Abstract
The reduced expression of nuclear factor of activated T cells-1 (NFAT1) protein in umbilical cord blood (UCB)–derived CD4+ T cells and the corresponding reduction in inflammatory cytokine secretion after stimulation in part underlies their phenotypic differences from adult blood (AB) CD4+ T cells. This muted response may contribute to the lower incidence and severity of high-grade acute graft-versus-host disease (aGVHD) exhibited by UCB grafts. Here we provide evidence that a specific microRNA, miR-184, inhibits NFAT1 protein expression elicited by UCB CD4+ T cells. Endogenous expression of miR-184 in UCB is 58.4-fold higher compared with AB CD4+ T cells, and miR-184 blocks production of NFAT1 protein through its complementary target sequence on the NFATc2 mRNA without transcript degradation. Furthermore, its negative effects on NFAT1 protein and downstream interleukin-2 (IL-2) transcription are reversed through antisense blocking in UCB and can be replicated via exogenous transfection of precursor miR-184 into AB CD4+ T cells. Our findings reveal a previously uncharacterized role for miR-184 in UCB CD4+ T cells and a novel function for microRNA in the early adaptive immune response.
Introduction
Numerous reports spanning 2 decades have confirmed umbilical cord blood (UCB) as a clinical source of hematopoietic progenitors for allogeneic transplantation in the treatment of hematologic malignancies.1,2 Despite the primary drawback of slower kinetics of myeloid engraftment as a result of limited graft cell dose,3 UCB has several advantages over bone marrow (BM) in a therapeutic setting, particularly the observed lowered incidence of acute graft-versus-host disease (aGVHD) despite the infusion of human leukocyte antigen (HLA) disparate grafts.4 aGVHD remains a major obstacle to the broader application of allogeneic stem cell therapy and is characterized by donor CD4+ T-cell activation in response to self-antigen presented by class II major histocompatibility complex (MHC) on host antigen-presenting cells (APCs). The clinical manifestation of aGVHD closely mimics the pathophysiology of autoimmune disorders with early secretion of proinflammatory cytokines including interferon γ (IFNγ), tumor necrosis factor α (TNFα),5 granulocyte macrophage colony-stimulating factor (GM-CSF), and interleukin (IL)-1, as well as later secretion of IL-2 by donor-derived T cells. These stimulate inflammatory cell proliferation, up-regulate MHC expression, natural killer (NK) and cytotoxic CD8+ T-cell recruitment, and widespread tissue damage particularly in the skin, large intestine, and liver.6,7
A key transcription factor in CD4+ T-cell activation and the downstream target of cyclosporine A (CsA) treatment, nuclear factor of activated T cells 1 (NFAT1, mRNA: NFATc2) influences the expression of a wide array of cytokines,8 surface receptors,9 and cell cycle regulators10 associated with normal and autoimmune responses.11 Tandem interactions with other transcription factors, particularly the AP1 (fos/jun) complex occur at adjacent DNA binding sites (WGGAAAWN for NFAT and TGAGTCA for AP1) located in the promoter regions of the genes encoding such factors as IFNγ, TNFα, IL-2, IL-4, IL-5, cytotoxic T-lymphocyte antigen 4 (CTLA-4), GM-CSF, and CD40L. Expression of many of these genes, specifically those associated with a Th2 or allergic response, is not severely diminished (and in some cases enhanced) in NFAT1-null mice, suggesting some level of redundancy among members of the NFAT family and their binding partners.12 However, NFAT1 has been shown to be required for the sustained production of IFN-γ,13 GM-CSF, IL-3, IL-4, IL-2 (with AP1),8,14 and TNF-α,8 indicating a critical role for NFAT1 in the initiation of a productive Th1 immune response. In the absence of AP1, NFAT1 heterodimers may activate an alternate, anergic repertoire of T-cell gene expression.15,16
Previous work by our laboratory has demonstrated significantly reduced NFAT1 protein expression in resting and stimulated UCB CD4+ T cells compared with adult blood (AB).17 This observation correlates with the severe reduction in IFNγ, TNFα, and other cytokine production by UCB after primary stimulation. Microarray gene expression profiling revealed markedly lower expression of nearly all NFAT1-associated transcripts, but importantly, not the mRNA for NFAT1 (NFATc2) itself. Notably, UCB CD4+ T cells express lower levels mRNA encoding GM-CSF, IFNγ, TNFα, IL-3, IL-4, IL-5, IL-13, macrophage inflammatory protein 1α (MIP-1α), as well as the inflammatory surface markers CD40L, CTLA-4, and the IL-2 receptor α chain (CD25).18 As secretion of the aforementioned cytokines is strongly associated with the non–self recognition of recipient APC by donor CD4+ T cells in aGVHD, the mechanisms by which expression of these factors is restrained in UCB may underlie peripheral tolerance exhibited by UCB CD4+ T cells and further the identification of risk factors or prophylactic treatments relevant to allogeneic transplantation. Our studies have led us to implicate a specific microRNA in the mechanism underlying restrained NFAT1 protein expression in UCB CD4+ graft T cells.
microRNA (miRNA) comprises a specialized subset of small cytoplasmic noncoding RNAs between 19 and 24 nucleotides in length. These highly processed RNAs most frequently bind to regulatory sequences in the 3′ untranslated region (UTR) of target mRNAs, thus blocking gene expression by mediating mRNA degradation or translational repression. The latter scenario is thought to involve the RNA-induced silencing complex (RISC) and accumulation in translation-deficient but nondegradative complexes known as P-bodies.19 mRNAs sequestered into these complexes can be later retargeted to translation initiation complexes or degraded through the decapping (Dcp) pathway,20 a choice thought to be in part dependent on sequence complementarity, RNA secondary structure, and other signals. The activity of specific miRNAs has been associated with a wide variety of cellular differentiation pathways, including hematopoiesis (reviewed in Baltimore et al21 ) and disease states such as cancer, diabetes,22,23 and neurodegenerative diseases.24,25 Notably, expression of miR-155 has been implicated in the regulation of mature T-cell lineage fate through c-Maf26,27 and is associated with FoxP3-mediated up-regulation in Treg cells.28
Here we provide evidence that a specific microRNA, miR-184, appears to contribute to the reduced NFAT1 protein in UCB-derived CD4+ T cells. Expression of miR-184 is increased in human UCB CD4+ T cells compared with AB controls, and specifically represses activity in luciferase reporter assays. A converse relationship is observed between miR-184 activity and NFAT1 protein expression in both antisense interference and miRNA precursor “knock-in” models. This study comprises the first known demonstration of miR-184 expression and activity in primary human lymphocytes and suggests a novel role for cellular RNA interference in regulating the CD4+ T-cell immune response in UCB.
Methods
Cell isolation and culture
Acquisition of both AB and UCB units conformed to and was approved under University Hospital Case Medical Center IRB protocol nos. 02-00-34/CASE 11Z05 (rev. 11/13/07). Whole blood was obtained from umbilical cords immediately after delivery or by venipuncture from healthy adult donors, and informed consent was obtained in accordance with the Declaration of Helsinki. Mononuclear cells were isolated after centrifugation through Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ) per the manufacturer's instructions. CD4+ T cells were then isolated via magnetic bead-labeling and separation (AutoMACS; Miltenyi Biotec, Auburn, CA) by first depleting the sample of CD14+ monocytes and then positively selecting for CD4+ cells, per the manufacturer's recommendations. CD45RA+ naive T cells were isolated via the Naive Human T-cell Isolation kit (Miltenyi Biotec). Purity as measured by flow cytometry was greater than 90%. CD4+ T cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (Gibco, Carlsbad, CA) supplemented with 2 mM l-glutamine (Gibco). Where applicable, T cells were stimulated in wells containing 1 μg/mL plate-bound αCD3 antibody and 5 μg/mL soluble αCD28 antibody (BD Biosciences, San Jose, CA).
Western blot analysis
Cells were pelleted and lysed with radioimmunoprecipitation assay (RIPA) buffer. After centrifugation, lysate supernatants were assayed for total protein content by modified Bradford assay (Bio-Rad Laboratories, Hercules, CA) per the manufacturer's instructions. The lysates were standardized for protein concentration, diluted with 4× sodium dodecyl sulfate (SDS) loading buffer containing β-mercaptoethanol and heated to 95°C for 5 minutes. Equal volumes of samples were loaded into the wells of a 7.5% polyacrylamide SDS gel and run per standard protocol. The gel was then transferred to an Immobilon-P polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA) via standard wet-transfer protocol. Membrane was blocked with 5% dry milk, cut, and probed with primary antibodies to NFAT1 (no. 610703; BD Transduction Laboratories, San Jose, CA), β-actin (no. A5441; Sigma-Aldrich, St Louis, MO), and horseradish peroxidase (HRP)-conjugated α-mouse secondary antibody (no. A9044; Sigma-Aldrich) per standard procedures. Bands were illuminated by ECL-Plus Visualization System (GE Healthcare) and exposed to film per standard procedure and digitized. Protein expression was quantified by integration of the relevant band intensity with ImageJ software (National Institutes of Health [NIH], Bethesda MD) and normalized to β-actin control.
miRNA candidate determination
The Sanger miRBase Targets v5 database was queried as described through the MicroCosm interface located online at http://microrna.sanger.ac.uk/.29 Complete computational protocol details are available therein and in the references. Briefly, this system uses the miRanda algorithm30 to determine and score sites of complementarity between mRNA 3′ UTR sequences and known human miRNA species. Predicted interactions are favored that exhibit a high degree of complementarity at the 5′ end of the miRNA, predicted thermodynamic stability by the Vienna RNA folding routines,31 and occur in UTR sequences conserved across multiple species.
Polymerase chain reaction and luciferase vector construction
RNA was obtained from UCB MNC or isolated CD4+ T cells with the PureLink Micro-to-Midi RNA Isolation kit (Invitrogen, Carlsbad, CA) and then DNase treated (DNA-free; Ambion, Austin, TX). After DNase removal, cDNA was generated from RNA as described below for reverse transcription–polymerase chain reaction (RT-PCR) and subjected to 30 rounds of PCR per standard protocol with the following primers to verify the presence of the predicted targeted sequence: 5′-TTACTATTTGGACGGAACACC-3′ (reverse, both reactions), 5′-TATGAAACAGAATGACTGTGATC-3′ (forward, NM_012340 reaction), and 5′-CTACTTGGATGATGTTAATGAAAT-3′ (forward, NM_173091 reaction). These PCRs additionally generated 3′ UTR sequences containing the full-length intervening sequence between the stop codon and the predicted miR-184 interacting site. This sequence (NM_012340 reaction only) was cloned into the pMIR-Report plasmid at the 3′ UTR position following the luciferase gene per the manufacturer's instructions (Ambion). In addition, short (38-mer) synthesized DNA sequences matching the plus and minus strand, including and surrounding the predicted targeted site were dimerized and cloned into the same vector. “Control” denotes pMIR-Report without either insert. Sequences of each vector are shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
CD4+ cell transfection
CD4+ T cells were transfected with luciferase vectors, synthetic precursor miRNA (no. 17000/17010; Ambion), and/or antisense miRNA inhibitor (no. 17100/17110; Ambion) via Amaxa Nucleofector (Amaxa, Gaithersburg, MD) per the manufacturer's protocol for unstimulated human T cells provided in the Human T-cell kit (no. VPA-1002) using program U-14. Approximately 1 μg plasmid or DNA sequence was transfected per 106 cells. Typical efficiencies in control green fluorescent protein (GFP) plasmid transfections were approximately 50%.
Luciferase assay
Cells were transfected with the aforementioned constructs alongside a consistent quantity of pGL4.71[hRlucP] plasmid (Promega, Madison, WI) to control for transfection efficiency. Cells were lysed and prepared per the manufacturer's instructions (Dual-Luciferase Reporter Assay System, no. E1910; Promega) and luciferase activity was measured via fluorescence on a NOVOStar plate reader (BMG Labtech, Durham, NC).
Quantitative RT-PCR
cDNA was generated from whole cellular RNA by MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA) per the manufacturer's instructions. TaqMan RT-PCR assays were prepared per the manufacturer's instructions (Applied Biosystems) using probes for either NFATc2 (no. Hs00234855_m1) or IL2 (Hs00174114_m1). Endogenous control for all relative mRNA quantifications was glyceraldehyde 3-phosphate dehydrogenase (GAPDH; no. Hs99999905_m1), and reactions were run in at least triplicate per experiment. qRT-PCR was carried out on an Applied Biosystems 7500 thermal cycler per the manufacturer's instructions. Relative expression was quantified from the amplification curves using the 7500 Fast System Software. For relative mature miRNA quantification, small RNA was enriched with the Mission Small RNA Isolation kit (Sigma-Aldrich) and qRT-PCR carried out using the TaqMan miRNA Reverse Transcription kit (Applied Biosystems) and primers for hsa-miR-184 (no. 4373113) or U47 snoRNA control (no. 4380911) and run per manufacturer's instructions.32
Statistical analysis
Quantitative RT-PCR error bars represent SEM assuming a 95% confidence interval. Luciferase error bars represent sample SD of the representative experiment. Quantified Western blot analysis error bars represent SEM for the stated number of experiments, and P values were calculated by Student t test and 1-way analysis of variance (ANOVA).
Results
UCB CD4+ T cells express significantly less NFAT1 protein but not mRNA compared with AB CD4+ T cells
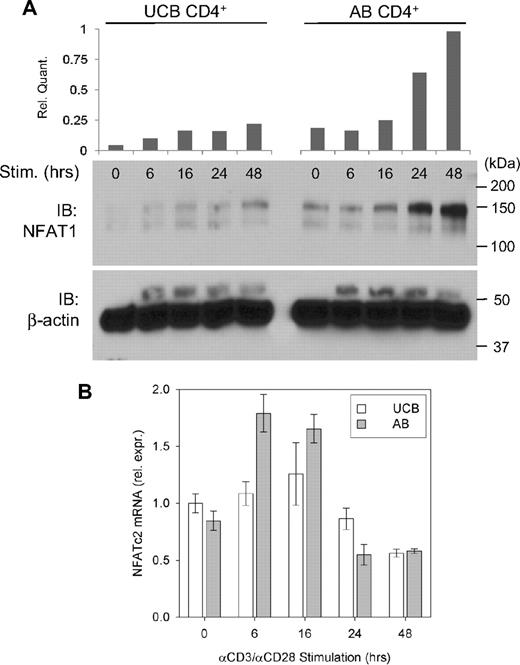
To determine the nature of the mechanism underlying reduced NFAT1 protein expression in UCB CD4+ T cells, an in vitro stimulation time course of UCB and AB CD4+ T cells was performed, and expression of NFAT1 protein and its mRNA transcript were compared. Western blot analysis comparing NFAT1 protein expression in UCB CD4+ T cells versus AB during primary stimulation confirmed reduced baseline expression and attenuated up-regulation in UCB. This discrepancy is evident throughout 48 hours of αCD3/αCD28 stimulation in vitro (Figure 1A). NFATc2 transcript levels from multiple donors were measured by qRT-PCR as described and compared between UCB and AB samples (Figure 1B). These findings reveal only modest differences in relative NFATc2 mRNA quantity, which are insufficient to account for the NFAT1 protein expression discrepancy in UCB versus AB CD4+ T cells.
Relative NFAT1 protein and mRNA expression in stimulated UCB and AB CD4+ T cells. (A) Approximately 2 × 106 isolated UCB and AB CD4+ T cells were stimulated in vitro as described for each of the designated time points and Western blot analysis performed. Data are representative of 5 independent experiments. (B) NFATc2 (NFAT1-encoding) mRNA was assayed in stimulated UCB and AB CD4+ T cells by qRT-PCR as described and normalized to UCB at 0 hours. Each bar indicates mean (± SEM) of 2 to 5 independent data points.
Relative NFAT1 protein and mRNA expression in stimulated UCB and AB CD4+ T cells. (A) Approximately 2 × 106 isolated UCB and AB CD4+ T cells were stimulated in vitro as described for each of the designated time points and Western blot analysis performed. Data are representative of 5 independent experiments. (B) NFATc2 (NFAT1-encoding) mRNA was assayed in stimulated UCB and AB CD4+ T cells by qRT-PCR as described and normalized to UCB at 0 hours. Each bar indicates mean (± SEM) of 2 to 5 independent data points.
Due to the dramatic differences in protein expression without significant corresponding differences in mRNA quantity, potential mechanisms of NFAT1 posttranscriptional regulation were investigated. Through proteasome inhibition and cellular fractionation experiments, translocation of the NFATc2 mRNA into polysomes by UCB CD4+ cells was observed to lag behind AB CD4+ T cells by at least 6 hours during in vitro stimulation (Figure S2). Because similar effects have been observed in other confirmed miRNA-mRNA interactions33,34 and our previous microarray studies had failed to reveal significant differences in translational initiation or elongation factors, we focused our subsequent work on identifying potential miRNA species that could specifically affect the translation of NFAT1 in UCB CD4+ T cells.
miR-184 is predicted to interact strongly with the 3′ UTR of the NFATc2 mRNA
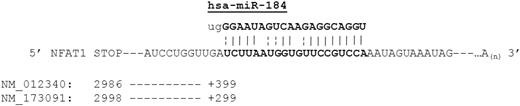
A search to determine specific miRNAs that may contribute to the observed differences in UCB CD4+ T-cell NFAT1 protein expression was conducted. Putative miRNA regulators were determined by querying the Sanger MicroCosm resource and miRBase targets registry. Many miRNA sequences predicted to bind to the 3′ UTR of NFAT1 were identified by this computational analysis (highest-scoring candidates shown in Table 1). Of the 58 and 35 predicted microRNA binders (for each transcript variant, National Center for Biotechnology Information [NCBI] accession nos. NM_012340 and NM_173091, respectively) identified by this query, the strongest predicted binder to the 3′ UTR (both variants) was miR-184, a recently characterized miRNA present in a variety of tissues35 and suggested to be of importance in DNA methylation pathways36 and a potential antagonist of miR-205.37 Conversely, the strongest predicted mRNA target of miR-184 (based on a reciprocal analysis of the miRNA sequence) was the previously identified sequence within the NFAT1 3′ UTR (Table 2). The NM_173091 variant lacks a 3′ exon containing the NM_012340 stop codon, but both variants show full homology downstream of that region. The complementary miR-184/NFATc2 sequences are diagrammed in Figure 2, with the predicted interaction occurring 399 and 299 nucleotides downstream of the stop codons, respectively.
Predicted NFATc2-interracting miRNAs
| Transcript variant (NCBI accession no.), microRNA ID . | Score . | Energy . | Base P . | Start . | End . |
|---|---|---|---|---|---|
| NM_012340 | |||||
| hsa-miR-184 | 19.7351 | −25.74 | 6.81 × 10−4 | 329 | 350 |
| hsa-miR-135a | 18.3064 | −14.69 | 7.16 × 10−3 | 89 | 111 |
| hsa-miR-494 | 18.2857 | −14.34 | 8.75 × 10−3 | 285 | 306 |
| hsa-miR-765 | 18.1568 | −25.71 | 2.32 × 10−2 | 159 | 179 |
| hsa-miR-23a | 17.8397 | −26.38 | 2.06 × 10−2 | 129 | 150 |
| hsa-miR-30b | 17.7282 | −23.31 | 3.38 × 10−2 | 173 | 194 |
| hsa-miR-29c | 17.6167 | −17.7 | 1.05 × 10−2 | 324 | 344 |
| hsa-miR-29a | 17.6167 | −17.64 | 1.01 × 10−2 | 324 | 344 |
| hsa-miR-342-5p | 17.5964 | −20.89 | 1.27 × 10−2 | 305 | 325 |
| hsa-miR-135b | 17.4188 | −13.41 | 1.57 × 10−2 | 89 | 111 |
| hsa-miR-30c-1 | 17.2822 | −18.77 | 3.15 × 10−2 | 173 | 194 |
| hsa-miR-30c-2 | 17.2822 | −17.22 | 2.72 × 10−2 | 173 | 194 |
| hsa-miR-302b | 17.1707 | −9.66 | 1.71 × 10−2 | 24 | 45 |
| hsa-miR-302d | 17.1707 | −13.13 | 1.34 × 10−2 | 24 | 45 |
| hsa-miR-29b | 17.086 | −16.53 | 1.85 × 10−2 | 323 | 344 |
| hsa-miR-29a | 17.0592 | −16.65 | 3.67 × 10−2 | 74 | 95 |
| hsa-miR-21 | 17.036 | −14.3 | 1.45 × 10−2 | 323 | 344 |
| hsa-miR-801 | 17.0057 | −26.53 | 1.09 × 10−2 | 196 | 219 |
| NM_173091 | |||||
| hsa-miR-184 | 20.1255 | −25.74 | 4.53 × 10−4 | 229 | 250 |
| hsa-miR-494 | 18.6473 | −14.34 | 6.44 × 10−3 | 185 | 206 |
| hsa-miR-765 | 18.5179 | −25.71 | 1.69 × 10−2 | 59 | 79 |
| hsa-miR-23a | 18.1925 | −26.38 | 1.51 × 10−2 | 29 | 50 |
| hsa-miR-30b | 18.0788 | −23.31 | 2.43 × 10−2 | 73 | 94 |
| hsa-miR-29c | 17.9651 | −17.7 | 7.70 × 10−3 | 224 | 244 |
| hsa-miR-29a | 17.9651 | −17.64 | 7.41 × 10−3 | 224 | 244 |
| hsa-miR-342-5p | 17.9463 | −20.89 | 8.91 × 10−3 | 205 | 225 |
| hsa-miR-30c-2 | 17.624 | −17.22 | 1.91 × 10−2 | 73 | 94 |
| hsa-miR-30c-1 | 17.624 | −18.77 | 2.22 × 10−2 | 73 | 94 |
| hsa-miR-29b | 17.4222 | −16.53 | 1.37 × 10−2 | 223 | 244 |
| hsa-miR-21 | 17.3748 | −14.3 | 1.06 × 10−2 | 223 | 244 |
| hsa-miR-801 | 17.3388 | −26.53 | 7.57 × 10−3 | 96 | 119 |
| hsa-miR-452 | 17.2829 | −13.26 | 1.75 × 10−2 | 236 | 257 |
| hsa-miR-369-5p | 17.2829 | −17.61 | 2.56 × 10−3 | 212 | 233 |
| hsa-miR-135a | 17.196 | −11.51 | 2.24 × 10−2 | 1 | 11 |
| hsa-miR-330-5p | 17.1692 | −19.06 | 4.48 × 10−2 | 195 | 217 |
| Transcript variant (NCBI accession no.), microRNA ID . | Score . | Energy . | Base P . | Start . | End . |
|---|---|---|---|---|---|
| NM_012340 | |||||
| hsa-miR-184 | 19.7351 | −25.74 | 6.81 × 10−4 | 329 | 350 |
| hsa-miR-135a | 18.3064 | −14.69 | 7.16 × 10−3 | 89 | 111 |
| hsa-miR-494 | 18.2857 | −14.34 | 8.75 × 10−3 | 285 | 306 |
| hsa-miR-765 | 18.1568 | −25.71 | 2.32 × 10−2 | 159 | 179 |
| hsa-miR-23a | 17.8397 | −26.38 | 2.06 × 10−2 | 129 | 150 |
| hsa-miR-30b | 17.7282 | −23.31 | 3.38 × 10−2 | 173 | 194 |
| hsa-miR-29c | 17.6167 | −17.7 | 1.05 × 10−2 | 324 | 344 |
| hsa-miR-29a | 17.6167 | −17.64 | 1.01 × 10−2 | 324 | 344 |
| hsa-miR-342-5p | 17.5964 | −20.89 | 1.27 × 10−2 | 305 | 325 |
| hsa-miR-135b | 17.4188 | −13.41 | 1.57 × 10−2 | 89 | 111 |
| hsa-miR-30c-1 | 17.2822 | −18.77 | 3.15 × 10−2 | 173 | 194 |
| hsa-miR-30c-2 | 17.2822 | −17.22 | 2.72 × 10−2 | 173 | 194 |
| hsa-miR-302b | 17.1707 | −9.66 | 1.71 × 10−2 | 24 | 45 |
| hsa-miR-302d | 17.1707 | −13.13 | 1.34 × 10−2 | 24 | 45 |
| hsa-miR-29b | 17.086 | −16.53 | 1.85 × 10−2 | 323 | 344 |
| hsa-miR-29a | 17.0592 | −16.65 | 3.67 × 10−2 | 74 | 95 |
| hsa-miR-21 | 17.036 | −14.3 | 1.45 × 10−2 | 323 | 344 |
| hsa-miR-801 | 17.0057 | −26.53 | 1.09 × 10−2 | 196 | 219 |
| NM_173091 | |||||
| hsa-miR-184 | 20.1255 | −25.74 | 4.53 × 10−4 | 229 | 250 |
| hsa-miR-494 | 18.6473 | −14.34 | 6.44 × 10−3 | 185 | 206 |
| hsa-miR-765 | 18.5179 | −25.71 | 1.69 × 10−2 | 59 | 79 |
| hsa-miR-23a | 18.1925 | −26.38 | 1.51 × 10−2 | 29 | 50 |
| hsa-miR-30b | 18.0788 | −23.31 | 2.43 × 10−2 | 73 | 94 |
| hsa-miR-29c | 17.9651 | −17.7 | 7.70 × 10−3 | 224 | 244 |
| hsa-miR-29a | 17.9651 | −17.64 | 7.41 × 10−3 | 224 | 244 |
| hsa-miR-342-5p | 17.9463 | −20.89 | 8.91 × 10−3 | 205 | 225 |
| hsa-miR-30c-2 | 17.624 | −17.22 | 1.91 × 10−2 | 73 | 94 |
| hsa-miR-30c-1 | 17.624 | −18.77 | 2.22 × 10−2 | 73 | 94 |
| hsa-miR-29b | 17.4222 | −16.53 | 1.37 × 10−2 | 223 | 244 |
| hsa-miR-21 | 17.3748 | −14.3 | 1.06 × 10−2 | 223 | 244 |
| hsa-miR-801 | 17.3388 | −26.53 | 7.57 × 10−3 | 96 | 119 |
| hsa-miR-452 | 17.2829 | −13.26 | 1.75 × 10−2 | 236 | 257 |
| hsa-miR-369-5p | 17.2829 | −17.61 | 2.56 × 10−3 | 212 | 233 |
| hsa-miR-135a | 17.196 | −11.51 | 2.24 × 10−2 | 1 | 11 |
| hsa-miR-330-5p | 17.1692 | −19.06 | 4.48 × 10−2 | 195 | 217 |
microRNAs predicted to interact with the 3′ UTR of each NFATc2 transcript. The Sanger microRNA database was queried for the NFATc2 transcript as described. (Last accessed July 15, 2008.) Table reflects all miRNAs scoring higher than 17.0.
Predicted miR-184 mRNA binding partners
| mRNA . | Score . | P . |
|---|---|---|
| NFATC2 | 20.1255 | 4.53 × 10−4 |
| SMPDL3B | 19.557 | 8.20 × 10−4 |
| GPBAR1 | 19.1021 | 1.05 × 10−3 |
| LMO1 | 19.0877 | 1.34 × 10−3 |
| MPL | 18.9884 | 1.48 × 10−3 |
| PSMA4 | 18.6473 | 1.19 × 10−4 |
| ABO | 18.5336 | 2.38 × 10−3 |
| THOP1 | 18.3249 | 5.10 × 10−5 |
| TPM3 | 18.1925 | 3.40 × 10−3 |
| GAS6 | 18.1856 | 3.42 × 10−3 |
| ANKRD54 | 18.1508 | 3.55 × 10−3 |
| CXYorf3 | 18.1496 | 8.49 × 10−4 |
| TCEAL4 | 18.0965 | 3.75 × 10−3 |
| GAS6 | 18.0921 | 3.77 × 10−3 |
| PZP | 18.0788 | 3.82 × 10−3 |
| C20orf196 | 18.056 | 3.92 × 10−3 |
| SIDT2 | 17.997 | 3.20 × 10−5 |
| ZBED3 | 17.9705 | 4.28 × 10−3 |
| FAM72B | 17.9651 | 4.30 × 10−3 |
| TFF3 | 17.9651 | 6.29 × 10−4 |
| mRNA . | Score . | P . |
|---|---|---|
| NFATC2 | 20.1255 | 4.53 × 10−4 |
| SMPDL3B | 19.557 | 8.20 × 10−4 |
| GPBAR1 | 19.1021 | 1.05 × 10−3 |
| LMO1 | 19.0877 | 1.34 × 10−3 |
| MPL | 18.9884 | 1.48 × 10−3 |
| PSMA4 | 18.6473 | 1.19 × 10−4 |
| ABO | 18.5336 | 2.38 × 10−3 |
| THOP1 | 18.3249 | 5.10 × 10−5 |
| TPM3 | 18.1925 | 3.40 × 10−3 |
| GAS6 | 18.1856 | 3.42 × 10−3 |
| ANKRD54 | 18.1508 | 3.55 × 10−3 |
| CXYorf3 | 18.1496 | 8.49 × 10−4 |
| TCEAL4 | 18.0965 | 3.75 × 10−3 |
| GAS6 | 18.0921 | 3.77 × 10−3 |
| PZP | 18.0788 | 3.82 × 10−3 |
| C20orf196 | 18.056 | 3.92 × 10−3 |
| SIDT2 | 17.997 | 3.20 × 10−5 |
| ZBED3 | 17.9705 | 4.28 × 10−3 |
| FAM72B | 17.9651 | 4.30 × 10−3 |
| TFF3 | 17.9651 | 6.29 × 10−4 |
Genes predicted to interact with miR-184 in humans. The Sanger microRNA database was queried for human transcripts with predicted complementary to miR-184. (Last accessed July 15, 2008.) Table reflects the top 20 results with duplicates removed.
Diagram of the predicted NFATc2 3′ UTR/hsa-miR-184 interaction. The sequence of miR-184 was retrieved from the Sanger database and the sequences for NFATc2 were retrieved from NCBI Entrez Nucleotide Sequence Viewer. Diagram indicates position of the stop codon relative to the first indexed base of each transcript and the position of the predicted 3′ UTR base pairing region relative to the stop codon.
Diagram of the predicted NFATc2 3′ UTR/hsa-miR-184 interaction. The sequence of miR-184 was retrieved from the Sanger database and the sequences for NFATc2 were retrieved from NCBI Entrez Nucleotide Sequence Viewer. Diagram indicates position of the stop codon relative to the first indexed base of each transcript and the position of the predicted 3′ UTR base pairing region relative to the stop codon.
miR-184 is more highly expressed in UCB than in AB CD4+ and decreases through early stimulation time points
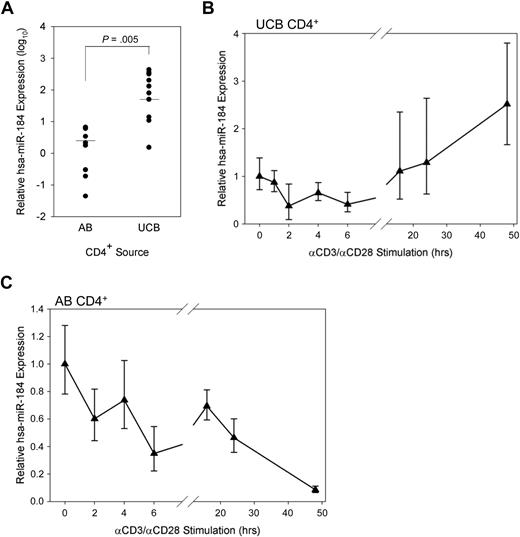
The expression of miR-184 in unstimulated UCB CD4+ T cells was on average quantified 58.4 times higher than in AB CD4+ T cells by qRT-PCR (Figure 3A). Notably, neonatal CD4+ T cells are known to contain a higher proportion of naive recent thymic emigrants than AB,38 although NFAT1 expression is lacking in both RA+ and RO+ subsets UCB and not expressed differently in either subset in AB.17 The expression of miR-184 was observed to be much more highly skewed toward the naive (CD45RA+) CD4+ subset in AB than in UCB (Figure S3).
miR-184 in UCB CD4+ T cells. (A) miR-184 was quantified in AB (n = 8) and UCB (n = 10) samples by qRT-PCR as described. P value obtained from unpaired, 2-tailed Student t test. (B) miR-184 was quantified in UCB CD4+ T cells stimulated in vitro as described for the designated time points (representative of 3 experiments). (C) miR-184 was quantified in AB CD4+ T cells stimulated in vitro as described for the designated time points (representative of 2 experiments).
miR-184 in UCB CD4+ T cells. (A) miR-184 was quantified in AB (n = 8) and UCB (n = 10) samples by qRT-PCR as described. P value obtained from unpaired, 2-tailed Student t test. (B) miR-184 was quantified in UCB CD4+ T cells stimulated in vitro as described for the designated time points (representative of 3 experiments). (C) miR-184 was quantified in AB CD4+ T cells stimulated in vitro as described for the designated time points (representative of 2 experiments).
The expression of miR-184 over a time course of in vitro simulation was also measured (Figure 3B). Subjecting isolated and stimulated UCB CD4+ T cells to the same qRT-PCR assay at early time points revealed a modest decline in miR-184 quantity (40% of original by 6 hours). Later time points at which the eventual up-regulation of NFAT1 protein expression is observable in UCB, but still dramatically lower compared with AB (Figure 1A) exhibit an eventual rebound in miRNA expression by 16 hours and modest up-regulation through 48 hours. Conversely, detectable miR-184 expression in AB CD4+ cells is observed to drop dramatically over the same time points after stimulation (Figure 3C).
miR-184 affects protein expression through its predicted binding site on the NFATc2 mRNA
To confirm the presence of the predicted binding site in the NFATc2 transcript, whole cell mRNA was isolated from UCB MNC and selected CD4+ T cells, transcribed into cDNA as described in the methods for qRT-PCR, and subjected to PCR using primers directed to sites adjacent to the stop codons and overlapping the 3′ end of the predicted binding sequence (Figure 4A). The gel bands verify the actual presence of the predicted target site within the 3′ UTR of both transcript variants. Complete sequencing of the insertions within our generated luciferase vectors further confirms this observation (Figure S1).
Interaction between the NFATc2 3′ UTR and miR-184. (A) PCR amplification of the predicted target region from UCB MNC and CD4+ cDNA. Sequences of primers are specified in the Methods section. (B) Expression of luciferase in transfected UCB CD4+ cells under the influence of minimal 3′ UTR (left column), the predicted miR-184 binding site from NFATc2 (middle column), or the cloned 3′ UTR from NFATc2 (right column) with antisense sequence to miR-184 or irrelevant control DNA sequence (representative of 3 independent experiments). (C) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, exogenous pre-miR-184, and antisense to miR-184 (representative of 3 independent experiments). N.D. indicates data not determined. (D) Expression of luciferase in transfected UCB CD4+ cells under the influence of the cloned NFATc2 3′ UTR (left column), the same UTR with the predicted miR-184 seed region (4 nucleotides [nt]) removed (middle column), and the same UTR with the entire predicted miR-184 binding site removed (right column) with and without antisense to miR-184 (representative of 2 independent experiments). (E) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, with and without precursor to miR-184 (representative of 2 independent experiments).
Interaction between the NFATc2 3′ UTR and miR-184. (A) PCR amplification of the predicted target region from UCB MNC and CD4+ cDNA. Sequences of primers are specified in the Methods section. (B) Expression of luciferase in transfected UCB CD4+ cells under the influence of minimal 3′ UTR (left column), the predicted miR-184 binding site from NFATc2 (middle column), or the cloned 3′ UTR from NFATc2 (right column) with antisense sequence to miR-184 or irrelevant control DNA sequence (representative of 3 independent experiments). (C) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, exogenous pre-miR-184, and antisense to miR-184 (representative of 3 independent experiments). N.D. indicates data not determined. (D) Expression of luciferase in transfected UCB CD4+ cells under the influence of the cloned NFATc2 3′ UTR (left column), the same UTR with the predicted miR-184 seed region (4 nucleotides [nt]) removed (middle column), and the same UTR with the entire predicted miR-184 binding site removed (right column) with and without antisense to miR-184 (representative of 2 independent experiments). (E) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, with and without precursor to miR-184 (representative of 2 independent experiments).
To determine whether miR-184 indeed interacts with the corresponding NFATc2 sequence as predicted (Figure 2), expression vectors designed to transcribe a luciferase-encoding mRNA containing either a short synthetically prepared sequence matching only the predicted miR-184 binding site from NFATc2 (“184-only”) or the NFATc2 (NM_012340 variant) 3′ UTR through and including the aforementioned sequence (“cloned UTR”) were constructed. The full-length NFATc2 3′ UTR has not been fully cloned or sequenced, and we wished to avoid the effects of any possible variations in UTR length between samples or cell types. These vectors were introduced into UCB (Figure 4B) and AB (Figure 4C) selected CD4+ T cells. Luciferase assays indicate only 38% and 60% expression compared with control when luciferase expression is influenced by the 184-only sequence and the cloned UTR, respectively (Figure 4B). This effect is almost completely reversed when a blocking antisense sequence to miR-184 is cotransfected. However in AB, the 184-only insertion has no effect on luciferase activity (middle column, white bar), whereas insertion of the cloned UTR results in a 58% reduction in expression (Figure 4C). Transfection of an exogenous precursor to miR-184 results in decreases of 61% and 23%, respectively (middle bars). This effect is again attenuated when a blocking antisense sequence is cotransfected with the miR-184 precursor (rightmost bars). These data indicate that miR-184 does indeed modulate protein expression through its predicted binding site and suggests significant endogenous miR-184 activity in UCB but not AB CD4+ T cells.
To further confirm these observations, we mutated the cloned UTR luciferase expression vector insert used previously to disrupt either the seed region (by removing the 4 3′-most nucleotides from its predicted binding site; “-seed region”) or the entire predicted binding site (“-184 site”). Base pair binding within the seed region has been shown to often be of importance to translational repression by miRNA.39 We observed a 4- and 3-fold induction, respectively, of luciferase expression in UCB CD4+ cells when either the seed region or the miR-184 binding site was removed (Figure 4D). However, we observed a comparatively slight (1.5-fold) increase in expression when the same vectors were transfected into AB CD4+ cells (Figure 4E). Failure to observe a significant change in luciferase expression with inhibition of miR-184 activity in UCB or introduction of miR-184 in AB (gray bars) confirmed the specificity of the observed effect.
miR-184 activity directly affects NFAT1 protein quantity in unstimulated CD4+ T cells
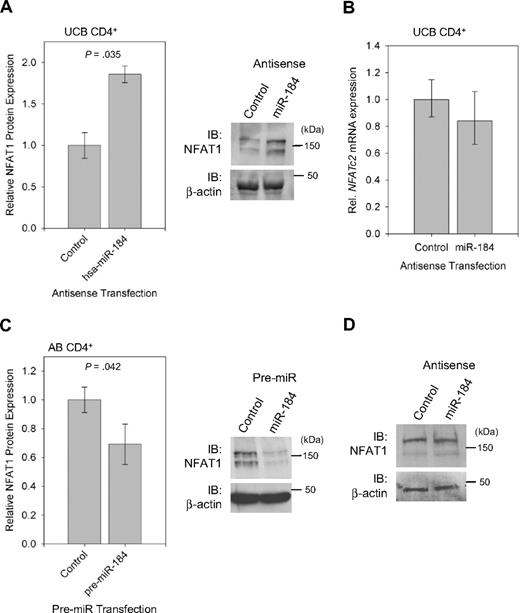
To determine whether endogenous miR-184 can directly repress NFAT1 protein expression in UCB CD4+ T cells, Western blot analysis for NFAT1 was performed on unstimulated selected CD4+ T cells after transfection with either control or blocking antisense to miR-184 (Figure 5A right). Band intensities were quantified and normalized to β-actin, and relative NFAT1 expression under the influence of each treatment was compared. Aggregate data (Figure 5A left) reveal an 86% increase in NFAT1 protein expression when the cells were treated with antisense to miR-184. NFATc2 mRNA levels (Figure 5B) were however unchanged between samples. Likewise, when unstimulated AB CD4+ T cells are transfected with a synthesized precursor to miR-184 (Figure 5C), NFAT1 protein levels as quantified by Western blot analysis are reduced by approximately 31%. However, interference with miR-184 in AB CD4+ cells failed to yield an observable change in NFAT1 protein expression (Figure 5D). This series of experiments indicates negative regulation of NFAT1 protein through the microRNA pathway by miR-184 in UCB CD4+ T cells and further suggests a nondegrading mechanism of action.
miR-184 negatively effects NFAT1 protein synthesis. (A) Quantification and representative blot of NFAT1 protein expression in UCB CD4+ T cells 16 hours after transfection with antisense to miR-184 (n = 3). (B) qRT-PCR analysis of samples in (A), confirming no significant change in NFATc2 mRNA quantity. (C) Quantification and representative blot of NFAT1 protein expression in AB CD4+ T cells under the following transfection with pre-miR-184 (n = 4). Blot bands were quantified using ImageJ software. (D) Western blot analysis of NFAT1 protein expression in AB CD4+ T cells 16 hours after transfection with antisense to miR-184.
miR-184 negatively effects NFAT1 protein synthesis. (A) Quantification and representative blot of NFAT1 protein expression in UCB CD4+ T cells 16 hours after transfection with antisense to miR-184 (n = 3). (B) qRT-PCR analysis of samples in (A), confirming no significant change in NFATc2 mRNA quantity. (C) Quantification and representative blot of NFAT1 protein expression in AB CD4+ T cells under the following transfection with pre-miR-184 (n = 4). Blot bands were quantified using ImageJ software. (D) Western blot analysis of NFAT1 protein expression in AB CD4+ T cells 16 hours after transfection with antisense to miR-184.
miR-184 activity conversely affects production of the NFAT-associated IL-2 transcript
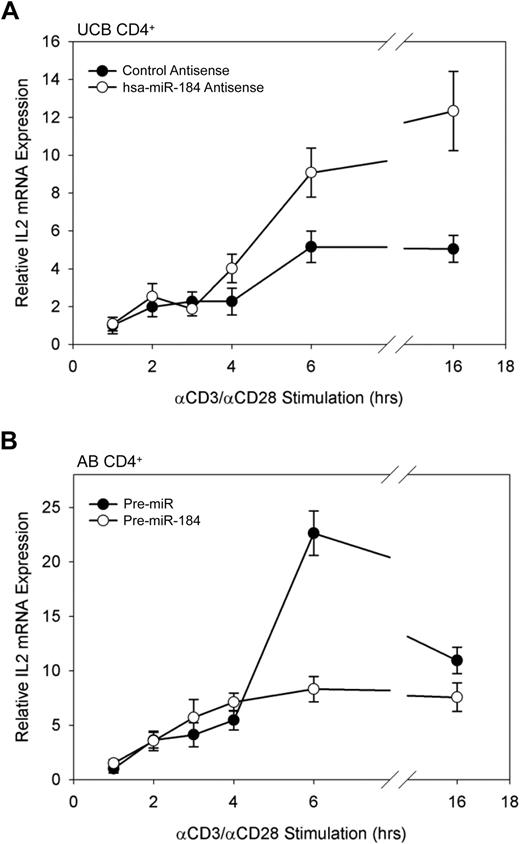
To determine whether manipulation of NFAT1 protein levels through interference with the activity of miR-184 is sufficient to result in an increase in transcription of NFAT1-target genes, the transcription of IL-2, a gene strongly activated by NFAT1 binding to its promoter after stimulation,14 was assayed by qRT-PCR. UCB CD4+ cells were transfected with blocking antisense to miR-184, assayed for up-regulated NFAT1 expression by Western blot analysis (data included in Figure 5A) after 16 hours, and then stimulated in vitro as described previously. Data reflect a significantly greater amount of IL-2 mRNA in the miR-184 antisense-treated samples at 6 hours of stimulation. This increase is maintained through 16 hours (Figure 6A). Conversely, AB CD4+ T cells transfected with an exogenous miR-184 precursor exhibited dramatically reduced IL-2 transcription through the same stimulation time points (Figure 6B). These findings indicate that miR-184 interference with NFAT1 in UCB CD4+ T cells is indeed sufficient to influence both NFAT1 protein levels and transcription of the known NFAT1 target gene, IL2.
IL2 transcription in stimulated CD4+ T cells. (A) IL2 transcription in stimulated UCB CD4+ T cells under the influence of miR-184 antisense. (B) IL2 transcription in stimulated AB CD4+ T cells after transfection of exogenous miR-184 precuror. Cells were stimulated in vitro 16 hours after transfection, and IL2 mRNA was assayed by qRT-PCR as described from 106 cells per data point. Data are representative of 2 independent experiments.
IL2 transcription in stimulated CD4+ T cells. (A) IL2 transcription in stimulated UCB CD4+ T cells under the influence of miR-184 antisense. (B) IL2 transcription in stimulated AB CD4+ T cells after transfection of exogenous miR-184 precuror. Cells were stimulated in vitro 16 hours after transfection, and IL2 mRNA was assayed by qRT-PCR as described from 106 cells per data point. Data are representative of 2 independent experiments.
Discussion
Our findings comprise the first observation of miR-184 activity in immune cells and a characterization of its activity on a key transcriptional regulator of inflammation specifically known to exhibit decreased activity in UCB CD4+ T cells. We identified miR-184 as a strong predicted regulator of NFAT1 and confirmed its interaction with the observed complementary binding site within the NFATc2 mRNA 3′ UTR. UCB CD4+ T cells were shown to exhibit significantly more miR-184 RNA and miR-184–mediated repressive activity than AB CD4+ T cells. We additionally confirmed through blocking and gain-of-function analyses that manipulation of miR-184 is sufficient to influence NFAT1 protein as well as its known downstream target, IL-2.
miRNA regulation of various myeloid and lymphoid lineage differentiation steps, as well as key signaling components including transcription factors within specific immune cells, has been recently described by multiple groups. Notable targets of miRNAs involved in lymphocyte activation include SHP-2 and multiple phosphatases associated with T cell receptor (TCR) signal transduction,40 TRAF6, IRAK1,41 and c-Myb.42 Additional studies have revealed dramatic immune phenotypes in specific miRNA-knockout mice, such as impaired T cell–dependent antibody response and Th2 skewing.27 Herein we present the first known evidence of NFAT targeting by a miRNA. Although the full implications of our findings with respect to immune cell function remain to be fully elucidated, particularly with respect to the naive and immunotolerant phenotype exhibited by UCB lymphocytes and the associated clinical observations for allogeneic stem cell transplantation in humans, it is clear that an understanding of miRNA targeting mechanisms will further a comprehensive understanding of immune cell activation.
miR-184 was first observed by Lagos-Quintana et al in the murine eye43 ; later, its expression was shown to be localized to basal and suprabasal regions of the corneal epithelium.44 miR-184 was later described in the human brain as being regulated by the methyl-CpG binding protein MeCP2.36 More recent studies have linked its overexpression in squamous cell carcinoma of the tongue and regulation of c-Myc in those cell lines,45 suggesting that a dysregulation of miR-184 leading to its overexpression may be associated with an increase in cellular proliferation. This hypothesis would agree with our earlier findings of increased in vitro proliferation of UCB CD4+ compared with AB CD4+ cells in response to antigen, as well as reports suggesting reduced apoptosis in UCB CD4+ T cells in response to primary stimulation.46 Yu et al have recently identified a role for miR-184 in antagonizing miR-205, leading to inhibition of blocking SHIP2 expression by miR-205,37 suggesting a possible proapoptotic role for miR-184 as well. However, the contribution of miR-184 to the proliferative phenotype observed in UCB CD4+ cells, either through NFAT1 inhibition or other mechanisms, remains to be investigated.
The mechanism we describe for miR-184 regulation of NFAT1 in UCB CD4+ T cells suggests a relatively high sustained expression of the miRNA in the absence of stimulation. miR-184 is located on chromosome 15, and how miR-184 expression is regulated remains unclear, however earlier reports in neural tissue link its expression to the release of promoter-bound MeCP2 due to phosphorylation after polarization in mice.36 Although DNA methylation patterns in the neonatal immune system are still largely unstudied, it is important to note that DNA methylation has been linked with the expression of many other miRNA species in humans.47-49 As expected, we observed a decrease in levels of miR-184 relative to NFATc2 mRNA at early time points after stimulation. Although this decrease is relatively modest, we observed a significant effect on both NFAT1 protein and downstream target gene expression after antisense blocking of miR-184. The mechanism underlying increased NFAT1 protein by 24 to 48 hours of stimulation in UCB in the midst of rebounding miR-184 expression at those time points remains an open question, although the relative increase in NFAT1 protein expression by these time points generally exceeds the relative observed increase in miR-184 expression and the kinetics of protein up-regulation are significantly delayed compared with AB CD4+ cells. Data from sucrose-gradient fractionation experiments (Figure S2) do suggest NFATc2 translocation to polysomal fractions by 16 hours. It is possible that miR-184 later assumes another function in the presence of NFAT1 protein and its downstream transcriptional targets, and it is likely that additional microRNAs predicted to interact with the NFATc2 mRNA 3′ UTR additionally play a role in these processes. Various groups have published data suggesting “rheostat” mechanisms for fine-tuning protein expression that are highly dependent on the expression of other miRNAs and other competitive mRNA targets over time (reviewed in Bartel and Chen50 ), resulting in dynamic changes in the relative regulatory function of each individual miRNA-mRNA interaction. Thus, it may be hypothesized that any one of several mechanisms may permit eventual up-regulation of NFAT1 protein, even in the midst of sustained miR-184 expression in UCB CD4+ cells. Multiple miRNA species are predicted to bind sites proximal to and overlapping with the miR-184 binding site, therefore a miRNA yet to be identified may modulate the initiation of NFAT1 protein translation at later time points. In AB CD4+ cells, it is foreseeable that the already higher expression of NFAT1 protein (Figure 1A) and the relatively lower expression of miR-184 at rest (Figure 3A) and its decrease in expression over stimulation (Figure 3C), combined with increasing expression of NFATc2 mRNA at early stimulation time points (Figure 1B) may be sufficient to permit efficient NFAT1 translation without miR-184 exhibiting a major regulatory role. Future investigations into the expression of additional UCB-associated miRNAs may elucidate potential miRNA expression thresholds in those cells with consequences for NFAT1 protein expression. In addition, although other pathways of posttranscriptional regulation have been implicated for NFAT1 in other cell lineages, specifically protein degradation through ubiquitin-mediated proteasome targeting in breast cancer cells,51 our work has not identified enhanced NFAT1 protein degradation to account for reduced expression in UCB CD4+ T cells.
We have not observed NFATc2 transcript degradation mediated by miR-184. Translational regulation of mRNA transcripts through microRNA interference as an alternate mechanism to transcript degradation is a subject of growing interest and inquiry. Multiple mechanisms have been suggested, including blocking of the initiation factor eIF4E binding to the 5′ m7G cap,52 cap-independent blocking of initiation,53 and cotranslational mechanisms,53-55 such as inhibition of ribosome recycling, elongation, favoring early termination, or influencing cotranslational degradation of the nascent protein. Importantly, recent studies have also revealed that miRNA may also positively regulate translation under specific conditions,56 further suggesting multiple complex mechanisms by which miRNA may play a role in translation. It is clear that some of these mechanisms involve “pseudopolysomes” and/or P-bodies, sites of reversible sequestration of targeted transcripts in association with miRNA and other ribonucleoprotein complexes.19 In light of these complex and sometimes disparate observations, our studies provide insight into the contribution of the specific miR-184/NFATc2 binding site on its own as well as in the context of the preceding UTR sequence. As is evident from our luciferase assays and other studies, the full 3′ UTR sequence likely contains multiple features that positively and negatively influence protein expression at rest.
Additional predicted miR-184–interacting transcripts include the transcription factor LMO1 associated with T-cell leukemia and the extensively characterized oncogene MPL, as well as a host of genes not yet characterized in immune cells but in some cases not tightly restricted to particular cell lineages (Table 2). We have observed MPL to be expressed significantly lower in UCB CD4+ cells compared with AB by microarray (S.K., B. Kaminski, R. Miller, unpublished data, May 2003); however, due to the complex and poorly understood nature of degradative versus nondegradative mechanisms involved in miRNA/mRNA interactions, it is clear that analyses based on simple mRNA quantification may fail to elucidate the true role of miRNA. It is often assumed that weaker complementarity in general results in a less degradative fate for the mRNA, but it is unlikely that a discrete binding affinity threshold exists for transcript degradation across miRNAs, transcripts, and cell types.
As specific miRNAs and miRNA families are currently being proposed as biomarkers for various cancers, future implications for this work may include graft selection, ex vivo graft manipulation, and targeted therapy of hematologic malignancies. Pharmacologic targeting of NFAT1 as a key regulator of the autoimmune/inflammatory response is well described: CsA and FK506 (tacrolimus), which separately disrupt the calcineurin/calmodulin signaling interaction preventing NFAT1 nuclear translocation, have been used as therapeutic prophylaxis and treatment for aGVHD for decades. However, these treatments exhibit toxic calcineurin/NFAT-independent side effects, particularly in the nephritic and circulatory systems, in addition to the traditional infection considerations of immunosuppressive therapy. GVHD remains the major limitation to successful allogeneic transplantation, and directed molecular T-cell targeting therapies could potentially alleviate some significant toxicities of pharmacologic immunosuppressive calcineurin inhibitors.
In summary, miR-184 is capable of regulating NFAT1 protein expression without causing transcript degradation through its predicted complementary binding site within the NFATc2 mRNA 3′ UTR. This process in UCB CD4+ T cells may comprise one mechanism underlying the relatively low levels of expressed NFAT1 protein compared with AB CD4+ cells at rest and early stimulation time points, resulting in their characteristically lower expression of proinflammatory cytokines upon activation. We have observed that UCB CD4+ T cells endogenously express significantly greater amounts of miR-184, and our studies have elucidated a previously uncharacterized role for miR-184 in the early adaptive immune response. Taken together, we have identified a key molecular difference between CD4+ T lymphocytes derived from adults and neonates with potential implications for our further understanding of autoimmunity and GVHD.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank W. Merrick, D. Baus, and the Case Western Reserve University Gene Expression and Genotyping Core for technical assistance. We also thank H. Meyerson, C. Harding, Y.-C. Yang, and A. Levine for their advice and review.
This work was supported by National Institute of Allergy and Infectious Diseases Grant R01-AI47289-01 (M.J.L.), National Cancer Institute Grant 5T32-CA059366-13 Research Oncology Training Grant (M.L.L.), the Gene Expression and Genotyping Facility of the Case Comprehensive Cancer Center 5P30-CA043703 (ClinicalTrials.gov identifier NCT00003335), the Abraham J. and Phyllis Katz Cord Blood Foundation, the Fannie E. Rippel Foundation, and the Dr Donald and Ruth Weber Goodman Philanthropic Fund (M.J.L.).
National Institutes of Health
Authorship
Contribution: R.P.W. designed and performed the experiments, analyzed data, and wrote the manuscript; M.L.L. and P.H. analyzed data and provided vital reagents; S.K. designed experiments and helped write the manuscript; P.L. designed experiments and provided vital reagents; and N.J.G and M.J.L. secured funding, designed experiments, supervised the research, and helped write the manuscript.
Conflict-of-interest disclosure: M.J.L. receives research support from the Abraham J. and Phyllis Katz Cord Blood Foundation. The remaining authors declare no competing financial interests.
Correspondence: Mary J. Laughlin, Case Western Reserve University, Cleveland Cord Blood Center, 10900 Euclid Ave, Wolstein Research Bldg (WRB) 2-129, Cleveland, OH 44106-7284; e-mail: Mary.Laughlin@case.edu.

![Figure 4. Interaction between the NFATc2 3′ UTR and miR-184. (A) PCR amplification of the predicted target region from UCB MNC and CD4+ cDNA. Sequences of primers are specified in the Methods section. (B) Expression of luciferase in transfected UCB CD4+ cells under the influence of minimal 3′ UTR (left column), the predicted miR-184 binding site from NFATc2 (middle column), or the cloned 3′ UTR from NFATc2 (right column) with antisense sequence to miR-184 or irrelevant control DNA sequence (representative of 3 independent experiments). (C) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, exogenous pre-miR-184, and antisense to miR-184 (representative of 3 independent experiments). N.D. indicates data not determined. (D) Expression of luciferase in transfected UCB CD4+ cells under the influence of the cloned NFATc2 3′ UTR (left column), the same UTR with the predicted miR-184 seed region (4 nucleotides [nt]) removed (middle column), and the same UTR with the entire predicted miR-184 binding site removed (right column) with and without antisense to miR-184 (representative of 2 independent experiments). (E) Expression of luciferase in transfected AB CD4+ cells under the influence of the aforementioned 3′ UTRs, with and without precursor to miR-184 (representative of 2 independent experiments).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/26/10.1182_blood-2008-09-181156/4/m_zh89990935860004.jpeg?Expires=1769940135&Signature=0ISdg687MIxajBKbXTRsZlHxUL83dKrsVb8j-mlkA83s9ldip2YuNm1TnnRXjyUyQmJjyReF4cfcIiC~~gjnCqxrtEZNL2SuSLvzthvMwOVTIjMQl16skDwbHhwRGC1tVfKZC3YFO7YUWsCQK1j-4BIjCwJDs6QWq1qmq9UQL9Lw0TUmqGhJ4LOI-oKINLeCUW-9i6YUZQqROFCzh1QzHpbMSiofO~ce~yAIsAFUXkA8o7SRR-v44pcUR6uQBhSD1Wx1Ei5VKBXkljrym2ehH2FuUi3TnGIA5IXJZz9MpDdTmAYObMBHcDONHBKq6idtzqA0EcQkccG7I9BfQ7AnFQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal