To the editor:
We read with interest the recent paper in Blood by Sampath et al1 proposing a key role for p73 in the induction of apoptosis induced by histone deacetylase inhibitors (HDACi) in chronic lymphocytic leukemia (CLL). The authors proposed that the HDAC-dependent induction of miR106b leads to translational inhibition of the Itch E3 ubiquitin ligase, and this in turn results in the up-regulation of the Itch target p73.2 The authors showed an inverse relationship between miR106b and Itch in CLL treated with the HDAC inhibitor, LBH589, and suggested a novel, p53-independent mechanism of CLL apoptosis, which would have important therapeutic implications.
We have previously shown that Itch is cleaved by caspases in CLL during apoptosis induced by various stimuli.3 Using HDACi, including LBH589 we confirmed the down-regulation of Itch during apoptosis. However, we also demonstrated the appearance of a 64 kDa Itch cleaved band due to the action of caspase-3, -6, and -7 on residue Asp240.3 This occurred during both HDACi-induced (Figure 1A) and spontaneous in vitro apoptosis (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article) of CLL cells. However, caspase inhibitors completely abrogated Itch down-regulation after HDACi treatment (please see Figures 1A and 3B in Rossi et al3 and Figure S1B). In this model, therefore, the down-regulation of Itch is a consequence, not a cause, of apoptosis, either spontaneously occurring in CLL cells in culture or triggered by LBH589.
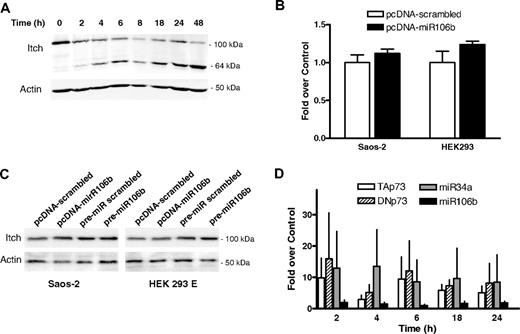
Regulation of Itch in chronic lymphocytic leukemia. (A) Western blotting of expression levels of protein Itch. Primary chronic lymphocytic leukemia (CLL) cells were incubated in presence of 10 nM LBH589 for the indicated times. Levels of actin were used as a loading control. Results from a representative experiment using CLL cells from 1 of the 12 analyzed patients are shown. There is a decrease in the 100 kDa band but with concomitant increase in 64 kDa cleaved product. See also Rossi et al3 . (B) Luciferase reporter assay of miR106b on 3′UTR-Itch in Saos-2 and HEK293 cells. The results were expressed as mean + SD from 3 independent experiments analyzed in triplicate. (C) Western blotting of endogenous levels of protein Itch in Saos-2 and HEK293 cells upon transfection. Cells were transfected either with pcDNA plasmids harboring the mature miR106b or a scrambled sequence, or with pre-miRs for miR106b or a scrambled sequence. Levels of actin were used as a loading control. MiR106b levels were assayed by qRT-PCR, to verify the transfection. (D) Expression levels of TAp73, DNp73, miR34a, and miR106b in primary CLL cells exposed for the indicated times to 10 nM LBH589 as analyzed by qRT-PCR. Expression levels relative to the untreated sample at the corresponding time points are shown. Expression of TAp73 and DNp73 was normalized to actin; expression of miR34a and miR106b was normalized to RNU6b. Statistical differences were determined by one-way analysis of variance (ANOVA) followed by Dunnet multiple comparison test, and the results were expressed as mean ± SEM from 12 independent CLL samples analyzed in triplicate.
Regulation of Itch in chronic lymphocytic leukemia. (A) Western blotting of expression levels of protein Itch. Primary chronic lymphocytic leukemia (CLL) cells were incubated in presence of 10 nM LBH589 for the indicated times. Levels of actin were used as a loading control. Results from a representative experiment using CLL cells from 1 of the 12 analyzed patients are shown. There is a decrease in the 100 kDa band but with concomitant increase in 64 kDa cleaved product. See also Rossi et al3 . (B) Luciferase reporter assay of miR106b on 3′UTR-Itch in Saos-2 and HEK293 cells. The results were expressed as mean + SD from 3 independent experiments analyzed in triplicate. (C) Western blotting of endogenous levels of protein Itch in Saos-2 and HEK293 cells upon transfection. Cells were transfected either with pcDNA plasmids harboring the mature miR106b or a scrambled sequence, or with pre-miRs for miR106b or a scrambled sequence. Levels of actin were used as a loading control. MiR106b levels were assayed by qRT-PCR, to verify the transfection. (D) Expression levels of TAp73, DNp73, miR34a, and miR106b in primary CLL cells exposed for the indicated times to 10 nM LBH589 as analyzed by qRT-PCR. Expression levels relative to the untreated sample at the corresponding time points are shown. Expression of TAp73 and DNp73 was normalized to actin; expression of miR34a and miR106b was normalized to RNU6b. Statistical differences were determined by one-way analysis of variance (ANOVA) followed by Dunnet multiple comparison test, and the results were expressed as mean ± SEM from 12 independent CLL samples analyzed in triplicate.
To investigate further the possible interaction of miR106b and Itch, we performed luciferase activity assay on different human cell lines with 3′UTR-Itch in presence of miR106b or a scrambled sequence (Figure 1B) and we assayed the endogenous levels of Itch in different human cell lines upon transfection with a plasmid expressing the mature miR106b, or with pre-miR106b (Figure 1C). In both cases we failed to detect any inhibition of Itch by miR106b. Moreover, we were not able to identify any significant increase in miR106b RNA levels in lymphocytes from 12 CLL patients treated in vitro with LBH589 (Figure 1D). Similarly, Sampath et al reported up-regulation of miR106b in only 16 patients of 47 studied.1 Figure 1D also shows that p73 was regulated at transcriptional level by LBH589; and not at the degradative level.
We also assessed the possibility that Itch could be a target of miR34a, expressed in CLL,4 as a high homology between the Itch 3′UTR and miR34a emerged from in silico analysis (Figure S1C) and miR34a was more consistently up-regulated by HDACi, even thought not reaching a strict statistical significance (Figure 1D). We did not observe any direct interaction between miR34a and Itch (Figure S1D,E).
Together, these data indicate that neither miR106b nor miR34a are involved in Itch down-regulation after exposure to HDACi, despite evidence of involvement of p73 in CLL, as suggested by Sampath et al.1
Authorship
Blood samples were obtained from CLL patients during routine diagnosis at the Leicester Royal Infirmary with patient consent in accordance with the Declaration of Helsinki and local ethical committee approval from the University of Leicester.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Gerry Melino or Martin J. S. Dyer, MRC-Toxicology Unit, Hodgkin Bldg, PO Box 138, University of Leicester, Lancaster Rd, Leicester, LE1 9HN United Kingdom; e-mail: gm89@le.ac.uk or mjsd1@le.ac.uk.
References
Author notes
*P.R.d.V.C. and P.T. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal