The molecular mechanism of autocrine regulation of vascular endothelial growth factor (VEGF) in chronic lymphocytic leukemia (CLL) B cells is unknown. Here, we report that CLL B cells express constitutive levels of HIF-1α under normoxia. We have examined the status of the von Hippel-Lindau gene product (pVHL) that is responsible for HIF-1α degradation and found it to be at a notably low level in CLL B cells compared with normal B cells. We demonstrate that the microRNA, miR-92-1, overexpressed in CLL B cells, can target the VHL transcript to repress its expression. We found that the stabilized HIF-1α can form an active complex with the transcriptional coactivator p300 and phosphorylated-STAT3 at the VEGF promoter and recruit RNA polymerase II. This is initial evidence that pVHL, without any genetic alteration, can be regulated by microRNA and explains the aberrant autocrine VEGF secretion in CLL.
Introduction
B-cell chronic lymphocytic leukemia (CLL) continues to be a more common leukemia with no obvious curative approaches.1,2 Elevated plasma vascular endothelial growth factor (VEGF) levels in CLL are associated with advancing disease, even in early-stage disease.3 CLL B cells have high endogenous levels of mRNA for VEGF and are able to spontaneously secrete VEGF.4,5 The tissue neovascularization has been shown to be elevated in the marrow6 and lymph nodes5 of patients with CLL, possibly related to the increased VEGF levels. Furthermore, the ability of VEGF to alter CLL B-cell apoptosis resistance is linked to the expression of VEGF receptors VEGF-R1, -R2, and neuropilin receptor-1, found on CLL B cells.7,8 Therefore, the advantage of understanding these pathways is that interference with critical components of these receptor-ligand/receptor-mediated pathways should induce CLL B-cell death or apoptosis.9 We believe the VEGF-based autocrine system that leads to increased apoptosis resistance in CLL B cells7 is an important pathway, as it lends itself to therapeutic exploitation given the increasing repertoire of anti-VEGF agents10 ; however, to date, the molecular mechanism of autocrine regulation of VEGF in CLL B cells is unknown.
In this study, we describe a plausible mechanism for activation of the VEGF-based autocrine pathway in CLL B cells. We found that HIF-1α, a known key transcription factor for VEGF,11,12 is overexpressed in leukemic CLL B cells under normoxia primarily because of notably reduced levels of von Hippel-Lindau (pVHL) protein without having any genetic alterations of the VHL gene. Importantly, HIF-1α then is able to form an active complex with the transcriptional coactivator p300 and phosphorylated-STAT3 as the VEGF promoter, and recruit RNA polymerase II. Finally, we present initial evidence that the microRNA, miR-92-1 (formerly known as miR-92a) known to be elevated in CLL B cells,13 is able to target the VHL transcript and repress translation. We believe this novel activation/regulation of the HIF-1α/VEGF axis in CLL B cells explains spontaneous VEGF secretion by these leukemic cells under normoxia.
Methods
Patient selection and purification of lymphocytes
Blood was collected from healthy donors or untreated patients with B-CLL. The study was approved by the Mayo Clinic Institutional Review Board and informed consent was obtained in accordance with the Declaration of Helsinki.
Cell culture
Peripheral blood mononuclear cells (PBMCs) from patients with CLL containing more than 90% CD5+/CD19+ B lymphocytes were used in this study and cultured in serum-free adoptive immunotherapy media-V (AIM-V) medium (Gibco, Carlsbad, CA). When needed, human tonsillar B cells or PBMCs, obtained from healthy subjects, were sorted using B cell–negative selection kits (StemCell Technologies, Vancouver, BC) and used in this study. The human embryonic kidney cell line (293T) and a human megakaryoblastic leukemia cell line (Meg-01) were used for transfection experiments in this report. All cells were maintained in appropriate media at 37°C in an atmosphere containing 95% air, 5% CO2.
Reagents
Antibodies to HIF-1α and actin were purchased from Novus Biologicals (Littleton, CO). Mouse monoclonal (Upstate Biotechnology, Lake Placid, NY) and rabbit polyclonal antibodies to p300 and Glut1 rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) were also purchased. Mouse monoclonal antibodies to VHL, phospho-STAT3 (S727), and rabbit polyclonal antibody to ORC2 (origin recognition complex) were purchased from BD Pharmingen (San Diego, CA). Cu/Zn SOD (superoxide dismutase) rabbit polyclonal antibody was purchased from Stressgen (Victoria, BC). Phospho-RNA polymerase II (ChIP grade) and phospho-STAT3 (S727) antibodies were purchased from GeneTex (San Antonio, TX) and Cell Signaling Technologies (Beverly, MA), respectively. All other chemicals and reagents were obtained from Sigma-Aldrich (St Louis, MO).
Immunohistochemistry
Sections were cut from paraffin-blocked bone marrow biopsies of patients with CLL or healthy subjects at 4 μm, mounted on slides, and immunostained according to the manufacturer's protocol (Dako, Carpinteria, CA) using the primary mouse monoclonal antibody to HIF-1α.
Transfection of cells
293T cells were transfected with the microRNA human miR-92-1 (Ambion, Austin, TX) at increasing doses using Lipofectamine2000 (Invitrogen, Carlsbad, CA). Transient transfection of the freshly isolated primary CLL B cells with anti–miR-92-1 inhibitor oligos (Ambion) was performed using the HiPerfect lipid reagent (QIAGEN, Valencia, CA). After 48 hours of transfection, cells were analyzed for pVHL levels by Western blot, as described in “Preparation of whole-cell extracts and immunoblot experiments,” using pVHL-specific antibody (BD Pharmingen).
Preparation of whole-cell extracts and immunoblot experiments
Primary CLL B cells were washed in phosphate-buffered saline (PBS) and lysed in ice-cold lysis buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 2 mM EGTA, 1% NP-40, 10 mM NaF, 1 mM Na3V04, and a cocktail of protease inhibitors). Protein content was determined and equal amounts of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto 0.45 μ nitrocellulose papers (Bio-Rad, Hercules, CA) followed by immunostaining with specific antibodies. Protein bands were detected using enhanced chemiluminescence detection kits (Pierce, Rockford, IL).
Immunoprecipitation
Freshly isolated primary leukemic cells from patients with B-cell CLL were used to prepare whole-cell extracts in lysis buffer, as described in “Preparation of whole-cell extracts and immunoblot experiments.” Endogenous HIF-1α or p300 was immunoprecipitated using specific antibodies or control mouse immunoglobulin (IgG; Sigma-Aldrich), followed by incubation overnight in 4°C with Protein A-Sepharose 4B beads (GE Healthcare, Piscataway, NJ). Beads were washed and analyzed by Western blot to detect the presence of p300, phospho-STAT3, or HIF-1α in the immunocomplex using a specific antibody to p300 (Santa Cruz Biotechnology), P-STAT3 (Cell Signaling), or HIF-1α (Novus), respectively. In some experiments, immunoprecipitated HIF-1α was analyzed for proline-hydroxylation by Western blot using a HIF-1α proline-hydroxylation–specific polyclonal antibody.14
Preparation of nuclear extracts from CLL cells
PBMCs isolated from patients with CLL were washed in PBS and once in a buffer containing 10 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and a cocktail of protease inhibitors. Cells were lysed in buffer B containing 10 mM HEPES, pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 0.1% NP-40, 0.5 mM DTT, and a cocktail of protease inhibitors on ice for 10 minutes. Lysates were centrifuged and collected in the cytosolic fraction. The pellet was washed 3 times in buffer A and resuspended in a hypertonic buffer (buffer C, containing 20 mM HEPES, pH 7.4, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, and a cocktail of protease inhibitors) and incubated on ice for 30 minutes. After centrifugation at 16 000g for 20 minutes, the supernatant was collected and used as the nuclear extract for immunoblot analyses. An ORC2 antibody was used in Western blot analyses to verify the purity of nuclear extract preparation.
Luciferase reporter gene assay
A 546-bp segment of the 3′ untranslated region (UTR) of the VHL gene (NM_000551) was amplified by polymerase chain reaction (PCR) from human genomic DNA and inserted into the pGL3 control vector (Promega) using the XbaI site immediately downstream from the stop codon of luciferase. We generated one insert with a deletion of 9 bp (3′M1) from the 5′ end of the miRNA site of complementarity using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA); wild-type and mutant inserts were confirmed by sequencing. Human megakaryocytic cell line, MEG-01, was cotransfected using siPORT neoFX (Ambion) according to the manufacturer's protocol with 0.4 μg of the firefly luciferase reporter vector and 0.08 μg of the control vector containing Renilla luciferase, pRL-TK (Promega). miR-92-1 or miR-16 microRNAs (Ambion) were also used (10 nM per well) in 2 independent experiments, each in triplicate. Firefly and Renilla luciferase activities were measured consecutively using the dual-luciferase assays (Promega, Madison, WI) 24 hours after transfection.
ChIP assay
Freshly isolated primary CLL B cells were cross-linked as described by Nelson et al.15 Cells were lysed, and isolated nuclei were washed and genomic DNA was sheared by sonication. Immunoprecipitation was performed by the addition of anti–HIF-1α (Novus), anti–P-STAT3 (S727; BD Pharmingen), anti–phospho-RNA polymerase II (GeneTex) antibodies, or control IgG (Sigma-Aldrich) to the precleared chromatin solution, followed by Protein A Sepharose beads, and the immunoprecipitated DNA was then extracted. The region −1386 to −1036 of the VEGF promoter was PCR amplified from the immunoprecipitated chromatin using the following primers: forward, 5′-CAGGTCAGAAACCAGCCAG-3′; reverse, 5′-CGTGATGATTCAAACCTACC-3′.16 The 350-bp PCR product was resolved on a 1.2% agarose gel and stained with ethidium bromide.
Measurement of miR-92-1 levels in CLL B cells
Expression levels of hsa-miR-92-1 microRNA in CLL B cells were measured by real-time reverse transcriptase–PCR (RT-PCR) using specific primers (Ambion). Total RNA was extracted from primary CLL B cells and purified CD19+ (98%) normal B lymphocytes using Trizol (Invitrogen). The single-tube TaqMan microRNA assays were used to quantify mature has-miR-92-1 using the 7500 Real Time PCR System (Applied Biosystems, Carlsbad, CA). Normalization was performed with RNU6B. Comparative real-time PCR was performed in triplicate. Relative expression was calculated using the comparative Ct method.17
Results
VEGF overexpression in CLL B cells is associated with elevated HIF-1α levels
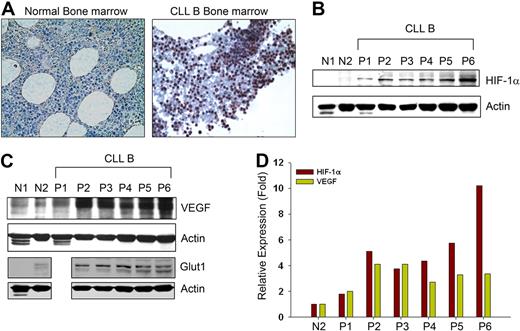
Spontaneous secretion of very high endogenous VEGF levels under normoxia by CLL B cells4 prompted us to determine HIF-1α levels in CLL B cells, as HIF-1α is a key transcription factor for VEGF expression.10,–12 First, HIF-1α expression in CLL B cells was studied in bone marrow by immunohistochemistry. Bone marrow sections from patients with CLL displayed high levels of HIF-1α expression in the nuclei, usually in a distinct cohort of CLL B cells. The HIF-1α levels were more evident in comparison to cells seen in the normal bone marrow sections (Figure 1A). In many solid tumors, HIF-1α is frequently elevated18 and often associated with adverse clinical outcomes; therefore, we decided to determine the protein levels for HIF-1α in primary blood CLL B cells. Figure 1B demonstrates that the protein levels of HIF-1α are elevated in CLL B cells compared with normal human CD19+ B cells or PBMCs under normoxia. We found that HIF-1α expression was associated with increased expression of VEGF in these patient cell samples (Figure 1C). In fact, densitometric analyses of the levels of HIF-1α and VEGF in CLL B cells suggest a general positive correlation of their expression when we calculated their relative values based on the normal (N2) CD19+ B-lymphocytes expression of these 2 proteins (Figure 1D). Moreover, CLL B cells were also found to overexpress the glucose transporter 1 (Glut1) gene, a known downstream target of HIF-1α,19 compared with normal B cells or PBMCs under normoxic conditions (Figure 1C). Together, the results suggest that HIF-1α is aberrantly expressed in CLL B cells under normoxic conditions as was evident from the bone marrow section evaluations and Western blot analysis of the PBMCs obtained from patients with CLL.
CLL B cells express constitutive levels of HIF-1α. (A) Immunohistochemistry of CLL and normal bone marrow for HIF-1α expression. Normal and CLL bone marrow sections were immunostained with a mouse monoclonal antibody to HIF-1α and counterstained with hematoxylin. Normal marrow cells show little or an undetectable level of HIF-1α while CLL bone marrow exhibit clusters of lymphocytes positive for HIF-1α expression with mostly nuclear staining. Figures shown are representative of 5 normal and 10 CLL bone marrows (magnification ×400). (B,C) CLL B cells express high levels of HIF-1α and its target genes under normoxia. Lysates prepared from CLL B cells (P1-6), normal PBMCs (N1), and purified normal B cells (N2) were analyzed for the expression of HIF-1α or VEGF by Western blot using specific antibodies. Expression of Glut1, another target gene of HIF-1α, is also shown in these samples (except CLL-P1) by Western blot analysis using a specific antibody to Glut1. Actin was used as the loading control. (D) Expression of HIF-1α and VEGF in CLL is positively correlated. Densitometric values of HIF-1α and VEGF expression in CLL B cells (B,C) were calculated and presented as relative expression based on the values obtained from the normal purified CD19+ B-cell lysates (panels B,C lane N2). Expression levels of HIF-1α and VEGF in normal B cells were arbitrarily chosen as one. Results demonstrate a generally positive association of HIF-1α and VEGF expression.
CLL B cells express constitutive levels of HIF-1α. (A) Immunohistochemistry of CLL and normal bone marrow for HIF-1α expression. Normal and CLL bone marrow sections were immunostained with a mouse monoclonal antibody to HIF-1α and counterstained with hematoxylin. Normal marrow cells show little or an undetectable level of HIF-1α while CLL bone marrow exhibit clusters of lymphocytes positive for HIF-1α expression with mostly nuclear staining. Figures shown are representative of 5 normal and 10 CLL bone marrows (magnification ×400). (B,C) CLL B cells express high levels of HIF-1α and its target genes under normoxia. Lysates prepared from CLL B cells (P1-6), normal PBMCs (N1), and purified normal B cells (N2) were analyzed for the expression of HIF-1α or VEGF by Western blot using specific antibodies. Expression of Glut1, another target gene of HIF-1α, is also shown in these samples (except CLL-P1) by Western blot analysis using a specific antibody to Glut1. Actin was used as the loading control. (D) Expression of HIF-1α and VEGF in CLL is positively correlated. Densitometric values of HIF-1α and VEGF expression in CLL B cells (B,C) were calculated and presented as relative expression based on the values obtained from the normal purified CD19+ B-cell lysates (panels B,C lane N2). Expression levels of HIF-1α and VEGF in normal B cells were arbitrarily chosen as one. Results demonstrate a generally positive association of HIF-1α and VEGF expression.
Hydroxylated HIF-1α is accumulated in CLL B cells
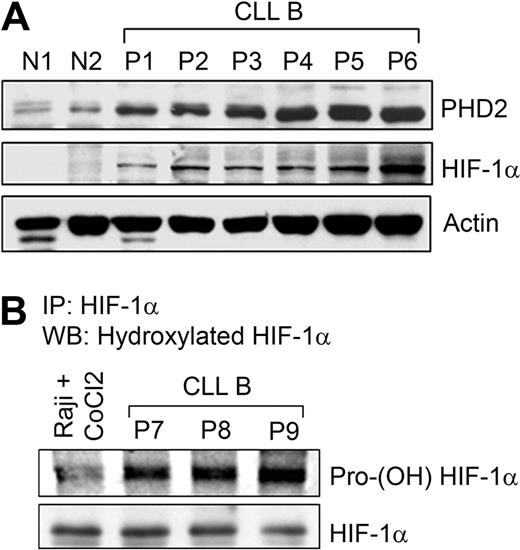
Three prolyl-4-hydroxylases (PHD1, 2, and 3) are able to hydroxylate HIF-1α at 2 prolyl residues (Pro402 and Pro564) in the presence of oxygen that mediates interactions with the von Hippel-Lindau (pVHL) for proteasome destruction.20,–22 PHD2, a direct target of HIF-1α,23 is the main isoform causing HIF-1α degradation in many cell lines, partly because it is most abundantly expressed24 and regarded as the main cellular oxygen sensor.23 As CLL B cells express constitutive levels of HIF-1α under normoxia, we examined PHD2 expression by Western blot analysis. Results indeed demonstrate increased levels of PHD2 in CLL B cells, another downstream target of HIF-1α (Figure 2A). This prompted us to examine the proline-hydroxylation status of HIF-1α in CLL B cells. In fact, we found heavily hydroxylated HIF-1α levels in primary CLL B-cell samples, suggesting that PHD2 in primary CLL B cells is catalytically active (Figure 2B). Given this finding, we wondered, why is HIF-1α constitutively elevated in CLL B cells under normoxia?
Prolyl hydroxylase enzyme is up-regulated and active in CLL B cells. (A) Constitutive levels of HIF-1α up-regulates PHD2 levels. Lysates prepared from CLL B cells (P1-6), normal PBMCs (N1), and purified normal B cells (N2) were analyzed for the expression of PHD2, a downstream target of HIF-1α, by Western blot using a specific antibody. Constitutive levels of HIF-1α in these samples are also shown. Actin was used as the loading control. (B) CLL B cells express heavily hydroxylated HIF-1α. CLL B-cell lysates (labeled as P7-9) were subjected to immunoprecipitation using a monoclonal antibody to HIF-1α. Immunoprecipitated protein was analyzed for posttranslational modification by Western blot using a polyclonal praline (564)–specific hydroxylated HIF-1α antibody. The blot was stripped and reprobed to detect immunoprecipitated HIF-1α. Raji cells pretreated with 100 μM CoCl2 for 6 hours were used as a negative control.
Prolyl hydroxylase enzyme is up-regulated and active in CLL B cells. (A) Constitutive levels of HIF-1α up-regulates PHD2 levels. Lysates prepared from CLL B cells (P1-6), normal PBMCs (N1), and purified normal B cells (N2) were analyzed for the expression of PHD2, a downstream target of HIF-1α, by Western blot using a specific antibody. Constitutive levels of HIF-1α in these samples are also shown. Actin was used as the loading control. (B) CLL B cells express heavily hydroxylated HIF-1α. CLL B-cell lysates (labeled as P7-9) were subjected to immunoprecipitation using a monoclonal antibody to HIF-1α. Immunoprecipitated protein was analyzed for posttranslational modification by Western blot using a polyclonal praline (564)–specific hydroxylated HIF-1α antibody. The blot was stripped and reprobed to detect immunoprecipitated HIF-1α. Raji cells pretreated with 100 μM CoCl2 for 6 hours were used as a negative control.
Accumulation of HIF-1α in CLL B cells is associated with low levels of pVHL
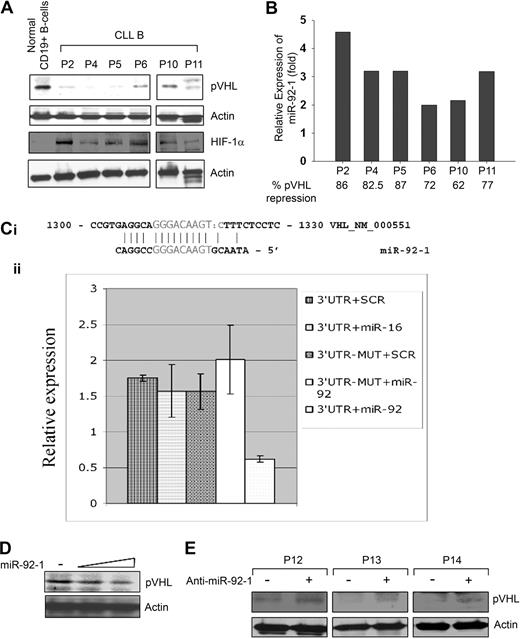
To begin to dissect the mechanism of HIF-1α elevation, we sequenced the VHL gene for the frequently occurring mutations at exon 1, 2, or 325 ; however, sequencing results indicate there are no mutations to the VHL gene of CLL B cells (data not shown). Next, we analyzed CLL B-cell lysates for the expression of pVHL by Western blot using a specific antibody. Results indicated that pVHL levels in CLL B cells were reduced significantly compared with normal CD19+ B cells (Figure 3A). These primary CLL B cells on Western blot analysis exhibited differential levels of HIF-1α elevation than control B cells (Figure 3A).
Endogenous prNL levels and regulation 6, miR 92-1. (A) CLL B cells express a low level of pVHL. CLL B-cell lysates (n = 6) were analyzed for endogenous expression of VHL by Western blot using an antibody to pVHL. Purified CD19+ B cells were used as control for comparison. Similarly, HIF-1α expression in these CLL B-cell lysates was also examined using a specific antibody to HIF-1α. Actin was used as a loading control. (B) CLL B cells overexpress miR-92-1. Expression levels of mature miR-92-1 in CLL B cells were measured by real-time RT-PCR using specific primers. Total RNA was extracted from the same primary CLL B cells (n = 6) used for the Western blot analysis of pVHL as indicated, and purified CD19+ (> 98%) normal B lymphocytes. The single-tube TaqMan microRNA assays were performed to quantify mature miR-92-1 and normalization was performed with RNU6B in triplicate. Relative expression (fold) was calculated using the comparative Ct method, where the expression of miR-92-1 in normal B cells was arbitrarily chosen as one. For comparison of the microRNA fold levels with the VHL protein expression in these samples, densitometric values of VHL from the CLL B-cell Western blot (panel A) were calculated based on the values from normal B cells (arbitrarily chosen as 100%) and are presented below the miR92-1 fold levels. (C) VHL is a target of miR-92-1. The 3′UTR of VHL enables miR-92-1 regulation. (i) The complementarity between VHL cDNA and miR-92-1 is conserved in human and mouse. (ii) Relative repression of firefly luciferase expression standardized to a transfection control, renilla luciferase. As expected, the mutant completely abolished the detected interaction between miR-92-1 and the 3′UTR of VHL as shown in the last column. Experiments were performed twice in triplicate (N = 6). Data are presented as mean ± 1 SE. (D) Introduction of miR-92-1 reduces pVHL level. 293T cells transfected with increasing amounts of miR-92-1 were analyzed for the expression of VHL by Western blot. Actin was used as the loading control. (E) Inhibitor of miR-92-1 up-regulates pVHL. Primary CLL B cells (n = 3) transfected with the antisense inhibitor of miR-92-1 were analyzed for pVHL by Western blot. Actin was used as the loading control. PBMCs from different patients with CLL are marked by numbers.
Endogenous prNL levels and regulation 6, miR 92-1. (A) CLL B cells express a low level of pVHL. CLL B-cell lysates (n = 6) were analyzed for endogenous expression of VHL by Western blot using an antibody to pVHL. Purified CD19+ B cells were used as control for comparison. Similarly, HIF-1α expression in these CLL B-cell lysates was also examined using a specific antibody to HIF-1α. Actin was used as a loading control. (B) CLL B cells overexpress miR-92-1. Expression levels of mature miR-92-1 in CLL B cells were measured by real-time RT-PCR using specific primers. Total RNA was extracted from the same primary CLL B cells (n = 6) used for the Western blot analysis of pVHL as indicated, and purified CD19+ (> 98%) normal B lymphocytes. The single-tube TaqMan microRNA assays were performed to quantify mature miR-92-1 and normalization was performed with RNU6B in triplicate. Relative expression (fold) was calculated using the comparative Ct method, where the expression of miR-92-1 in normal B cells was arbitrarily chosen as one. For comparison of the microRNA fold levels with the VHL protein expression in these samples, densitometric values of VHL from the CLL B-cell Western blot (panel A) were calculated based on the values from normal B cells (arbitrarily chosen as 100%) and are presented below the miR92-1 fold levels. (C) VHL is a target of miR-92-1. The 3′UTR of VHL enables miR-92-1 regulation. (i) The complementarity between VHL cDNA and miR-92-1 is conserved in human and mouse. (ii) Relative repression of firefly luciferase expression standardized to a transfection control, renilla luciferase. As expected, the mutant completely abolished the detected interaction between miR-92-1 and the 3′UTR of VHL as shown in the last column. Experiments were performed twice in triplicate (N = 6). Data are presented as mean ± 1 SE. (D) Introduction of miR-92-1 reduces pVHL level. 293T cells transfected with increasing amounts of miR-92-1 were analyzed for the expression of VHL by Western blot. Actin was used as the loading control. (E) Inhibitor of miR-92-1 up-regulates pVHL. Primary CLL B cells (n = 3) transfected with the antisense inhibitor of miR-92-1 were analyzed for pVHL by Western blot. Actin was used as the loading control. PBMCs from different patients with CLL are marked by numbers.
VHL is posttranscriptionally modulated in CLL B cells
Because the pVHL levels were lower in CLL B cells but with no clear evidence for aberrant pVHL fragments or any mutations of the VHL gene, we looked for other explanations for the lower level of pVHL in CLL B cells. As microRNAs are emerging as potential regulators of many tumor suppressor genes/oncogenes, we interrogated the microRNA genes known to be overexpressed in CLL B cells for potential gene targets, and found that the VHL 3′UTR contains a miR-92-1 target sequence.13,26 In an effort to examine the expression levels of miR-92-1 in CLL B cells and any correlation with pVHL levels, we quantified the relative expression of mature miR-92-1 by real-time RT-PCR. As reported previously,13,26 we found differential levels of up-regulated (2- to 4.5-fold) mature miR-92-1 in CLL B cells obtained from various patients with B-cell CLL (Figure 3B), who had notably low levels of VHL (Figure 3A). Of interest, results obtained from densitometric analyses of the pVHL levels in these samples (shown in Figure 3A), displayed an apparent inverse relationship between the expression of mature miR-92-1 and VHL protein in CLL (Figure 3B). Together, these findings prompted us to hypothesize that the VHL gene could be a possible target of miR-92-1 in CLL.
To explore the possibility of whether miR-92-1 can target the VHL 3′UTR, we first performed an in vitro luciferase-reporter gene assay wherein a 546-bp segment of the 3′UTR of the VHL gene containing the miR-92-1 target sequence was inserted into the pGL3 vector immediately downstream from the stop codon of luciferase. We found a direct effect of miR-92-1 on VHL 3′UTR with significant repression of luciferase activity (approximately 60%) by miR-92-1 compared with control vectors, a mutated target mRNA sequence, or a miRNA that has no interaction site with VHL (miR-16; Figure 3C). This result indicated that wild-type miR-92-1 was able to interact directly with the 3′UTR sequence of VHL and subsequently repress the expression of the VHL gene. To support our observation with the in vitro reporter assay, we transfected a human embryonic kidney cell line (293T) with increasing doses of miR-92-1 and performed Western blot analysis for the expression of endogenous pVHL. Here, we chose to use the 293T cell line, as it expresses constitutive levels of pVHL and is of a nonmalignant phenotype. As expected, we observed a dose-dependent down-regulation of pVHL level in 293T cells with the exogenous introduction of the microRNA (Figure 3D).
To further validate our hypothesis that miR-92-1 regulates VHL expression, we introduced an antisense oligo-targeting miR-92-1 sequence (anti–miR-92-1; Ambion) into primary CLL B cells. Here, Western blot analysis demonstrated an up-regulation of pVHL expression in CLL B cells upon introduction of anti–miR-92-1 (Figure 3E). Together, these observations suggest that overexpression of miR-92-1 can regulate the pVHL expression in CLL B cells, at least in part. To our knowledge, this is the first report demonstrating that the tumor suppressor gene VHL is posttranscriptionally regulated by a microRNA.
HIF-1α accumulates in the nucleus and forms a complex with p300 and STAT3
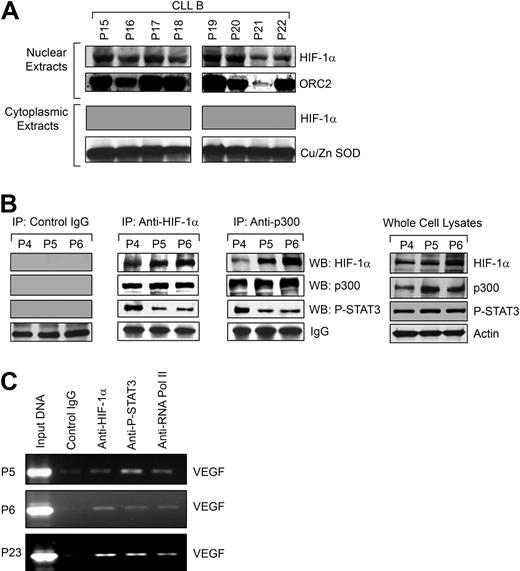
We hypothesized that as HIF-1α is stabilized in CLL B cells under normoxic conditions, it would be able to translocate to the nucleus and then complex with the transcriptional coactivator p300/CBP known to be related to VEGF gene promoter activity.16 We observed accumulation of HIF-1α in the nuclei of the CLL B cells under normoxia (Figure 4A), which confirmed our earlier finding that when examined by immunohistochemistry, bone marrow of patients with CLL demonstrates most of the HIF-1α is in the nuclei of CLL B cells (Figure 1A); in addition, we could not detect HIF-1α in isolated cytoplasmic fractions of primary CLL B cells (Figure 4A).
HIF-1α is functionally active in CLL B cells. (A) HIF-1α accumulates in the nuclei of CLL B cells. Nuclear extracts prepared from freshly isolated CLL B cells (n = 8) were analyzed for nuclear accumulation of HIF-1α by Western blot. An antibody to ORC2 was used for the purity of nuclear preparation. Analysis of the cytoplasmic fractions for HIF-1α was also performed by Western blot. An antibody to Cu/Zn SOD was used to indicate the purity of the cytoplasmic preparations. (B) HIF-1α forms a complex with p300 and P-STAT3. Freshly isolated CLL B-cell lysates (P4-6) were used to immunoprecipitate HIF-1α or p300 using specific mouse monoclonal antibodies. Mouse whole immunoglobulin (IgG) was used as control antibody for immunoprecipitation. The immunoprecipitated complexes were subjected to Western blot analyses using specific antibodies to p300, phospho-STAT3, or HIF-1α, as indicated. Endogenous expression levels of HIF-1α, p300, and phospho-STAT3 in the CLL B-cell lysates used in these experiments are shown by Western blot analyses using specific antibodies. Actin was used as the loading control. (C) HIF-1α and P-STAT3 form an active complex at the VEGF promoter in CLL B cells. Chromatin immunoprecipitation assay was performed with an antibody to HIF-1α, phospho-STAT3, or phospho-RNA polymerase II or control mouse IgG using the cross-linked nuclear extracts from CLL B cells (P5, P6, and P23) as described in “ChIP assay.” The immunoprecipitated DNA was purified and the region from −1386 to −1036 bp of the human VEGF promoter was amplified by PCR. Input DNA is indicated.
HIF-1α is functionally active in CLL B cells. (A) HIF-1α accumulates in the nuclei of CLL B cells. Nuclear extracts prepared from freshly isolated CLL B cells (n = 8) were analyzed for nuclear accumulation of HIF-1α by Western blot. An antibody to ORC2 was used for the purity of nuclear preparation. Analysis of the cytoplasmic fractions for HIF-1α was also performed by Western blot. An antibody to Cu/Zn SOD was used to indicate the purity of the cytoplasmic preparations. (B) HIF-1α forms a complex with p300 and P-STAT3. Freshly isolated CLL B-cell lysates (P4-6) were used to immunoprecipitate HIF-1α or p300 using specific mouse monoclonal antibodies. Mouse whole immunoglobulin (IgG) was used as control antibody for immunoprecipitation. The immunoprecipitated complexes were subjected to Western blot analyses using specific antibodies to p300, phospho-STAT3, or HIF-1α, as indicated. Endogenous expression levels of HIF-1α, p300, and phospho-STAT3 in the CLL B-cell lysates used in these experiments are shown by Western blot analyses using specific antibodies. Actin was used as the loading control. (C) HIF-1α and P-STAT3 form an active complex at the VEGF promoter in CLL B cells. Chromatin immunoprecipitation assay was performed with an antibody to HIF-1α, phospho-STAT3, or phospho-RNA polymerase II or control mouse IgG using the cross-linked nuclear extracts from CLL B cells (P5, P6, and P23) as described in “ChIP assay.” The immunoprecipitated DNA was purified and the region from −1386 to −1036 bp of the human VEGF promoter was amplified by PCR. Input DNA is indicated.
Next, we examined whether HIF-1α forms a complex with p300 in CLL B cells by coimmunoprecipitation experiments using an antibody to HIF-1α or p300 or control mouse IgG. The immunoprecipitated complex obtained from freshly isolated CLL B cells was analyzed for the presence of HIF-1α or p300 by Western blot. We found that HIF-1α was able to bind to the nuclear coactivator p300 (Figure 4B), indicating that HIF-1α is likely a constitutively active transcription factor in CLL B cells. Previously, we have shown that CLL B cells express constitutively active STAT3, which is localized in the nuclei,8 and STAT3 activation has been shown to mediate VEGF transcription.27,28 In addition, both HIF-1α and STAT3 bind the transcriptional coactivator CBP/p300, suggesting that if simultaneous occupancy of the VEGF promoter occurs they may be part of a single transcriptional complex.29,30 Therefore, we examined whether HIF-1α, phospho-STAT3, and p300 coimmunoprecipitate from the CLL B-cell extracts. We found that HIF-1α, p300, and phospho-STAT3 are present in the same immunocomplexes when immunoprecipitated with either an antibody to HIF-1α or p300. These complexes were, however, not detectable in the immunocomplexes obtained using a control antibody (Figure 4B). Endogenous expression levels of HIF-1α, p300, and phospho-STAT3 in the CLL B-cell lysates used in these experiments are also shown by Western blot analyses using specific antibodies (Figure 4B). Together, these results suggest that these 2 DNA-binding transcription factors, HIF-1α and STAT3, are in a complex that could be cooperating in the promotion of VEGF synthesis by their simultaneous binding to the VEGF promoter.
HIF-1α and STAT3 bind simultaneously to the VEGF promoter in CLL B cells
HIF-1α and STAT3 have been implicated in mediating VEGF transcription,27 and simultaneous binding of each transcription factor to the VEGF promoter has been demonstrated in human cancer cells.16 As HIF-1α, p300, and phospho-STAT3 were found to be in the same immunocomplex, we sought to investigate whether HIF-1α and STAT3 are bound to the VEGF promoter in CLL B cells by chromatin immunoprecipitation (ChIP) assay. As shown in Figure 4C, we found that both HIF-1α and phospho-STAT3 were simultaneously bound to the VEGF promoter, but control mouse immunoglobulin showed no reactivity. We observed that phospho-RNA polymerase II was recruited by the HIF-1α, p300, and phospho-STAT3 complex at the VEGF promoter as evident from the PCR-amplified VEGF promoter region from the immunoprecipitated DNA. The latter was brought down with a specific antibody to phospho-RNA polymerase II. Together, these results suggest that HIF-1α and STAT3 form a functional complex in CLL B cells and both are likely capable of activating VEGF transcription by binding directly to the VEGF promoter.
Discussion
Here, we addressed why the VEGF pathway is active in CLL B cells under normoxia. This study indicates that abnormal elevation of HIF-1α, the key upstream regulator of VEGF,31 in CLL B cells is a primary cause for the increased levels of VEGF secretion in CLL B cells. We found that not only is HIF-1α overexpressed and translocated in the nuclei of CLL B cells, but is able to form a complex with the transcriptional coactivator p300 and is likely functional as a transcription factor32 in CLL B cells. Based on our work, the mechanism for the high-resting level of HIF-1α in CLL B cells appears to be in part related to the ability of the HIF-1α to escape from pVHL-mediated degradation, despite the presence of elevated levels of PHD2, a downstream target of HIF-1α. In fact, we found HIF-1α is heavily hydroxylated at proline residues in primary CLL B cells. pVHL is the physiologic regulator of the HIF-1α activity by virtue of targeting it to the proteasome for degradation under normoxia. Although pVHL is reported to be the main regulator of HIF-1α protein stability under normoxia, we recognize that other mechanisms may be operative in CLL B cells in the regulation of HIF. For example activation of AKT or STAT3, which are reported to be constitutively activated in CLL B cells,33,34 can also increase HIF-1α levels either by enhancing translation or transcription, respectively.35,36 However, hypoxic cells, or cells lacking pVHL because of genetic alterations, accumulate high levels of HIF, which can result in significantly elevated levels of VEGF among other HIF-regulated genes.10 To date, these latter factors are the primary known reasons why tumor cells have elevated levels of VEGF.
Our present work uncovered an important and unique mechanism for the constitutive HIF-1α levels in CLL B cells. We report that VHL gene is a target of microRNA-mediated regulation. Our findings in CLL regarding the low level of pVHL can be explained, at least in part, by the direct interaction between miR-92-1, known to be overexpressed in CLL B cells, and the VHL 3′UTR (Figure 3C). The less-than-complete suppression of pVHL by miR-92-1 in the reporter assays is not surprising given that we do not see complete absence of the pVHL protein and because other microRNAs can probably target the pVHL gene and/or its protein expression. However, introduction of miR-92-1 in 293T cells reduced their endogenous pVHL levels. Importantly, introduction of the antisense inhibitor of miR-92-1 displayed a subtle but definite up-regulation of pVHL levels in primary CLL B cells. Relevant to these latter findings, it has recently been shown that the effects of microRNAs on protein levels are usually quite modest, changing their expression levels by less than 2-fold.37 To our knowledge, this is the first report where pVHL expression has been shown to be regulated after transcription by a microRNA rather than by the common genetic alterations previously well characterized in certain human cancers, including renal carcinomas. This finding fits well with the recently proposed roles of microRNAs in CLL.26
The VEGF promoters from different species share a lot of homology regarding the transcription factors consensus binding sites including Sp1/Sp3, AP1/AP2, Egr-1, STAT3, and HIF. The mouse promoter is the only one to contain an additional NF-κB consensus site between −90 and −185, but its functionality has not been proven.38 However, the involvement of NF-κB in VEGF production in cells of human origin has been investigated with variable and conflicting results.39 Previous studies have suggested that HIF-1α or STAT3 alone is capable of activating VEGF transcription.27,28,40,41 It has been shown that the binding of both STAT3 and HIF-1α to the VEGF promoter is required for maximum transcription of VEGF after hypoxia.16 In fact, our ChIP analyses suggest that both HIF-1α and constitutively active STAT3 are bound simultaneously to the VEGF promoter in primary CLL B cells, indicating that HIF-1α and STAT3 are components of a large transcription complex with p300 governing VEGF expression in CLL B cells. Gene activation by p300 is controlled in part by its ability to bind upstream transcription factors, such as those in the composite VEGF regulatory element, and coordinate this complex with the basal transcription machinery.42,–44 Relevant to this point, we also found that RNA polymerase II is recruited at the VEGF promoter in CLL B cells as evident from the ChIP analyses (Figure 4C). These observations suggest that the transcription complex of HIF-1α, STAT3, and p300 are functional and up-regulate VEGF transcription, and consequently VEGF secretion in CLL B cells. Figure 5 summarizes and details our current model for VEGF secretion using the data we have obtained in our studies of the biology of VEGF in CLL B cells.
A model for the mechanism on transcription of human VEGF gene by HIF-1α associated with STAT3 in CLL B cells. Expression of HIF-1α in CLL B cells is stabilized due to a reduced level of pVHL, at least in part, through posttranscriptional modification by the overexpressed miR-92-1. Stabilized HIF-1α localizes in the nucleus and heterodimerizes with HIF-1β. Active HIF-1 heterodimer then forms a complex with constitutively active STAT3 and p300 by physical association. In CLL B cells, this active transcriptional complex binds the human VEGF promoter through the HIF-1– and STAT3-specific DNA-binding elements present on the promoter. DNA-bound HIF-1/STAT3/p300 complex then recruits RNA polymerase II transcription machinery at the promoter to transcribe the VEGF gene.
A model for the mechanism on transcription of human VEGF gene by HIF-1α associated with STAT3 in CLL B cells. Expression of HIF-1α in CLL B cells is stabilized due to a reduced level of pVHL, at least in part, through posttranscriptional modification by the overexpressed miR-92-1. Stabilized HIF-1α localizes in the nucleus and heterodimerizes with HIF-1β. Active HIF-1 heterodimer then forms a complex with constitutively active STAT3 and p300 by physical association. In CLL B cells, this active transcriptional complex binds the human VEGF promoter through the HIF-1– and STAT3-specific DNA-binding elements present on the promoter. DNA-bound HIF-1/STAT3/p300 complex then recruits RNA polymerase II transcription machinery at the promoter to transcribe the VEGF gene.
We believe these findings highlight some novel therapeutic opportunities in CLL. Elevated HIF-1α levels in solid tumors have been shown to be associated with more aggressive tumors.18,45,46 The important issue is in relation to CLL: does HIF-1α overexpression relate to CLL B-cell progression? It has been shown that pancreatic cells overexpressing HIF-1α have increased apoptosis resistance induced by hypoxia and being deprived of nutrition.47 In aggressive prostate cancer, increased expression of HIF-1α is accompanied by loss of p53 transcription, insensitivity to p21, and the ability to proliferate in hypoxia.48 Interestingly, more aggressive CLL disease is strongly associated with a genetic defect involving p53. Could HIF-1α overexpression be the crucial biologic feature that helps make the disease more aggressive in patients with CLL with an abnormal p53 defect?2,49 We speculate that drugs or drug conjugates that can down-regulate HIF-1α levels or perhaps block the interaction between miR-92-1 and the VHL gene in CLL B cells will be beneficial.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mr Edson Spencer for his continued philanthropic support (N.E.K.).
This work was supported by the National Institutes of Health, National Cancer Institute (Rockville, MD) grant R01 CA116237 (N.E.K.) and grants P01CA76259 and P01CA81534 (C.M.C.), and by a Kimmel Scholar award by CLL Global Research Foundation (Houston, TX; G.A.C.).
National Institutes of Health
Authorship
Contribution: A.K.G. designed and performed the experiments, analyzed the data, and wrote the manuscript; T.D.S. analyzed the data; A.C., C.T., S.V., C.-g.L., G.A.C., and C.M.C. designed and performed experiments and analyzed the data; D.A.C. and A.J.G. performed experiments and analyzed data; C.S., L.E.W., Y.K.L. performed experiments; and D.M. and N.E.K. designed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Neil E. Kay, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: kay.neil@mayo.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal