Familial platelet disorder (FPD), a rare autosomal dominant disorder characterized by quantitative and qualitative platelet abnormalities, is considered as a model of genetic predisposition to acute myeloid leukemia (AML). So far, monoallelic RUNX1 germline mutations have been found in 19 of 20 families with reported FPD, and the analysis of blast cells from only 5 patients at acute leukemia (AL) stage has shown no additional RUNX1 abnormality. Here, we performed RUNX1 analysis at constitutional and somatic levels in 8 persons with FPD who developed AL from 4 independent families. In addition to the germline RUNX1 mutation, we identified a second RUNX1 alteration in 6 AML cases (acquired point mutations in 4 cases and duplication of the altered RUNX1 allele associated with acquired trisomy 21 in 2 other cases). Although haploinsufficiency of RUNX1 causes FPD, our findings suggest that a second genetic event involving RUNX1 is often associated with progression to AML.
Introduction
Germline mutations are rare in hematologic malignancies. Familial platelet disorder (FPD) is an autosomal dominant disorder characterized by quantitative and qualitative platelet abnormalities and predisposition to acute myeloid leukemia (AML; FPD/AML OMIN no. 601399). Deleterious mutations in the RUNX1 gene, located at 21q22, including missense, nonsense, frameshift mutations, and intragenic deletions, have been identified in the 19 of 20 families reported so far,1,,,,,–7 but considerable genotypic and phenotypic heterogeneity are observed.8 No RUNX1 mutation was found in the remaining family, but exons 7 and 8 were not analyzed in this study.6 Molecular analysis of leukemic cells from only 5 persons with FPD, 4 cases at AML stage1,6 and 1 at myelodysplasia (MDS) stage,6 has been performed to date, and no additional molecular abnormality, including second RUNX1 alteration, has been detected, raising the question of how haploinsufficiency of RUNX1 may contribute to the development of acute leukemia (AL).
Methods
Patients and samples
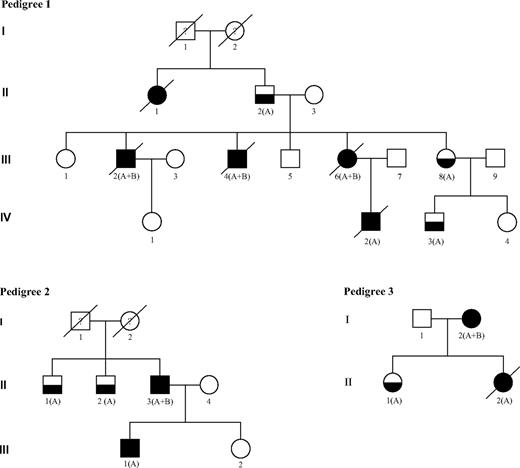
We studied 15 persons with FPD from 4 independent families identified from 2005 to 2007 through a national cooperative network supported by the French National Cancer Institute. This study was approved in October 2007 by the scientific committee of the Lille Tumorotheque (Lille IRB). Ten of 16 patients with FPD progressed to AL: 7 developed AML (patients III:2, III:4, III:6, IV:2 in pedigree 1; II:3 in pedigree 2; I:2 in pedigree 3; and patient I:1 in pedigree 4), 1 had a T-acute lymphoblastic leukemia (T-ALL; III:1 in pedigree 2), and another had a T-ALL followed by AML (II:2 in pedigree 3). Patient II:1 in pedigree 1 died of uncharacterized leukemia (Figure 1). The main clinicobiologic features of the 8 patients with characterized AL and for whom frozen material was available for molecular analysis are indicated in Table 1. No other tumor was observed in these families except for patient I:2 in pedigree 3 who developed a breast cancer 2 years after AML was diagnosed.
FPD/AML pedigrees. Squares indicate male and circles indicate female. Open symbols represent unaffected persons, half-filled symbols represent persons affected by FPD, and closed symbols represent persons affected by FPD who developed acute leukemia. The letters A and B indicate that a germline RUNX1 mutation and an acquired RUNX1 alteration could have been identified, respectively. Person III-1 in pedigree 2 developed T-ALL and person II-2 in pedigree 3 developed T-ALL followed by AML 5 years later. Both are also denoted with a closed symbol.
FPD/AML pedigrees. Squares indicate male and circles indicate female. Open symbols represent unaffected persons, half-filled symbols represent persons affected by FPD, and closed symbols represent persons affected by FPD who developed acute leukemia. The letters A and B indicate that a germline RUNX1 mutation and an acquired RUNX1 alteration could have been identified, respectively. Person III-1 in pedigree 2 developed T-ALL and person II-2 in pedigree 3 developed T-ALL followed by AML 5 years later. Both are also denoted with a closed symbol.
Characteristics of patients with FPD who progressed to acute leukemia and types of RUNX1 gene alterations
| Pedigree/patient . | Age at AL diagnosis, y . | Sex . | Type of AL . | Karyotype at AL stage . | Germline RUNX1 alteration . | Acquired RUNX1 alteration at AL stage . | Loss of the acquired RUNX1 alteration in CR . | Biallelic RUNX1 mutations . | Other gene mutations detected . | Relapse . | ABMT with related donor* (donor) . | Status at last follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/III:2 | 25 | M | AML-M5 | ND | p.Ala129Glu mutation | p.Glu395_Arg400>HisfsX172 mutation | ND | ND | None | Yes† (after ABMT) | Yes (brother III:5) | Death |
| 1/III:4 | 25 | M | AML-M5 | ND | p.Ala129Glu mutation | p.Arg135Ser mutation | ND | Yes | None | NA (refractory disease) | No | Death |
| 1/III:6 | 42 | F | AML-M0 | 46,XX, +21[14] | p.Ala129Glu mutation | Duplication of the chromosome carrying the mutated allele | ND | NA | None | Yes‡ | No | Death |
| 2/II:3 | 60 | M | AML-M4 | 46,XY [20] | p.Arg177Gln mutation | p.Ala160Thr mutation | Yes | Yes | FLT3-ITD | No | No | Alive in CR |
| 2/III:1 | 28 | M | T-ALL | 46,XY [20] | p.Arg177Gln mutation | Absence | NA | NA | None | No | Yes (sister III:2) | Alive in CR |
| 3/I:2 | 55 | F | AML-M5 | 46,XX [20] | p.Gln308ArgfsX259 mutation | p.Gly138ProfsX12 mutation | Yes | ND | None | No | Yes (sister) | Alive in CR |
| 3/II:2 | 24 | F | AML-M2§ | 46,XX,t(1;3)(p36;q26)[15]/46,XX[5] | p.Gln308ArgfsX259 mutation | Absence | NA | NA | K-RAS mutation (p.Gly12Asp) | No | No | Death |
| 4/I:1 | 12 | F | AML-M6 | 48,XXXc,+21[28]/47,XXXc[3] | Complete deletion of RUNX1 gene | Duplication of the chromosome carrying the deleted allele | Yes | NA | None | Yes†‖ (after ABMT) | Yes (sister) | Alive in relapse |
| Pedigree/patient . | Age at AL diagnosis, y . | Sex . | Type of AL . | Karyotype at AL stage . | Germline RUNX1 alteration . | Acquired RUNX1 alteration at AL stage . | Loss of the acquired RUNX1 alteration in CR . | Biallelic RUNX1 mutations . | Other gene mutations detected . | Relapse . | ABMT with related donor* (donor) . | Status at last follow-up . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/III:2 | 25 | M | AML-M5 | ND | p.Ala129Glu mutation | p.Glu395_Arg400>HisfsX172 mutation | ND | ND | None | Yes† (after ABMT) | Yes (brother III:5) | Death |
| 1/III:4 | 25 | M | AML-M5 | ND | p.Ala129Glu mutation | p.Arg135Ser mutation | ND | Yes | None | NA (refractory disease) | No | Death |
| 1/III:6 | 42 | F | AML-M0 | 46,XX, +21[14] | p.Ala129Glu mutation | Duplication of the chromosome carrying the mutated allele | ND | NA | None | Yes‡ | No | Death |
| 2/II:3 | 60 | M | AML-M4 | 46,XY [20] | p.Arg177Gln mutation | p.Ala160Thr mutation | Yes | Yes | FLT3-ITD | No | No | Alive in CR |
| 2/III:1 | 28 | M | T-ALL | 46,XY [20] | p.Arg177Gln mutation | Absence | NA | NA | None | No | Yes (sister III:2) | Alive in CR |
| 3/I:2 | 55 | F | AML-M5 | 46,XX [20] | p.Gln308ArgfsX259 mutation | p.Gly138ProfsX12 mutation | Yes | ND | None | No | Yes (sister) | Alive in CR |
| 3/II:2 | 24 | F | AML-M2§ | 46,XX,t(1;3)(p36;q26)[15]/46,XX[5] | p.Gln308ArgfsX259 mutation | Absence | NA | NA | K-RAS mutation (p.Gly12Asp) | No | No | Death |
| 4/I:1 | 12 | F | AML-M6 | 48,XXXc,+21[28]/47,XXXc[3] | Complete deletion of RUNX1 gene | Duplication of the chromosome carrying the deleted allele | Yes | NA | None | Yes†‖ (after ABMT) | Yes (sister) | Alive in relapse |
AL indicates acute leukemia; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; FPD, familial platelet disorder; ITD, internal tandem duplication; ND, not determined; NA, not applicable; ABMT, allogeneic bone marrow transplantation; and CR, complete remission.
The HLA-matched related donor of each patient who underwent ABMT was screened for RUNX1 alterations and was found to have a wild-type status.
RUNX1 status identical at diagnosis and at relapse.
RUNX1 mutations could not be tested at relapse.
AML after T-ALL was probably therapy related.
Complex cytogenetic aberrations and FLT3-ITD mutation acquired at relapse.
Peripheral blood samples were collected from patients with FPD or their parents and from unaffected family members after informed consent was obtained in accordance with the Declaration of Helsinki. For patients who progressed to AL, peripheral blood samples were collected during complete remission (CR), and blast cells were collected at the time of diagnosis when available. Genomic DNA was extracted with the use of standard procedures.
Cytogenetic analysis
Cytogenetic G-banding analysis was performed according to standard methods. The definition of a cytogenetic clone and descriptions of karyotypes followed the International System for Human Cytogenetic Nomenclature.9
Fluorescence in situ hybridization analysis
Fluorescence in situ hybridization (FISH) analysis with the use of the RUNX1 probe (Vysis, Woodcreek, IL) was performed on metaphases at the time of AML diagnosis and at CR in patient from pedigree 4.
Comparative genomic hybridization array analysis
Genomic DNA from leukemic cells of patient 4/I:1 was subjected to comparative genomic hybridization (CGH) array analysis, using 105K Agilent chips (Agilent Technologies, Santa Clara, CA).
Gene mutations screening
RUNX1 mutations detection was performed by direct sequencing. Exons 3 through 8 of RUNX1 were amplified from genomic DNA with the use of the intronic forward and reverse primers shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The purified polymerase chain reaction (PCR) products were sequenced in both directions with the use of the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and analyzed on the Applied Biosystems 3130xl Genetic Analyzer. NPM1, CEBPA, FLT3 internal tandem duplication (FLT3-ITD), FLT3-D835/I836, N- and K-RAS, and c-KIT mutations were detected as previously described.10,,,,,–16
Results and discussion
In each family with FPD studied, a distinct RUNX1 germline alteration was found to segregate with the disease (Table 1; Figure 1).
In pedigree 1, a c.386C>A, p.Ala129Glu RUNX1 germline mutation was identified in 7 members. The 3 patients analyzed who developed AML harbored an additional somatic event that involved RUNX1: a second point mutation (c.1183_1199del17, p.Glu395_Arg400>HisfsX172 in patient III:2 and c.405G>C, p.Arg135Ser in patient III:4) and an acquired trisomy 21 associated with duplication of the mutated allele in patient III:6. The comparison of 30 PCR product subclones between leukemic and germline DNA from patient III:6 confirmed the third RUNX1 allele was the mutated one (21 of 30 versus 14 of 30 mutated clones). For patient III:4, the germline and the somatic RUNX1 mutation were found to be in different subclones, indicating that these mutations are not located on the same allele (Table 1).
In pedigree 2, a germline mutation in RUNX1 exon 4 (c.530G>A, p.Arg177Gln) was identified in 4 members (II:1, II:2, II:3, and III:1). Patient II:3 who developed AML had a somatic RUNX1 mutation localized also in exon 4 of RUNX1 (c. 478G>A, p.Ala160Thr), and the analysis of 30 PCR product subclones confirmed that each mutation involved a different RUNX1 allele. In contrast, patient III:1 who developed T-ALL harbored no somatic RUNX1 mutation, and no cytogenetic or molecular abnormality was found at leukemia stage.
In pedigree 3, a germline mutation in RUNX1 exon 8 (c.922_923insGCGCC, p.Gln308ArgfsX259) was identified in 3 members (I:2, II:1, II:2). Patient I:2 who developed AML presented a somatic RUNX1 mutation (c.411_412 insCC, p.Gly138ProfsX12). Conversely, no second RUNX1 alteration was detected in the blast cells of her daughter (patient 3/II:2) who developed AML 5 years after a T-ALL.
The last observation (patient 4/I:1) is a child with thrombocytopenia diagnosed at 2 years of age who developed refractory anemia with excess of blasts 10 years later that soon progressed to M6-AML. This patient was found to have a complete germline deletion of RUNX1. Constitutional karyotype was normal, whereas bone marrow karyotype performed at MDS and AML stages showed an acquired trisomy 21. In both situations, FISH analysis showed only one spot corresponding to RUNX1, suggesting the deletion of one RUNX1 allele and subsequently the duplication of the chromosome 21 carrying the germline RUNX1-deleted allele. This finding was ascertained by CGH Array on leukemic DNA, which showed a 1.8-Mb deletion on the long arm of the duplicated chromosome 21 including RUNX1.
Overall, 19 families have previously been reported with inherited RUNX1 mutations causing FPD/AML.1,,,,,–7 In these cases, there was a strong predominance of mutations in the Runt homology domain (RHD), because only 5 families had a germline mutation located outside this domain. In the current study, we also described a family with a germline mutation occurring in sequences C-terminal to the RHD of RUNX1 (Figure S1). However, most mutations in FPD/AML and de novo AML involve DNA-contacting residues in the RHD and are predicted to disrupt DNA binding, which would lead to the functional inactivation of RUNX1. Complete deletion of RUNX1 was reported in one family with FPD1 and in 4 sporadic cases associated with thrombocytopenia,7,17 similarly to patient 4/I:1 in our study. The deletion was very variable but included the RUNX1 gene in each case. Remarkably, the deleted genes in patient 4/I:1 were the same as in the case reported by Shinawi et al.17
Development of AL in the context of FPD is very heterogeneous and, in the families reported here, 10 of 16 patients with FPD developed AL. Surprisingly, not only AML was observed because 2 patients developed T-ALL. These data are in agreement with the recent study by Owen et al6 who described a case of T-ALL after FPD. These 3 observations are very interesting because RUNX1 plays a major role in megakaryopoiesis, myeloid differentiation, but also T-cell development.18,–20 Rare occurrence of RUNX1 mutations in ALL has been reported in the literature,21 but our finding suggests that RUNX1 mutation screening may be of importance in T-ALL, especially in the case of family history of leukemia.
At MDS or AML stage, karyotype was available in 22 cases reported in the literature with abnormalities in 2 of 3 of them. A trisomy 21 was found in a total of 4 patients, including our 2 cases. In the 2 previously published cases, 1 with a RUNX1 mutation affecting the RHD and the other with a germline RUNX1 deletion, RUNX1 analysis was not performed on leukemic cells. However, Shinawi et al17 clearly demonstrated by CGH Array that, at leukemic stage, the duplicated chromosome 21 carried the germline RUNX1-deleted allele. In the present study, the same mechanism was observed in patients 4/I:1 and 1/III:6 who harbored a duplication of the chromosome carrying the RUNX1-deleted or -mutated allele, respectively. In patient 1/III:6, we can speculate that the modified wild-type/mutated ratio is sufficient to induce leukemia. In patient 4/I:1, progression to AML may not only be explained by a dosage effect of RUNX1 gene but also by other genes located on chromosome 21, a situation close to that observed in the Down syndrome.22 Furthermore, the implication of other unknown genetic alterations in AML progression cannot be excluded.
At the molecular level, only 5 patients with FPD have been studied at leukemic stage, and no additional somatic RUNX1 abnormality was reported so far, but RUNX1 mutations screening was performed at MDS stage in one case,6 and the percentage of blasts was not mentioned in the other cases with AML.1,6 However, cytogenetic abnormalities involving chromosome 5 (monosomy 5 and 5q deletion) were detected in 2 of 5 AML cases in the context of FPD.1,6 These data are discordant with our findings because we detected a somatic RUNX1 mutation in 4 AML cases after FPD. It is important to note that, except for patient 2/II:3 with a FLT3-ITD, no other cytogenetic abnormality, and no NPM1, CEBPA, FLT3, N- and K-RAS, or c-KIT mutation was associated with the somatic RUNX1 mutation in the 3 remaining patients. Finally, in patient 3/II:2, the single patient with AML for which no somatic RUNX1 alteration was found, the major hypothesis is a therapy-related AML after ALL. Two additional somatic alterations were identified in this patient at AML stage: a K-RAS mutation and a t(1;3)(p36;q26) inducing a PRDM16 overexpression. Recently, Roche-Lestienne et al23 confirmed that PRDM16 is an important partner of RUNX1 in the progression of chronic myeloid leukemia to AL.23 Furthermore, an association, possibly indicating cooperation, has been observed between point mutations of RUNX1 and RAS, especially in therapy-related AML.24
In this study, 4 patients with AL underwent allogeneic bone marrow transplantation (ABMT) from an HLA-matched related donor. The selected donors were screened for RUNX1 alterations in the context of family genetic studies and were found to have a wild-type status (Table 1). As emphasized in a recent report,6 FPD/AML may be more prevalent than previously appreciated. Consequently, testing patients with MDS/AL for RUNX1 mutations would be interesting before considering sibling hematopoietic stem cell transplantation.
In conclusion, although haploinsufficiency of RUNX1 causes FPD, a very novel result from our study is that a second genetic event involving RUNX1 is often associated with progression to AML. This concept resembles what has been recently observed in families with germline CEBPA mutations and familial AML, in which additional somatic CEBPA mutations represent a frequent second event in AML pathogenesis.25 However, because tumor development is a complex multistep process, other somatic alterations may also contribute to the development of leukemia. Another important issue we can draw from preliminary evidence is that germline RUNX1 mutations also predispose to ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all patients, families, and clinicians for their participation.
This work was supported by the French National Cancer Institute (INCa), the North-West Canceropole (Onco-Hematology axis), the PACA Canceropole, and the University of Aix Marseille II.
Authorship
Contribution: C.P. and H.S. conceptualized the idea, designed the research, analyzed data, and wrote the paper; A.R. analyzed data and wrote the paper; V.B. performed research and contributed to writing the paper; N.P. performed research; C.R.-L. contributed to writing the paper; and N.B., N.D., J.-M.A., P.C.-L., M.-J.M., and A.B. provided samples and data and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hagay Sobol, Department of Genetic Oncology, Inserm CIC-P 9502, Paoli Calmettes Institute, Oncology and Immunology Institute of Marseille (IFR 137), 232 Sainte Marguerite Blvd, 13273 Marseille Cedex 9, France; e-mail: hagay.sobol@inserm.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal