Abstract
Our previous studies with genomic minigenes have demonstrated that an engineered small nuclear RNA-U1 (U1+5a) partially rescued coagulation factor VII (FVII) mRNA processing impaired by the 9726+5G>A mutation. Here, to evaluate the U1+5a effects on FVII function, we devised a full-length FVII splicing-competent construct (pSCFVII-wt). This construct drove in COS-1 cells the synthesis of properly processed FVII transcripts and of secreted functional FVII (23 ± 4 ng/mL), which were virtually undetectable upon introduction of the 9726+5G>A mutation (pSCFVII-9726+5a). Cotransfection of pSCFVII-9726+5a with pU1+5a resulted in a partial rescue of FVII splicing and protein biosynthesis. The level increase in medium was dose dependent and, with a molar excess (1.5×) of pU1+5a, reached 9.5% plus or minus 3.2% (5.0 ± 2.8 ng/mL) of FVII-wt coagulant activity. These data provide the first insights into the U1-snRNA–mediated rescue of donor splice sites at protein level, thus further highlighting its therapeutic implications in bleeding disorders, which would benefit even from tiny increase of functional levels.
Introduction
The elucidation of molecular mechanisms underlying aberrant mRNA processing, a frequent cause of all inherited human disorders,1,2 has provided the rationale for RNA-based correction strategies that offer several advantages over the gene replacement therapy methods, including maintenance of the proper transcriptional control of the disease gene.
The vast majority of RNA-based approaches have exploited, in vitro and in vivo, antisense sequences to either mask natural splice sites, to induce skipping of defective exons,3-5 or newly generated cryptic sites,3,6-8 to favor the use of the canonical ones. Only a few attempts9-12 have been made to restore gene expression impaired by mutations at canonical donor splice sites (5'ss), which are the most frequent targets in human disease genes,1 including coagulation factor genes.13-18 Although we10-12 and others9 have partially restored correct splicing using the small nuclear RNA U1 (U1-snRNA),19 the experimental settings did not allow the assessment of rescue at protein and function levels, the key issue for the evaluation of the therapeutic potential.
Here, by exploiting a splicing-competent full-length construct, and thus a novel cellular model of severe coagulation factor VII (FVII) deficiency caused by the IVS7 9726+5G>A mutation,20 we demonstrated that the U1-snRNA–mediated rescue of FVII mRNA processing eventually results in an appreciable secretion of functional FVII.
Methods
Creation of vectors
Expression vectors for the secreted human FVII21 and for the parental (pU1-wt) and mutated (pU1+5a) human U1-snRNA11 were available in the laboratory.
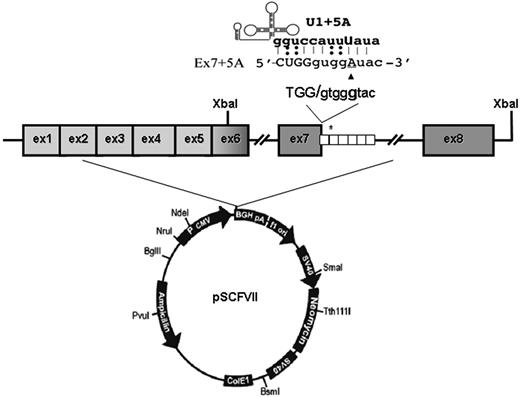
To create the full-length splicing-competent constructs, the FVII gene region spanning exons 6 through 8 (nt's 8926-11157)22 from our previously prepared wild-type and mutated minigene constructs11 was amplified with primers 5′GCATCTTTCTGACTTTTGTT3′ (forward) and 5′ATATTACTCTAGAACAGGCCAGGGCTGCTG3′ (reverse; including a XbaI site, underlined) and subcloned through the XbaI site in exon 6 downstream the FVII coding region spanning exons 1 to 6 in pcDNA3 (Figure 1). Plasmids were named pSCFVII-wt and pSCFVII-9726+5a (SC, splicing competent).
Transfection and studies at the mRNA and protein level
COS-1 (African green monkey kidney fibroblasts) cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) into 30-mm plates11,21 with 3 μg of each FVII expression vector and, in complementation assays, with 1×, 1.5×, 2×, and 2.5× molar excess of pU1-snRNA vectors. Culture medium (OptiMEM; Invitrogen) was supplemented with 5 μg/mL vitamin K (Konakion; Roche, Welwyn Garden City, United Kingdom) to allow proper FVII biosynthesis. Lysis of cells11 and collection of medium, for mRNA and protein studies, were conducted 72 hours after transfection. Ten independent experiments were performed for each transfection condition.
mRNA studies.
Reverse-transcription–polymerase chain reaction (RT-PCR), fluorescent labeling of product, and denaturing capillary electrophoresis were carried out as previously described.11
FVII antigen determination.
FVII protein levels in medium were measured by a commercially available enzyme-linked immunosorbent assay (ELISA) kit (FVII-EIA; Affinity Biologicals, Ancaster, ON). Besides the standard curve with serial dilution of pooled normal plasma, a reference curve with the recombinant FVII-wt in conditioned medium was also made.
FVII activity determination.
The ability of FVII in medium to activate its physiologic substrate, factor X, was assessed by exploiting a specific FXa fluorogenic substrate.21,23 FVII coagulant activity was also tested in prothrombin time (PT)–based assays.24
Assays were standardized using serial dilution of FVII-wt in medium from cells transfected with the gutted pcDNA3, not encoding FVII, used as negative control. All assays have been conducted in duplicate in each sample.
Results and discussion
Expression studies with genomic minigenes represent a well-established strategy to detail splicing mechanisms. Through the use of FVII constructs including the genomic region from exons 6 through 8, we have recently demonstrated that an appropriately engineered U1-snRNA partially restored the correct FVII mRNA processing impaired by the IVS7 9726+5G>A mutation.11 However, this approach has not allowed the evaluation of effects on secreted FVII protein levels, the key indicator of the intervention efficacy.
To address this issue, we devised a chimeric expression cassette in which introns 6 and 7 have been inserted within the entire FVII coding sequence (pSCFVII-wt; Figure 1).
FVII minigene construct and modified U1-snRNAs. Schematic representation of the expression vector pSCFVII. The 37-bp repeats in the IVS7 are indicated by white boxes, and the asterisk represents the 5'ss cryptic site in the second repeat. The 5'ss consensus sequence and the 9726+5G>A change (▴) are shown at the top. The top part shows the complementary between the sequence of the mutated exon 7-IVS7 junction and the 5′ tail of the engineered U1-snRNA.
FVII minigene construct and modified U1-snRNAs. Schematic representation of the expression vector pSCFVII. The 37-bp repeats in the IVS7 are indicated by white boxes, and the asterisk represents the 5'ss cryptic site in the second repeat. The 5'ss consensus sequence and the 9726+5G>A change (▴) are shown at the top. The top part shows the complementary between the sequence of the mutated exon 7-IVS7 junction and the 5′ tail of the engineered U1-snRNA.
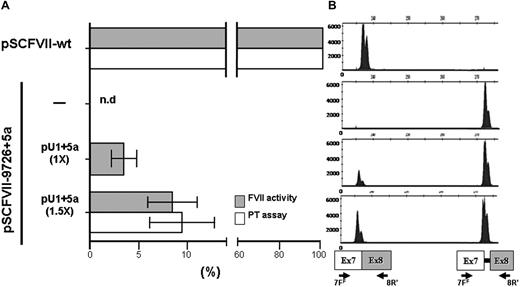
In transient transfection experiments in COS-1 cells, successfully exploited to express and characterize several FVII protein variants,25 this construct drove the synthesis of correct FVII transcripts (Figure 2B), validated by sequencing, and the secretion of functional FVII molecules. This was indicated by the appreciable FVII activity detected by fluorogenic FXa generation assays in conditioned medium (Figure 2A). FVII protein levels were also clearly detectable by ELISA (23 ± 4 ng/mL).
Rescue of FVII splicing and FVII function by the modified U1+5a in COS-1 cells. (A) FVII-mediated FXa generation (grey histograms) and FVII coagulant (white histograms) activity in conditioned medium from COS-1 cells transfected with pSCFVII-wt or the pSCFVII-9726+5a without and with equimolar concentration (1×) or an excess (1.5×) of pU1+5a. Mean and SD are shown. The PT-based assays have been performed on an ACL TOP automated coagulometer (Instrumentation Laboratory, Milan, Italy). Briefly, 50 μL conditioned medium was mixed with 50 μL FVII-depleted plasma (Instrumentation Laboratory) and incubated for 30 seconds at 37°C. RecombiPlasTin 2G (100 μL; Instrumentation Laboratory), as source of recombinant human tissue factor, calcium, and phospholipids were then added and coagulation time recorded. nd indicates not detectable. (B) Separation on a denaturing capillary system (automated ABI-3100) of fluorescently labeled RT-PCR products obtained from total RNA of cells transfected as indicated in panel A. A representative example is shown. The scheme of transcripts, and of primers used (7FF, fluorescently labeled; 8R),12 is depicted at the bottom. Separation of 1 μL of 1:100 diluted RT-PCR reaction is shown. As expected, the fragment sizes of the normal and aberrant transcripts were 236 bp and 273 bp, respectively.
Rescue of FVII splicing and FVII function by the modified U1+5a in COS-1 cells. (A) FVII-mediated FXa generation (grey histograms) and FVII coagulant (white histograms) activity in conditioned medium from COS-1 cells transfected with pSCFVII-wt or the pSCFVII-9726+5a without and with equimolar concentration (1×) or an excess (1.5×) of pU1+5a. Mean and SD are shown. The PT-based assays have been performed on an ACL TOP automated coagulometer (Instrumentation Laboratory, Milan, Italy). Briefly, 50 μL conditioned medium was mixed with 50 μL FVII-depleted plasma (Instrumentation Laboratory) and incubated for 30 seconds at 37°C. RecombiPlasTin 2G (100 μL; Instrumentation Laboratory), as source of recombinant human tissue factor, calcium, and phospholipids were then added and coagulation time recorded. nd indicates not detectable. (B) Separation on a denaturing capillary system (automated ABI-3100) of fluorescently labeled RT-PCR products obtained from total RNA of cells transfected as indicated in panel A. A representative example is shown. The scheme of transcripts, and of primers used (7FF, fluorescently labeled; 8R),12 is depicted at the bottom. Separation of 1 μL of 1:100 diluted RT-PCR reaction is shown. As expected, the fragment sizes of the normal and aberrant transcripts were 236 bp and 273 bp, respectively.
The introduction of the 9726+5G>A20 mutation at +5 position of the consensus sequence of the IVS7 donor splice site (Figure 1) resulted in the exon 7 skipping and, to a lesser extent, in the inclusion of the first 37-bp IVS7 repeat, thus providing a pattern of exon 7 definition virtually indistinguishable from that observed with the exons 6 to 8 genomic minigene.11 As expected, the FVII protein and activity levels in medium were virtually undetectable (Figure 2A), in accordance with the plasma coagulation phenotype observed in the homozygous patients.20
This observation provided us with an appropriate experimental model to evaluate the rescue of secreted FVII levels by the previously designed U1-snRNA (U1+5a), having the 5′ tails engineered to bind the mutated FVII IVS7 5'ss (Figure 1) and able to redirect recognition of the mutated donor splice site. Cotransfection of COS-1 cells with the pSCFVII-9726+5a vector and increasing concentrations of pU1+5a resulted in the appreciable synthesis of normally spliced transcripts (Figure 2B). Because of the sequence complementarity between transcripts, which makes their specific quantification virtually impossible, we estimated the rescue by evaluating the relative amount of the correct form over that including the additional 37 bp. In the presence of a molar excess (1.5×) of pU1+5a, the levels of correct transcript accounted for 48% plus or minus 4% of the +37-bp aberrant form, in accordance with our previous findings.11 Densitometric analysis of RT-PCR bands (not shown) indicates that the third mRNA form, caused by the exon 7 skipping, accounts for approximately 60% of all transcripts. Therefore we estimate that the correctly processed FVII form would represent approximately 20% of all FVII mRNAs.
Noticeably, coexpression of the U1+5a was also responsible for a significant and dose-dependent increase of FVII activity in medium (Figure 2A). In particular, the FVII activity levels in medium from cells cotransfected with equimolar (1×) or higher (1.5×) concentrations of pU1+5a, measured by sensitive fluorogenic FXa generation assays, were 3.1% plus or minus 1.4% and 8.2% plus or minus 2.6% of that obtained with the pSCFVII-wt, respectively.
To corroborate these findings, we assessed by PT-based assays the FVII coagulant activity in FVII-depleted plasma. The addition of medium from cells cotransfected with 1.5× excess of pU1+5a resulted in PTs (100 ± 6 seconds) significantly shorter than those produced by mock medium (119.1-119.6 seconds). Compared with a standard curve, the activity upon rescue corresponded to 9.5% plus or minus 3.2% of that of 25 ng/mL FVII-wt (Figure 2A).
Secreted FVII protein levels were also measured to substantiate the evidence for rescue of FVII biosynthesis. In medium from cells cotransfected with an excess of pU1+5a, the FVII antigen levels were 5.0 plus or minus 2.8 ng/mL. Further overexpression of the U1+5a did not produce additional effects on FVII levels. On the other hand, coexpression of the U1-wt did not result in any appreciable increase of FVII activity levels.
Taken together, our data in a cellular model of FVII deficiency demonstrated for the first time that the U1-snRNA–mediated rescue of splicing impaired by mutations at 5'ss results in an appreciable increase of functional protein. Because bleeding disorders would significantly benefit even from a tiny increase in coagulation factor levels, the correction efficacy observed has potential therapeutic implications.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Rosella Mari for her assistance in the evaluation of FVII coagulant activity by PT-based assays.
M.P., D.B., L.R., and F.B. were supported by grants from the Fondazione Cassa di Risparmio di Ferrara (CARIFE), Telethon (GGP 05214), and the University of Ferrara. F.P. was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) and Italian Cystic Fibrosis Foundation.
Authorship
Contribution: M.P., F.P., and F.B. conceived the study, analyzed and interpreted data, and wrote the paper; D.B. and L.R. created FVII constructs and performed expression studies and functional assays; and I.M. performed capillary electrophoresis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mirko Pinotti, Department of Biochemistry and Molecular Biology, University of Ferrara, Via Fossato di Mortara 74, 44100 Ferrara, Italy; e-mail: pnm@unife.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal