a consequence of TFR1 mRNA destabilization,16 and possibly by iron sequestration into translationally derepressed ferritin.15
The microcytic anemia in zebrafish with glutaredoxin 5 deficiency,28 to which a human counterpart has recently been described,29 has been attributed to an aberrantly high IRE-binding activity of IRP1 lacking iron-sulfur clusters in erythroid cells, leading to IRE-mediated repression of the translation of eALAS,30 the rate-limiting enzyme in erythroid heme biosynthesis. These data also illustrate the importance of the IRP/IRE regulatory network for local iron management in erythroid cells. Of note, IRPs can also potentially influence red cell physiology indirectly by their ability to repress the translation of HIF2α,31 the transcription factor that positively regulates erythropoietin synthesis. However, mice that are deficient for only one of the IRPs do not display the polycythemic phenotype that would be predicted from derepression of HIF2α translation, consistent with the high degree of functional redundancy that has been observed.13,16
Role of IRP2 in duodenal enterocytes
Ireb2Δ/Δ and Ireb2VilCre(+) mice display duodenal iron accumulation associated with ferritin up-regulation. We suggest that ferritin is translationally derepressed as a direct consequence of IRP2 deficiency and that the increased ferritin expression leads to some degree of duodenal iron retention and hence loading.12,16 However, Ireb2VilCre(+) mice exhibit normal systemic iron parameters (this paper) and duodenal 59Fe transfer in Ireb2Δ/Δ mice is comparable with wild type.16 Thus, it appears as if the translational derepression of ferritin synthesis in the IRP2-deficient intestine is limited by the presence of IRP1, and not sufficient to impair duodenal iron transport.
Interestingly, 2-week-old mice with selective ablation of both IRP1 and IRP2 in the intestinal mucosa display dramatic ferritin up-regulation in the duodenum, associated with DMT1 down-regulation and increased FPN1 expression.13 Surprisingly, these mice exhibit normal plasma iron levels and hepatic iron stores. These data showed that the IRP/IRE system is essential for the control of iron absorption molecules in the duodenum; the data also suggested that although the opposite changes in DMT1 and FPN1 expression may balance each other, even a marked increase in duodenal ferritin content appears not to detectably impact on systemic iron metabolism, at least up to 2 weeks of age. Hence, even if IRP2 appears to be important for the control of ferritin synthesis in the duodenum, the limited impact of mucosal IRP2 deficiency on the synthesis of intestinal iron transporters (ie, DMT1 and FPN1) and on steady-state systemic iron metabolism most likely reflects the ability of IRP1 to partially substitute for IRP2.
Role of IRP2 in hepatocytes
The detailed mechanisms underlying iron accumulation in the liver of Ireb2Δ/Δ and Ireb2AlfpCre(+) mice remain unknown. The iron accumulation is unlikely to be explained by increased TFR1-mediated iron uptake, as the liver of IRP2-deficient mice displays normal TFR1 mRNA levels.16 Non–transferrin-bound iron acquisition via DMT1 and/or ZIP14/solute carrier family 39 member 1432 could also be augmented. However, the IRE-less ZIP14 mRNA is not a direct IRP target. In addition, hepatic DMT1 mRNA levels in mice with total and constitutive IRP2 deficiency are not detectably changed,16 and FPN1 protein expression is also unaltered.16 Perhaps, more plausibly, hepatic ferritin up-regulation may not be secondary to iron loading but due to translational derepression as a direct consequence of IRP2 deficiency; similar to the consideration regarding duodenal enterocytes, the increased liver iron content would reflect the partial retention of hepatic iron in ferritin. As such, it might not be sensed as iron overload. This would also plausibly explain the absence of increased Hamp expression in the liver of Ireb2Δ/Δ and Ireb2AlfpCre(+) mice. Alternatively or additionally, hepatocytic Hamp expression has recently been shown to be regulated by transferrin saturation.33 Hence, a reason for why Ireb2Δ/Δ and Ireb2AlfpCre(+) mice with iron-loaded livers do not up-regulate Hamp could be that they have normal plasma iron and transferrin saturation values (Table S1).
As previously reported for a distinct IRP2-deficient line,9,15 Ireb2Δ/Δ mice exhibit a marked increase in plasma ferritin levels (Table S2). As ferritin can be secreted by hepatocytic cell lines,34,35 the elevated plasma ferritin of Ireb2Δ/Δ mice could originate from their hepatocytes that display ferritin overexpression. However, Ireb2AlfpCre(+) animals exhibit normal plasma ferritin values (Table S2), indicating that the source of increased ferritin in the plasma of Ireb2Δ/Δ mice is unlikely to be hepatocytic; similarly, Ireb2VilCre(+) mice have normal plasma ferritin concentrations (Table S2), implying that the source is also unlikely to be enterocytic.
The normal plasma iron values in Ireb2Δ/Δ animals suggest that the diversion of hepatic iron fluxes into translationally derepressed ferritin has no detectable impact on systemic iron availability. This could be due to a compensatory increase in duodenal iron absorption and/or increased release of iron from splenic macrophages. However, animals with total and constitutive IRP2 deficiency exhibit normal duodenal iron transfer rates16 and Ireb2AlfpCre(+) mice with comparable liver iron loading have normal splenic iron levels (this paper). It is also possible that, similar to the duodenum, the retention of iron in the liver of Ireb2Δ/Δ and Ireb2AlfpCre(+) animals is quantitatively not large enough to affect systemic iron availability since the liver accounts only for approximately 8% of plasma iron turnover in humans.36 It would thus be interesting to further derepress hepatic ferritin translation by ablation of both IRP1 and IRP2. However, coablation of both IRPs in hepatocytes using the albumin/α-fetoprotein Cre-deletor strain appears to be embryonically lethal (B.G, D.F.-A., M.W.H., unpublished data, 2007).
IRP2 function in macrophages
In contrast to the duodenum and the liver, selective IRP2 ablation in macrophages does not recapitulate the splenic phenotype of mice with total and constitutive IRP2 deficiency. The most trivial explanation of this is that this is for technical reasons, due to insufficient removal of IRP2 in macrophages; the low abundance of the recombined Ireb2 allele in the spleen of Ireb2LysMCre(+) mice (Figure 1B) may support such a notion. However, this more likely reflects the small fraction of macrophages (2%–5%) in this tissue.19,29 Tuckerman et al directly determined that Cre-mediated recombination occurs in 70% of splenic macrophages in the LysozymeM-Cre deletor line, as assessed with a tk-LoxP–enhanced GFP reporter (RA/EG) strain.37 As splenic macrophages of mice with total IRP2 deficiency display an approximately 4-fold decrease in ferritin and ferroportin levels as assessed by Western blotting,16 a relatively inefficient ablation of IRP2 in only approximately 50% of the macrophages should still suffice to trigger a detectable change (∼ 2-fold) in ferritin and ferroportin expression in the spleen, if it occurred. In addition, tissue immunostaining confirmed the lack of detectable FPN down-regulation in splenic macrophages from Ireb2LysMCre(+) mice (Figure S4). Moreover, the observed ablation of IRP2 in highly enriched preparations of peritoneal and bone marrow–derived macrophages is effective and in good agreement with the reported recombination efficiency of the LysozymeM-Cre deletor strain19 and, although the various macrophage populations of the organism are not strictly identical,38 is expected to be in the same range in macrophages of the spleen. Hence, we consider inefficient IRP2 ablation in macrophages of Ireb2LysMCre(+) mice to be an unlikely explanation of the absence of splenic iron metabolism alterations. Unfortunately, a more direct assessment of the recombination efficiency in splenic macrophages by, for example, IRP2 immunofluorescence has failed for technical reasons.
An alternative explanation could be that IRP1 compensates for the lack of IRP2 in macrophages of Ireb2LysMCre(+) animals. However, we observed no compensatory up-regulation of IRP1 protein levels and no increase in the IRE-binding activity of IRP1 in the macrophages of Ireb2LysMCre(+) mice. Interestingly, Meyron-Holtz et al reported increased ferritin and decreased TFR1 expression in macrophages derived from a distinct IRP2-deficient line,39 and we observed similar changes in bone marrow–derived macrophages from our Ireb2Δ/Δ line (B.G., D.F.-A., M.W.H., unpublished data, 2005). Although both of these findings correspond to the expected pattern in IRP deficiency, the discrepancy in ferritin expression between in vivo and ex vivo macrophages further supports the notion that the iron deficiency and the down-regulation of ferritin and ferroportin in splenic macrophages of Ireb2Δ/Δ mice is unlikely to represent a direct consequence of their IRP2 deficiency. Altogether these data suggest that splenic iron mismanagement in Ireb2Δ/Δ mice is dependent on an IRP2-deficient environment.
Because Ireb2VilCre(+) and Ireb2AlfpCre(+) mice exhibit normal steady-state iron metabolism parameters in the spleen, splenic iron mismanagement in Ireb2Δ/Δ mice cannot be explained by the alterations of iron metabolism in the duodenum or in the liver, respectively; hence it results from the consequences of IRP2 deficiency in another cell type (eg, the microcytic red blood cells), or to combined IRP2 deficiency in several organs. We observed no significant change in HAMP mRNA expression in the liver of Ireb2Δ/Δ mice that could account for posttranslational FPN1 down-regulation3 ; we did not detect any local increase in HAMP mRNA levels in the spleen either (data not shown). At this stage, the mechanisms underlying ferritin and FPN1 down-regulation in splenic macrophages of Ireb2Δ/Δ mice remain unknown.
Interestingly, Ireb2LysMCre(+) mice display slightly elevated plasma ferritin levels (Table S2), possibly reflecting ferritin secretion from macrophages.40
In conclusion, our work dissects the tissue-specific functions of IRP2 in organs that play a central role in body iron homeostasis. It shows that IRP2 is intrinsically required to secure physiologic iron metabolism in enterocytes, hepatocytes, and likely erythroid cells. By contrast, iron management in macrophages seems to involve IRP2 functions in one or several distinct tissues and could hence be dependent on the systemic context. This study also shows that the IRP/IRE system exerts its effects mostly by local, cell-autonomous, or organ-autonomous interventions, by contrast to HAMP that acts as a hormone.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank G. Schütz and I. Förster, respectively, for the albumin/α-fetoprotein- and the LysozymeM-Cre strains. We are grateful to M. Vujic (Molecular Medicine Partnership Unit, Heidelberg, Germany) and F. Cannone-Hergaux (Centre National de la Recherch Scientifique, Gif sur Yvette, France), respectively, for their advice on the preparation of peritoneal and bone marrow–derived macrophages. We thank the staff of the EMBL Laboratory of Animal Resources for their dedicated animal care.
This work was supported in part by a European Economic Community FP6 grant (LSHM-CT-2006-037296 Euroiron1; M.W.H.).
Authorship
Contribution: D.F.-A. performed experiments and analyzed the data; M.W.H. conceived the study, analyzed the data, and wrote the paper; and B.G. conceived the study, performed experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthias W. Hentze, EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany; e-mail: hentze@embl.de; or Bruno Galy, EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany; e-mail: galy@embl.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal