Abstract
We report on prognostic implications for postrelapse survival (PRS) of a gene expression profiling (GEP)–defined risk score at relapse available in 120 myeloma patients previously enrolled in tandem transplantation trial Total Therapy 2. Among the 71 patients with additional GEP baseline information, 3-year PRS was 71% in 40 patients with low risk present both at baseline and relapse contrasting with only 17% in 28 patients with high risk at relapse, 12 of whom with baseline low-risk status fared better than the remainder (P = .08). On multivariate analysis of relapse parameters available in 104 patients, high risk conferred short PRS (hazard ratio = 4.00, P < .001, R2 = 33%), whereas relapse hyperdiploidy predicted long PRS (hazard ratio = 0.37, P = .022, cumulative R2 = 41%). In case the initial partial response lasted less than 2 years, relapse low-risk identified 26 patients with superior 3-year PRS of 61% versus 9% among 32 with relapse high-risk (P < .001). Based on its PRS predictive power, GEP analysis should be an integral part of new agent trials in search of better therapy for high-risk myeloma.
Introduction
We have previously reported on the discriminatory power of gene expression profiling (GEP) of highly purified plasma cells from patients with multiple myeloma. Thus, developed in Total Therapy 2 (TT2) and validated in Total Therapy 3 (TT3),1 the approximately 15% of patients presenting with GEP-defined high-risk multiple myeloma had a vastly inferior overall survival and event-free survival than their low-risk counterparts. These prognostic implications have since been validated in high-dose therapy programs by the Mayo Clinic group2 and in a salvage trial using dexamethasone or bortezomib.3 We are now reporting on the prognostic implications of such GEP studies performed at the time of relapse in patients initially enrolled in TT2.
Methods
The details of the TT2 regimen and clinical outcomes have been reported previously.4,5 Here we focus on 120 of a total of 276 relapsed patients for whom GEP studies were performed at the time of relapse; 71 of the 120 had such data also available before initiation of TT2, compared with 51 in our previous report,6 with an additional 15 months of follow-up. In keeping with the original study design, the current evaluations had been preplanned, and all patients had signed an informed consent in keeping with institutional, Food and Drug Administration, and Helsinki Declaration guidelines. The protocol had been approved by the Institutional Review Board of the University of Arkansas for Medical Sciences. A federally accredited audit team had reviewed the charts of approximately two-thirds of all patients enrolled for protocol adherence, response, and toxicity.
Kaplan-Meier methods were used to generate survival distribution graphs,7 and comparisons were made using the log-rank test.8 Running log-rank tests were used to determine optimal cut points for durations of response.9 Multivariate analyses applied stepwise selection and Cox proportional hazard regression modeling.10
Results and discussion
Of 668 patients initially enrolled in TT2, 405 had an event and 287 had died as of January 25, 2009. GEP data were available in 120 at relapse, of whom 71 also had such investigations performed at baseline. Their characteristics at relapse and at baseline are depicted in Table 1. Considering all 120 patients, relapse features included greater proportions of patients with cytogenetic abnormalities, older age (≥ 65 years), and low platelet counts (< 150 000/μL), whereas low serum albumin (< 3.5 g/dL), high C-reactive protein (≥ 8 mg/L), and low hemoglobin levels (< 10 g/dL) were underrepresented at relapse. Some of these differences also extended to the 71 patients for whom, in addition to standard laboratory variables, GEP characteristics were available. Among the latter, the high-risk category was overrepresented at relapse (Table 1). Subgroup designations were maintained in all 5 CD-1, 3 of 4 CD-2, all 3 MF, 14 of 15 MS, and 15 of 21 hyperdiploidy subgroup patients. Seven of 8 patients with baseline PR annotation relapsed with this characteristic; an additional 10 evolved from other subgroups, including 3 from the hyperdiploidy subgroup. TP53 deletion status, gleaned from expression levels,11 was observed more frequently at relapse than at baseline (23% vs 8%, P = .014 when all patients were considered; 19% vs 8%, P = .067 among the 71 with paired observations).
Patient characteristics at baseline and at relapse
| Factor . | Overall . | Paired samples . | ||||
|---|---|---|---|---|---|---|
| Baseline, n/N (%) . | Relapse, n/N (%) . | P . | Baseline, n/N (%) . | Relapse, n/N (%) . | P . | |
| Age ≥ 65 y | 25/120 (21) | 39/120 (33) | .041 | 13/71 (18) | 23/71 (32) | .054 |
| Albumin < 3.5 g/dL | 28/120 (23) | 9/115 (8) | .001 | 14/71 (20) | 6/70 (9) | .058 |
| B2M ≥ 3.5 mg/L | 55/120 (46) | 41/103 (40) | .365 | 35/71 (49) | 25/63 (40) | .264 |
| B2M ≥ 5.5 mg/L | 23/120 (19) | 16/103 (16) | .476 | 15/71 (21) | 11/63 (17) | .592 |
| CRP ≥ 8 mg/L | 45/117 (38) | 6/105 (6) | < .001 | 20/69 (29) | 4/63 (6) | < .001 |
| LDH ≥ 190 U/L | 36/120 (30) | 37/108 (34) | .491 | 20/71 (28) | 18/63 (29) | .959 |
| Hb < 10 g/dL | 41/120 (34) | 21/112 (19) | .008 | 27/71 (38) | 12/67 (18) | .009 |
| Platelets < 150 × 109/μL | 30/120 (25) | 64/112 (57) | < .001 | 18/71 (25) | 39/67 (58) | < .001 |
| Cytogenetic abnormalities | 50/118 (42) | 72/106 (68) | < .001 | 36/70 (51) | 51/68 (75) | .004 |
| GEP high-risk | 15/71 (21) | 46/120 (38) | .014 | 15/71 (21) | 28/71 (39) | .018 |
| GEP CD-1 | 5/71 (7) | 5/104 (5) | .532* | 5/71 (7) | 5/67 (7) | .924 |
| GEP CD-2 | 4/71 (6) | 6/104 (6) | .970* | 4/71 (6) | 3/67 (4) | .757* |
| GEP HY | 21/71 (30) | 21/104 (20) | .153 | 21/71 (30) | 17/67 (25) | .581 |
| GEP LB | 3/71 (4) | 0/104 (0) | .034* | 3/71 (4) | 0/67 (0) | .089* |
| GEP MF | 3/71 (4) | 5/104 (5) | .856* | 3/71 (4) | 3/67 (4) | .942* |
| GEP MS | 17/71 (24) | 26/104 (25) | .873 | 17/71 (24) | 17/67 (25) | .846 |
| GEP MY | 9/71 (13) | 9/104 (9) | .390 | 9/71 (13) | 5/67 (7) | .311 |
| GEP PR | 9/71 (13) | 32/104 (31) | .006 | 9/71 (13) | 17/67 (25) | .057 |
| TP53 deletion (Affymetrix expression < 727) | 6/71 (8) | 24/106 (23) | .014 | 6/71 (8) | 13/68 (19) | .067 |
| Factor . | Overall . | Paired samples . | ||||
|---|---|---|---|---|---|---|
| Baseline, n/N (%) . | Relapse, n/N (%) . | P . | Baseline, n/N (%) . | Relapse, n/N (%) . | P . | |
| Age ≥ 65 y | 25/120 (21) | 39/120 (33) | .041 | 13/71 (18) | 23/71 (32) | .054 |
| Albumin < 3.5 g/dL | 28/120 (23) | 9/115 (8) | .001 | 14/71 (20) | 6/70 (9) | .058 |
| B2M ≥ 3.5 mg/L | 55/120 (46) | 41/103 (40) | .365 | 35/71 (49) | 25/63 (40) | .264 |
| B2M ≥ 5.5 mg/L | 23/120 (19) | 16/103 (16) | .476 | 15/71 (21) | 11/63 (17) | .592 |
| CRP ≥ 8 mg/L | 45/117 (38) | 6/105 (6) | < .001 | 20/69 (29) | 4/63 (6) | < .001 |
| LDH ≥ 190 U/L | 36/120 (30) | 37/108 (34) | .491 | 20/71 (28) | 18/63 (29) | .959 |
| Hb < 10 g/dL | 41/120 (34) | 21/112 (19) | .008 | 27/71 (38) | 12/67 (18) | .009 |
| Platelets < 150 × 109/μL | 30/120 (25) | 64/112 (57) | < .001 | 18/71 (25) | 39/67 (58) | < .001 |
| Cytogenetic abnormalities | 50/118 (42) | 72/106 (68) | < .001 | 36/70 (51) | 51/68 (75) | .004 |
| GEP high-risk | 15/71 (21) | 46/120 (38) | .014 | 15/71 (21) | 28/71 (39) | .018 |
| GEP CD-1 | 5/71 (7) | 5/104 (5) | .532* | 5/71 (7) | 5/67 (7) | .924 |
| GEP CD-2 | 4/71 (6) | 6/104 (6) | .970* | 4/71 (6) | 3/67 (4) | .757* |
| GEP HY | 21/71 (30) | 21/104 (20) | .153 | 21/71 (30) | 17/67 (25) | .581 |
| GEP LB | 3/71 (4) | 0/104 (0) | .034* | 3/71 (4) | 0/67 (0) | .089* |
| GEP MF | 3/71 (4) | 5/104 (5) | .856* | 3/71 (4) | 3/67 (4) | .942* |
| GEP MS | 17/71 (24) | 26/104 (25) | .873 | 17/71 (24) | 17/67 (25) | .846 |
| GEP MY | 9/71 (13) | 9/104 (9) | .390 | 9/71 (13) | 5/67 (7) | .311 |
| GEP PR | 9/71 (13) | 32/104 (31) | .006 | 9/71 (13) | 17/67 (25) | .057 |
| TP53 deletion (Affymetrix expression < 727) | 6/71 (8) | 24/106 (23) | .014 | 6/71 (8) | 13/68 (19) | .067 |
n indicates number with factor; N, number with valid data for factor; B2M, β2-microglobulin; CRP, C-reactive protein; and LDH, lactic dehydrogenase.
Sample size assumption for χ2 test is suspect.
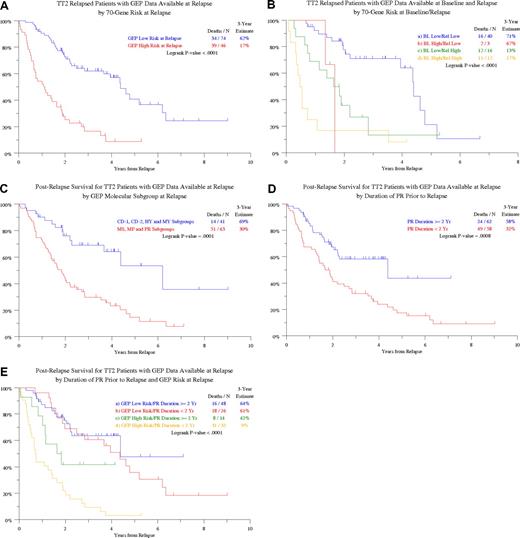
Postrelapse survival (PRS) was similar among the patients with and without GEP data at relapse (data not shown). The 3-year PRS estimate was significantly lower (17% vs 62%, P < .001) among the 46 patients relapsing with high-risk compared with the 74 relapsing with low-risk GEP features (Figure 1A). Among the 71 patients with both baseline and relapse GEP data, the best outcome was observed in the 40 patients with low-risk designation at both time points with a 3-year PRS estimate of 71% (Figure 1B). Among the 28 patients with high-risk values at relapse, the 16 with low-risk myeloma at baseline fared better than the 12 with high-risk disease before initial therapy (P = .08). Two of only 3 patients with baseline high-risk myeloma who relapsed with a low-risk score have died. In the context of molecular subgroup designation at relapse, available for 104 of the 120 patients with relapse GEP data, PR (n = 32), MS (n = 26), and MF assignments (n = 5) together constituted a poor-risk category of 63 patients with 3-year PRS of 30% as opposed to 69% in the remaining 41 patients (P < .001; Figure 1C).
Postrelapse survival is plotted from the time of relapse according to various features present at baseline or relapse. (A) Postrelapse survival (PRS) by GEP-defined risk at relapse. PRS was shorter among patients with GEP-defined high-risk status at relapse. (B) PRS by GEP-defined risk in the context of GEP data at both baseline before therapy and at relapse. PRS was superior in the absence of high risk at both observation times. Among the 28 patients relapsing with high-risk status, the presence of high risk also at baseline identified 12 patients with very poor prognosis. P values are as follows: a versus b, P = .029; b versus c, P = .808; a versus c, P = .004; b versus d, P = .199; a versus d, P < .001; c versus d, P = .079. (C) PRS by GEP-defined molecular subgroup designation at relapse. PRS was significantly shorter among patients with MS, MF, and PR (proliferation) designations. (D) PRS according to the duration of the preceding partial response (PR). PRS is significantly shorter in case the duration of preceding partial response was shorter than 2 years. (E) PRS according to the duration of the preceding partial response (PR) in the context of GEP-derived risk designation at relapse. P values are as follows: a versus b, P = .474; b versus c, P = .098; a versus c, P = .052; b versus d, P < .001; a versus d, P < .001; c versus d, P = .026.
Postrelapse survival is plotted from the time of relapse according to various features present at baseline or relapse. (A) Postrelapse survival (PRS) by GEP-defined risk at relapse. PRS was shorter among patients with GEP-defined high-risk status at relapse. (B) PRS by GEP-defined risk in the context of GEP data at both baseline before therapy and at relapse. PRS was superior in the absence of high risk at both observation times. Among the 28 patients relapsing with high-risk status, the presence of high risk also at baseline identified 12 patients with very poor prognosis. P values are as follows: a versus b, P = .029; b versus c, P = .808; a versus c, P = .004; b versus d, P = .199; a versus d, P < .001; c versus d, P = .079. (C) PRS by GEP-defined molecular subgroup designation at relapse. PRS was significantly shorter among patients with MS, MF, and PR (proliferation) designations. (D) PRS according to the duration of the preceding partial response (PR). PRS is significantly shorter in case the duration of preceding partial response was shorter than 2 years. (E) PRS according to the duration of the preceding partial response (PR) in the context of GEP-derived risk designation at relapse. P values are as follows: a versus b, P = .474; b versus c, P = .098; a versus c, P = .052; b versus d, P < .001; a versus d, P < .001; c versus d, P = .026.
We next examined whether PRS was affected by the duration of response before relapse. Applying running log-rank statistical methodology, PRS was significantly longer when partial response status was maintained for at least 2 years as opposed to an earlier relapse scenario (Figure 1D). Considering both partial response duration before relapse and GEP risk designation at relapse, PRS was similar in the 74 patients with low-risk features regardless of prior response duration. However, for the 46 with high-risk myeloma, the 14 with prerelapse response duration of at least 2 years had a 3-year PRS estimate of 42% compared with 9% among the 32 patients, in whom a partial response status had not been maintained for this duration (P = .026; Figure 1E).
Among 11 variable categories examined at baseline before initiation of the TT2 program, only the high-risk GEP feature was associated with inferior PRS (Table 2). Consideration of the same 11 parameter categories at relapse revealed adverse implications, on univariate analysis, for renal insufficiency (creatinine > = 2 mg/dL), anemia (hemoglobin < 10 g/dL), cytogenetic abnormalities, GEP-defined high-risk, and the MF/MS/PR subgroup combination, whereas the hyperdiploidy designation was prognostically favorable. Sustaining partial response status for at least 2 years was an additional favorable variable for PRS. The observed differences in PRS among patients relapsing with low-risk versus high-risk myeloma were unrelated to initial CR rate (53% vs 41%, P = .224) and to the salvage regimens used, except for more frequent use in high-risk relapse of DT-PACE or VTD-PACE combination chemotherapy (Table 3): PACE indicates cisplatin 10 mg/m2, doxorubicin 10 mg/m2, cyclophosphamide 400 mg/m2, etoposide 40 mg/m2, all daily for 4 consecutive days but continuous intravenous infusion; DT-PACE, with added dexamethasone 20 to 40 mg/day for 4 days and thalidomide 100 to 200 mg/day for 4 days; VTD-PACE, plus bortezomib 1.0 to 1.3 mg/m2 on days 1 and 4. More of the high-risk than low-risk relapses had been initially randomized to thalidomide (57% vs 36%, P = .032).
Univariate and multivariate analyses of baseline and relapse features as well as prior response and its duration on PRS
| Variable . | n/N (%) . | HR (95% CI) . | P* . | Cumulative R2 . |
|---|---|---|---|---|
| Univariate analysis | ||||
| Baseline: GEP high-risk | 15/71 (21) | 4.57 (2.28,9.15) | < .001 | — |
| Relapse: creatinine ≥ 2 mg/dL | 10/114 (9) | 2.83 (1.38,5.77) | .004 | — |
| Relapse: Hb < 10 g/dL | 21/112 (19) | 2.42 (1.40,4.18) | .002 | — |
| Relapse: LDH ≥ 190 U/L | 37/108 (34) | 1.30 (0.78,2.17) | .309 | — |
| Relapse: cytogenetic abnormalities | 72/106 (68) | 2.03 (1.16,3.54) | .013 | — |
| Relapse: GEP high-risk | 46/120 (38) | 4.01 (2.47,6.49) | < .001 | — |
| Relapse: GEP CD-1 | 5/104 (5) | 0.87 (0.21,3.56) | .844 | — |
| Relapse: GEP CD-2 | 6/104 (6) | 0.65 (0.23,1.81) | .412 | — |
| Relapse: GEP HY | 21/104 (20) | 0.37 (0.16,0.87) | .022 | — |
| Relapse: GEP MF | 5/104 (5) | 2.12 (0.85,5.32) | .109 | — |
| Relapse: GEP MS | 26/104 (25) | 1.11 (0.65,1.92) | .698 | — |
| Relapse: GEP MY | 9/104 (9) | 0.25 (0.06,1.02) | .054 | — |
| Relapse: GEP MF/MS/PR | 63/104 (61) | 3.01 (1.66,5.44) | < .001 | — |
| Relapse: TP53 deletion (TP53 < 727) | 24/106 (23) | 0.56 (0.30,1.05) | .072 | — |
| CR to initial therapy | 58/120 (48) | 1.13 (0.71,1.80) | .610 | — |
| PR duration ≥ 2 y | 62/120 (52) | 0.44 (0.27,0.72) | .001 | — |
| Randomization to thalidomide | 53/120 (44) | 1.56 (0.97,2.51) | .064 | — |
| Multivariate analysis with GEP | ||||
| Relapse: GEP high-risk | 38/104 (37) | 4.00 (2.38,6.73) | < .001 | 32.835 |
| Relapse: GEP HY | 21/104 (20) | 0.37 (0.16,0.87) | .022 | 40.821 |
| Multivariate analysis without GEP | ||||
| PR duration ≥ 2 y | 51/101 (50) | 0.51 (0.29,0.87) | .014 | 15.045 |
| Relapse: cytogenetic abnormalities | 69/101 (68) | 2.57 (1.41,4.70) | .002 | 24.322 |
| Relapse: creatinine ≥ 2 mg/dL | 9/101 (9) | 2.89 (1.31,6.40) | .009 | 31.399 |
| Variable . | n/N (%) . | HR (95% CI) . | P* . | Cumulative R2 . |
|---|---|---|---|---|
| Univariate analysis | ||||
| Baseline: GEP high-risk | 15/71 (21) | 4.57 (2.28,9.15) | < .001 | — |
| Relapse: creatinine ≥ 2 mg/dL | 10/114 (9) | 2.83 (1.38,5.77) | .004 | — |
| Relapse: Hb < 10 g/dL | 21/112 (19) | 2.42 (1.40,4.18) | .002 | — |
| Relapse: LDH ≥ 190 U/L | 37/108 (34) | 1.30 (0.78,2.17) | .309 | — |
| Relapse: cytogenetic abnormalities | 72/106 (68) | 2.03 (1.16,3.54) | .013 | — |
| Relapse: GEP high-risk | 46/120 (38) | 4.01 (2.47,6.49) | < .001 | — |
| Relapse: GEP CD-1 | 5/104 (5) | 0.87 (0.21,3.56) | .844 | — |
| Relapse: GEP CD-2 | 6/104 (6) | 0.65 (0.23,1.81) | .412 | — |
| Relapse: GEP HY | 21/104 (20) | 0.37 (0.16,0.87) | .022 | — |
| Relapse: GEP MF | 5/104 (5) | 2.12 (0.85,5.32) | .109 | — |
| Relapse: GEP MS | 26/104 (25) | 1.11 (0.65,1.92) | .698 | — |
| Relapse: GEP MY | 9/104 (9) | 0.25 (0.06,1.02) | .054 | — |
| Relapse: GEP MF/MS/PR | 63/104 (61) | 3.01 (1.66,5.44) | < .001 | — |
| Relapse: TP53 deletion (TP53 < 727) | 24/106 (23) | 0.56 (0.30,1.05) | .072 | — |
| CR to initial therapy | 58/120 (48) | 1.13 (0.71,1.80) | .610 | — |
| PR duration ≥ 2 y | 62/120 (52) | 0.44 (0.27,0.72) | .001 | — |
| Randomization to thalidomide | 53/120 (44) | 1.56 (0.97,2.51) | .064 | — |
| Multivariate analysis with GEP | ||||
| Relapse: GEP high-risk | 38/104 (37) | 4.00 (2.38,6.73) | < .001 | 32.835 |
| Relapse: GEP HY | 21/104 (20) | 0.37 (0.16,0.87) | .022 | 40.821 |
| Multivariate analysis without GEP | ||||
| PR duration ≥ 2 y | 51/101 (50) | 0.51 (0.29,0.87) | .014 | 15.045 |
| Relapse: cytogenetic abnormalities | 69/101 (68) | 2.57 (1.41,4.70) | .002 | 24.322 |
| Relapse: creatinine ≥ 2 mg/dL | 9/101 (9) | 2.89 (1.31,6.40) | .009 | 31.399 |
HR indicates hazard ratio; CI, confidence interval; and —, not applicable.
Variables considered for the multivariate model with GEP data were: baseline 70-gene defined high-risk, relapse: creatinine ≥ 2 mg/dL, relapse: Hb < 10 g/dL, relapse: platelets < 150 × 109/μL, relapse: cytogenetic abnormalities, relapse: 70-gene defined HiR, relapse: HY GEP molecular subgroup, relapse: MY GEP molecular subgroup, Relapse: MF/MS/PR GEP molecular subgroups, PR duration of at least 2 years. Variables considered for the multivariate model without GEP data were: relapse: β2-microglobulin ≥ 5.5 mg/L, relapse: creatinine ≥ 2 mg/dL, relapse: Hb < 10 g/dL, relapse: platelets < 150 × 109/μL, relapse: cytogenetic abnormalities and PR duration of at least 2 years.
P value from Wald χ2 test in Cox regression. NS2, multivariate results not statistically significant at .05 level. All univariate P values reported regardless of significance. Multivariate model uses stepwise selection with entry level 0.1 and variable remains if meets the 0.05 level. A multivariate P value greater than .05 indicates variable forced into model with significant variables chosen using stepwise selection.
Salvage regimens used according to GEP-derived risk status at relapse
| Salvage regimens used after TT2 relapse . | Low risk at relapse, n (%) . | High risk at relapse, n (%) . | P . |
|---|---|---|---|
| Thalidomide alone or in combination | 50/74 (68) | 27/46 (59) | .324 |
| Lenalidomide alone or in combination | 5/74 (7) | 3/46 (7) | .960 |
| Bortezomib alone or in combination | 37/74 (50) | 23/46 (50) | > .999 |
| BTD or BLD with or without chemotherapy (eg, PACE), | 29/74 (39) | 15/46 (33) | .467 |
| DT-PACE, or VTD-PACE | 8/74 (11) | 13/46 (28) | .014 |
| Further transplant | 15/74 (20) | 13/46 (28) | .314 |
| Salvage regimens used after TT2 relapse . | Low risk at relapse, n (%) . | High risk at relapse, n (%) . | P . |
|---|---|---|---|
| Thalidomide alone or in combination | 50/74 (68) | 27/46 (59) | .324 |
| Lenalidomide alone or in combination | 5/74 (7) | 3/46 (7) | .960 |
| Bortezomib alone or in combination | 37/74 (50) | 23/46 (50) | > .999 |
| BTD or BLD with or without chemotherapy (eg, PACE), | 29/74 (39) | 15/46 (33) | .467 |
| DT-PACE, or VTD-PACE | 8/74 (11) | 13/46 (28) | .014 |
| Further transplant | 15/74 (20) | 13/46 (28) | .314 |
PACE indicates cisplatin 10 mg/m2, doxorubicin 10 mg/m2, cyclophosphamide 400 mg/m2, etoposide 40 mg/m2, all daily for 4 consecutive days but continuous intravenous infusion; DT-PACE, with added dexamethasone 20 to 40 mg/day for 4 days and thalidomide 100 to 200 mg/day for 4 days; VTD-PACE, plus bortezomib 1.0 to 1.3 mg/m2 on days 1 and 4.
A multivariate analysis of both baseline and relapse parameters along with response duration revealed, in the context of GEP data, only 2 GEP-derived variables to be independently significantly associated with PRS: high-risk designation, pertinent to 37%, predicted short PRS (hazard ratio [HR] = 4.00, P < .001, R2 = 32.8%), whereas the hyperdiploidy subgroup designation, present in 20%, imparted prolonged PRS (HR = 0.37, P = .022, for a cumulative R2 value of 40.8%). In the absence of GEP parameters, the presence of cytogenetic abnormalities and elevated creatinine levels (≥ 2 mg/dL), present in 68% and 9%, respectively, predicted short PRS (HR = 2.57, 2.89; P = .002, .009), whereas partial response duration of at least 2 years, pertaining to 50%, favored longer PRS (HR = 0.51, P = .014).
Thus, as in other malignancies,12 short duration of disease control in myeloma implied short PRS. Our data extend the powerful prognostic implications of GEP-defined risk to the PRS setting where, univariately, both relapse and baseline high-risk annotation imparted poor PRS. Adjusting for all variables that impacted PRS individually, GEP-derived high-risk designation at relapse was a poor and hyperdiploidy a favorable prognostic parameter. In the context of prior response duration, high-risk GEP status at relapse tended to shorten PRS among the longer prerelapse responders but was a powerful discriminator of PRS among patients with shorter initial response. Results presented here indicate that the predictive power of GEP-defined risk, developed in TT2 and validated in another front-line setting of TT3, extends to PRS after TT2 unrelated to the salvage therapies used. Recognizing that GEP risk status increases at relapse in a sizeable proportion of patients, GEP analysis should be repeated on treatment failure so that salvage strategy can be selected according to this powerful risk-discriminating tool. Whereas translocation-pertinent molecular subgroup designation was largely maintained at relapse, the hyperdiploidy group was the major contributor to the increased occurrence of PR designation at relapse. Toward advancing the lot of patients with high-risk myeloma, we recommend that GEP analysis should be an integral part of novel drug testing as already executed in the case of bortezomib.3,13
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the National Cancer Institute (NCI; Program Project grant CA55819, Bethesda, MD).
National Institutes of Health
Authorship
Contribution: B.N., J.D.S., F.v.R., E.A., and B.B. conceptualized the project; B.N., F.v.R., E.A., Y.A., S.W., K.H., and B.B. enrolled and treated patients; N.P. and M.A.-C. collected data; Y.Z., M.A.-C., J.S., J.D.S., P.Q., and A.H. analyzed data; and B.N., J.D.S., and B.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bart Barlogie, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 West Markham St, Slot 816, Little Rock, AR 72205; e-mail: barlogiebart@uams.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal