Abstract
Venous thromboembolism (VTE) is a complex disease that has a major genetic component of risk. To identify genetic factors that may modify the risk of VTE, we conducted a genome-wide association study by analyzing approximately 317 000 single nucleotide polymorphisms (SNPs) in 453 VTE cases and 1327 controls. Only 3 SNPs located in the FV and ABO blood group genes were found associated with VTE at a genome-wide significant level of 1.7 × 10−7. Detailed analysis of these SNPs in additional cohorts of more than 1700 cases and 1400 controls revealed that the association observed at the FV locus was the result of the increased risk mediated by the FV Leiden mutation, whereas O and A2 blood groups were found to be at lower risk for VTE. Apart from the FV and ABO loci, no other locus was found strongly associated with VTE. However, using this large cohort of subjects, we were able to replicate the mild effects of 2 nonsynonymous SNPs, rs1613662 in GP6 and rs13146272 in CYP4V2, recently suspected to be associated with VTE.
Introduction
Venous thromboembolism (VTE) is a frequent disease with an annual incidence of 1 to 3 per 1000 in the general population.1 VTE is responsible for a substantial public health burden, with an estimated mortality rate resulting from disease of approximately 10% in patients with deep vein thrombosis (DVT) and 15% in patients with pulmonary embolism (PE). More than 10 000 deaths annually in France are attributed to PE,2 which is also the principal cause of death during pregnancy in industrialized countries. VTE usually arises in situations where external risk factors such as surgery, hospitalization, pregnancy, puerperium, or oral contraception are present.1 However, family and twin studies indicate that the disease also has a major genetic component of risk.3,4 It is estimated that 30% of patients with idiopathic VTE (ie, without acquired risk factors) have an identified coagulation genetic defect such as coagulation inhibitor deficiencies, factor V (FV) Leiden or factor II (FII) 20210A mutations.5 Coagulation inhibitor (protein C [PC], protein S [PS], or antithrombin [AT]) deficiencies are found in less than 1% of the population and represent only a few percent of patients with VTE. These deficiencies increase the risk of VTE approximately 10-fold in heterozygous carriers, with the highest risk observed for antithrombin deficiency. FV Leiden is present in 4% to 6% of the general population, and heterozygosity for this mutation leads to a 3-fold increase in relative risk of VTE. FII 20210A is less common (found in 2% of whites) and increases the risk of VTE approximately 3-fold. In addition to these defects, ABO locus is also a known susceptibility locus for VTE6 as non-O blood group carriers share a higher risk of VTE than O carriers. Non-O blood group carriers also have higher levels of von Willebrand factor (VWF) and factor VIII (FVIII), 2 known risk factors for VTE,7 and this is probably the mechanism by which blood group is related to the risk of VTE.8 During the last 10 years, genetic research in the field of VTE has focused mainly on a limited number of candidate genes. However, it has not provided clear and consistent results that could have led to the identification of new genetic variants involved either in the coagulation/fibrinolysis cascade and/or in other pathways that contribute to VTE risk.9
The growing knowledge of human gene sequence variability and the availability of new technologies, allowing genotyping hundreds of thousand single nucleotide polymorphisms (SNPs) at low cost, have led to the initiation of the Genome Wide Association Study (GWAS) approach to identify genetic variants that are associated with multifactorial diseases. This approach has been successfully applied to give rise to the identification of novel susceptibility loci in many multifactorial diseases10 but, to our knowledge, has not been yet applied to VTE. A large-scale association scan on VTE has recently been performed,11 but it was restricted to only 19 682 SNPs located in known genes. In this report, we described the results of (what we think is) the first genome-wide association analysis of VTE using a panel of approximately 317 000 SNPs.
Methods
Studies
All subjects were whites of Northern European origin, and informed written consent was obtained in accordance with the Declaration of Helsinki. All studies were approved by the local Ethics Committee at La Timone Hospital (Marseille, France).
GWAS data
The GWAS screening was performed in a sample composed of 453 patients with early age of onset of VTE (< 50 years) recruited in 4 different French centers (Grenoble, Marseille, Montpellier, and Paris) between 1999 and 2006, and 1327 healthy French persons from the Suvimax study.12 Criteria for patient inclusion in the study were (1) European origin; (2) early age of onset of first VTE (< 50 years); (3) DVT and/or PE as first event (documented by venography, Doppler ultrasound, angiography, and/or ventilation/perfusion lung scan); (4) lack of acquired risk factors at the time of VTE (including surgery, hospitalization, pregnancy, puerperium, oral contraception, cancer, and autoimmune disease); and (5) lack of strong known genetic risk factors, including AT, PC, or PS deficiencies, and homozygosity for FV Leiden or FII 20210A (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Criteria for inclusion of the 1327 studied controls were European origin, no chronic conditions, and no regular medicines.
MARTHA study
The MARseille THrombosis Association study (MARTHA) is a case-control study that includes 1150 cases and 801 controls. The case sample was composed of unrelated consecutively recruited whites with a documented history of VTE and without strong known risk factors, including AT, PC, or PS deficiency, homozygosity for FV Leiden or FII 20210A, and lupus anticoagulant who attended the Thrombophilia Center of La Timone Hospital between January 1994 and October 2005. All study subjects were interviewed by a physician about their medical history, which emphasized manifestations of DVT and PE using a standardized questionnaire. The date of occurrence of every episode of VTE and the presence of precipitating factors (such as surgery, trauma, prolonged immobilization, pregnancy or puerperium, and oral contraceptive intake) were collected (Table S2). VTE was classified as secondary when occurring within 3 months after exposure to exogenous risk factors, including surgery, trauma, immobilization for 7 days or more, oral contraceptive use, pregnancy, and puerperium. In the absence of these risk factors, VTE was defined as primary. In all cases, history of VTE was recorded before the laboratory diagnosis was made. The thrombotic events were documented by venography, Doppler ultrasound, spiral computed tomographic scanning angiography, and/or ventilation/perfusion lung scan.
As the MARTHA study was initially designed to study the interaction between common genetic risk factors (heterozygous FV Leiden and FII 2021A), the control sample was composed of 2 groups. One included 475 healthy French whites without a personal history of cardiovascular disease (including VTE) from the Marseilles area. The second one consisted of 326 healthy French white heterozygotes for the FV Leiden or FII 20210A mutations selected from the national health examination centers of French Social Security in collaboration with the Hemostasis and Thrombosis Study Group.
FARIVE study
The FARIVE study is a multicenter case-control study involving consecutive subjects treated as inpatients or outpatients for a first episode of proximal DVT and/or PE. Recruitment began in January 2003 and is still ongoing. The protocol methods for diagnosing DVT are venography and compression-ultrasonography, and those for PE are spiral computed tomography, high probability ventilation-perfusion lung scan, pulmonary angiography, and compatible physical findings in a patient with proven DVT. Patients are not eligible for the study if they are younger than 18 years, have already had VTE, have a diagnosis of active cancer or a history of malignancy less than 5 years previously, or have a short life expectancy because of other causes. The control group was composed during the same period and consists of age- and sex-matched inpatients and outpatients free of venous and arterial thrombotic disease. Potential control subjects with cancer, liver or kidney failure, or a history of venous and/or arterial thrombotic disease are ineligible. After enrollment, demographic and clinical characteristics were recorded during a face-to-face interview conducted by a medical doctor or a research assistant, using a standardized questionnaire covering age, educational level, medication history, personal and familial history of thrombotic disease, and acquired risk factors for VTE, including pregnancy, postpartum, use of oral contraceptives or estrogen replacement therapy, trauma or surgery less than 3 months previously, prolonged immobilization, or travel lasting more than 5 hours. Patients were classified as having secondary (provoked) VTE if they had one or more of these acquired risk factors. All other patients were considered to have an unprovoked episode of VTE (Table S3).
Genotyping
Persons for the initial GWAS analysis were typed with the Illumina Sentrix HumanHap300 Beadchip (Eindhoven, The Netherlands) containing 317 139 SNPs. SNPs were excluded if any of the following quality criteria was met: (1) test for deviation from Hardy-Weinberg equilibrium in the control groups with P value less than 10−5; (2) minor allele frequency (MAF) less than 5% in the patient group or less than 1% in the control group; or (3) missing genotype frequency more than or equal to 3% per SNP. These criteria left 291 872 SNPs tested for association with VTE. Subjects were excluded entirely if they failed specific criteria, including genotyping success rates less than 95%, close relationship to other samples as detected with a pairwise identical by sharing (IBS) analysis, and non-European ancestry as detected by principal component analysis through the use of the Eigenstrat program.13 This resulted in the exclusion of 34 cases and 99 controls. In MARTHA and FARIVE studies, genotyping was performed with TaqMan technology (Applied Biosystems, Foster City, CA).
Statistical analysis
GWAS screening was performed with the 2-sided Cochran-Armitage trend test.14 To correct for variance inflation resulting from genotyping errors or subtle subpopulation structure, if any, the observed χ2 statistic was adjusting using the genomic control (GC) approach based on the median test statistic.15 A quantile-quantile (Q-Q) plot analysis was also carried out to check whether the distribution of the inflation corrected P values deviated from the expected distribution under the null hypothesis of no genetic association. Population stratification was also investigated through principal component analysis.13 All quality checked SNPs showing GC-corrected P values significant at the genome-wide level of α = 1.71 × 10−7 were further investigated for association with VTE in the replication studies using Cochran-Armitage trend test. Logistic regression analyses were also used to estimate age- and sex-adjusted odds ratio (OR). Homogeneity of genetic effects across studies was assessed through the use of Mantel-Haenszel method16 that provided, when appropriate, combined estimates.
Linkage disequilibrium (LD) analysis was carried out by use of the Haploview software.17 For assessing the effects of variants not represented on the Illumina Sentrix HumanHap300 Beadchip, variants were imputed using the MACH 1.0 software18 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) according to the haplotype information in the CEU (European) HapMap population.
Results
GWAS analysis
In the first step of the study, 291 872 SNPs were tested for association in a sample of 419 patients with early age of onset of first VTE and 1228 controls. Although a quantile-quantile plot of the association test P values did not visually show obvious deviation from what was expected under the assumption of no genetic association, except in the extreme right tail of the distribution (Figure 1), the GC parameter was 1.22, raising the possibility of stratification between cases and controls. To correct for this possibility, we adjusted for significant principal components and obtained a new GC value of 1.00. Three SNPs reached the genome-wide significance at α = 1.71 × 10−7 after Bonferroni correction for the number of tested SNPs. One of these 3 SNPs, rs2420371, was located at the FV locus on chromosome 1q23 and the other 2, rs505922 and rs657152, were located in the ABO locus on chromosome 9q34.1-q34.2. These 3 SNPs along with an additional 2 SNPs, 1 at each locus, that were just under the threshold of genome-wide significance, were then tested for replication in the MARTHA study. All strongly replicated with P values less than 3.0 × 10−5 (Table 1).
Q-Q plot analysis of 291 872 SNPs between 1228 controls and 419 VTE cases. The Q-Q plot did not indicate any evidence for systematic increase in false positives as the observed distribution did not deviate from what was expected, except in its extreme right tail. However, the GC inflation factor λ was estimated as 1.22, indicating a systematic inflation of the test statistics.
Q-Q plot analysis of 291 872 SNPs between 1228 controls and 419 VTE cases. The Q-Q plot did not indicate any evidence for systematic increase in false positives as the observed distribution did not deviate from what was expected, except in its extreme right tail. However, the GC inflation factor λ was estimated as 1.22, indicating a systematic inflation of the test statistics.
Suggested VTE-associated SNPs identified in the GWAS and their replication in independent samples
| Chromosome, gene . | Position . | rsID . | Alleles* . | Study† . | MAF . | Allelic association‡ OR (95% confidence interval) . | P . | |
|---|---|---|---|---|---|---|---|---|
| Controls . | Cases . | |||||||
| 1, F5 | 167,695,568 | rs1208134 | A/G | GWAS | 0.05 | 0.12 | 2.29 (1.58-3.32) | 3.47 × 10−7 |
| MARTHA | 0.13 | 0.19 | 1.57 (1.31-1.88) | 2.89 × 10−7 | ||||
| FARIVE | 0.07 | 0.09 | 1.31 (0.97-1.78) | 7.90 × 10−3 | ||||
| 167,758,179 | rs2420371 | A/G | GWAS | 0.07 | 0.15 | 2.27 (1.62-3.18) | 8.08 × 10−10 | |
| MARTHA | 0.16 | 0.21 | 1.39 (1.17-1.64) | 3.00 × 10−5 | ||||
| FARIVE | 0.07 | 0.10 | 1.44 (1.07-1.93) | 1.80 × 10−3 | ||||
| 9, ABO | 135,129,086 | rs657152 | C/A | GWAS | 0.38 | 0.54 | 1.89 (1.51-2.36) | 2.22 × 10−13 |
| MARTHA | 0.37 | 0.51 | 1.75 (1.51-2.03) | 1.20 × 10−13 | ||||
| FARIVE | 0.36 | 0.47 | 1.58 (1.34-1.87) | 5.19 × 10−8 | ||||
| 135,139,050 | rs505922 | T/C | GWAS | 0.35 | 0.52 | 1.91 (1.53-2.39) | 1.48 × 10−14 | |
| MARTHA | 0.35 | 0.49 | 1.81 (1.56-2.11) | 3.72 × 10−15 | ||||
| FARIVE | 0.34 | 0.46 | 1.65 (1.39-1.95) | 7.25 × 10−9 | ||||
| 135,139,543 | rs630014 | G/A | GWAS | 0.47 | 0.37 | 0.64 (0.51-0.80) | 2.0 × 10−7 | |
| MARTHA | 0.49 | 0.38 | 0.66 (0.57-0.76) | 1.21 × 10−8 | ||||
| FARIVE | 0.49 | 0.38 | 0.63 (0.53-0.74) | 5.01 × 10−8 | ||||
| Chromosome, gene . | Position . | rsID . | Alleles* . | Study† . | MAF . | Allelic association‡ OR (95% confidence interval) . | P . | |
|---|---|---|---|---|---|---|---|---|
| Controls . | Cases . | |||||||
| 1, F5 | 167,695,568 | rs1208134 | A/G | GWAS | 0.05 | 0.12 | 2.29 (1.58-3.32) | 3.47 × 10−7 |
| MARTHA | 0.13 | 0.19 | 1.57 (1.31-1.88) | 2.89 × 10−7 | ||||
| FARIVE | 0.07 | 0.09 | 1.31 (0.97-1.78) | 7.90 × 10−3 | ||||
| 167,758,179 | rs2420371 | A/G | GWAS | 0.07 | 0.15 | 2.27 (1.62-3.18) | 8.08 × 10−10 | |
| MARTHA | 0.16 | 0.21 | 1.39 (1.17-1.64) | 3.00 × 10−5 | ||||
| FARIVE | 0.07 | 0.10 | 1.44 (1.07-1.93) | 1.80 × 10−3 | ||||
| 9, ABO | 135,129,086 | rs657152 | C/A | GWAS | 0.38 | 0.54 | 1.89 (1.51-2.36) | 2.22 × 10−13 |
| MARTHA | 0.37 | 0.51 | 1.75 (1.51-2.03) | 1.20 × 10−13 | ||||
| FARIVE | 0.36 | 0.47 | 1.58 (1.34-1.87) | 5.19 × 10−8 | ||||
| 135,139,050 | rs505922 | T/C | GWAS | 0.35 | 0.52 | 1.91 (1.53-2.39) | 1.48 × 10−14 | |
| MARTHA | 0.35 | 0.49 | 1.81 (1.56-2.11) | 3.72 × 10−15 | ||||
| FARIVE | 0.34 | 0.46 | 1.65 (1.39-1.95) | 7.25 × 10−9 | ||||
| 135,139,543 | rs630014 | G/A | GWAS | 0.47 | 0.37 | 0.64 (0.51-0.80) | 2.0 × 10−7 | |
| MARTHA | 0.49 | 0.38 | 0.66 (0.57-0.76) | 1.21 × 10−8 | ||||
| FARIVE | 0.49 | 0.38 | 0.63 (0.53-0.74) | 5.01 × 10−8 | ||||
Common/minor alleles observed in controls.
In the GWAS study, the number of controls and cases were 1228 and 419. In MARTHA and FARIVE, corresponding numbers were 801 and 1150, and 607 and 607, respectively.
Allelic OR (95% confidence interval) associated with the minor allele. In GWAS, P values were obtained after correction for stratification using Eigenstrat. In MARTHA and FARIVE, reported P values were obtained using the Cochran-Armitage trend test.
The 2 FV SNPs, rs1208134 and rs2420371, were in strong LD with each other (D′ = + 0.73; r2 = 0.44; P < 10−4) in MARTHA, with similar amplitude in controls and cases. They were also in strong LD with the FV Leiden, D′ = 0.80 and r2 = 0.50 (P < 10−4) for the rs1208134, and D′ = 0.96 and r2 = 0.54 (P < 10−4) for the rs2420371. In MARTHA, carrying the FV Leiden was associated with an OR of 2.01 (1.63-2.48; P = 9.91 × 10−11) that was slightly higher than those associated with the rs1208134 and rs2420371 (Table 1). After adjusting for FV Leiden, these 2 FV SNPs were no longer associated with VTE risk, confirming that their effects were the result of their LD with the FV Leiden (Table S4). In FARIVE, the rs1208134 and rs2420371 showed some trends of association with VTE (P = .079 and P = .018, respectively; Table 1). These associations disappeared (P = .443 and P = .545, respectively) after adjusting for FV Leiden that was associated with an OR for VTE of 2.46 (1.55-3.93; P = 1.50 × 10−4).
With respect to the ABO locus, it has recently been suggested that SNPs located at this locus but not involved in the determination of the ABO blood group could modify the activity of the glycosyltransferase encoded by this locus.19 Therefore, to verify that the effects observed at the ABO SNPs were completely explained by their LD with the ABO blood group, we genotyped 3 additional SNPs exactly tagging the ABO blood group rs8176750 (A2), rs8176746 (B), and rs8176719 (O), in MARTHA. The 3 ABO SNPs identified in the GWAS were indeed in strong LD with the SNPs tagging the ABO blood group (Figure 2). As expected, O blood group persons were at lower risk of VTE than non-O blood group with an OR of 0.33 (0.26-0.42; P = 1.70 × 10−18), whereas the A2 blood group was independently associated with decreased risk of VTE (OR = 0.53; 0.38-0.74; P = 2.46 × 10−4). After adjusting for ABO blood group, none of the rs657152, rs505922, or rs630014 remained associated with VTE, confirming that their effects were the result of LD with the SNPs defining the ABO blood group. Note that directly typing the ABO blood group provided stronger statistical association than the ABO SNPs detected from the GWAS (Table 1). These results were confirmed in the FARIVE study where the 6 ABO SNPs were also genotyped. In FARIVE, O and A2 blood groups were independently associated with decreased risk of VTE, OR = 0.53(0.41-0.69; P = 2.21 × 10−6) and OR = 0.73 (0.49-1.10; P = .13), respectively, and none of the rs657152, rs505922, or rs630014 remained significantly associated with VTE after adjusting for ABO blood group.
Linkage disequilibrium matrix (expressed in terms of D′) observed at the ABO locus in the case-control MARTHA samples.
Linkage disequilibrium matrix (expressed in terms of D′) observed at the ABO locus in the case-control MARTHA samples.
Candidate gene analysis
From a literature search, we identified 25 candidate genes involved in coagulation and fibrinolysis pathways. A total of 172 SNPs overlapping these 25 candidate genes were available in the array used for the GWAS scan (Table S5). Among those, only 2 displayed promising associations (P < 10−3) with VTE. The first one, rs6825454, is located within the FGA/FGB cluster on chromosome 4, whereas the second one, rs867186, is a nonsynonymous (Ser219Gly) polymorphism located in the EPCR gene on chromosome 20. Only one SNP within the F2 (prothrombin) gene, rs5896, was available in the array but was not associated with VTE. This SNP was not in LD (r2 = 0.00) with the FII 20210A mutation, and its minor allele frequency did not differ between cases with and without the mutation (0.069 vs 0.105; P = .389).
As 3 SNPs, rs2227589 in SERPINC1 gene, rs1613662 in GP6 gene, and rs13146272 in CYP4V2 gene, have very recently been suggested to be associated with VTE using a scan of approximately 20 000 gene-centric SNPs,11 we genotyped them in the MARTHA and FARIVE studies. Unlike the rs1613662 and rs13146272, which were available in the arrays used for the GWAS scan, the rs2227589 was not and was therefore substituted by imputed genotype data in the GWAS data. Whereas no evidence of association was observed for the rs2227589 SNP, we partly confirmed the effects of the rs1613662 and rs13146272 SNPs (Table 2). Combining the results of the 3 studies, carrying the rs1613662-C allele was associated with a decreased risk of VTE (OR = 0.80; 0.70-0.92; P = .002). A similar trend was observed for the rs13146272-C allele (OR = 0.84; 0.74-0.95; P = .007). No evidence of heterogeneity across studies was observed (P = .70 and P = .59 for the rs1613662 and rs13146272, respectively).
Association of rs13146272, rs1613662, and rs2227589 with VTE
| Chromosome, gene . | Position . | rsID . | Alleles* . | Study† . | MAF . | OR‡ (95% confidence interval) . | P . | |
|---|---|---|---|---|---|---|---|---|
| Controls . | Cases . | |||||||
| 1, SERPINC1 | 172,152,839 | rs2227589§ | G/A | GWAS | 0.10 | 0.12 | 1.22 (0.95-1.55) | .112 |
| MARTHA | 0.11 | 0.11 | 0.99 (0.80-1.22) | .918 | ||||
| FARIVE | 0.10 | 0.10 | 0.98 (0.74-1.28) | .868 | ||||
| All | 1.05 (0.92-1.21) | .454 | ||||||
| 4, CYP4V2 | 187,357,205 | rs13146272 | A/C | GWAS | 0.37 | 0.33 | 0.77 (0.61-0.96) | .020 |
| MARTHA | 0.38 | 0.36 | 0.86 (0.68-1.09) | .200 | ||||
| FARIVE | 0.39 | 0.37 | 0.89 (0.74-1.08) | .236 | ||||
| All | 0.84 (0.74-0.95) | .007 | ||||||
| 19, GP6 | 60,228,407 | rs1613662 | T/C | GWAS | 0.15 | 0.13 | 0.76 (0.59-0.99) | .039 |
| MARTHA | 0.15 | 0.14 | 0.86 (0.70-1.06) | .157 | ||||
| FARIVE | 0.16 | 0.13 | 0.76 (0.59-0.99) | .041 | ||||
| All | 0.80 (0.70-0.92) | .002 | ||||||
| Chromosome, gene . | Position . | rsID . | Alleles* . | Study† . | MAF . | OR‡ (95% confidence interval) . | P . | |
|---|---|---|---|---|---|---|---|---|
| Controls . | Cases . | |||||||
| 1, SERPINC1 | 172,152,839 | rs2227589§ | G/A | GWAS | 0.10 | 0.12 | 1.22 (0.95-1.55) | .112 |
| MARTHA | 0.11 | 0.11 | 0.99 (0.80-1.22) | .918 | ||||
| FARIVE | 0.10 | 0.10 | 0.98 (0.74-1.28) | .868 | ||||
| All | 1.05 (0.92-1.21) | .454 | ||||||
| 4, CYP4V2 | 187,357,205 | rs13146272 | A/C | GWAS | 0.37 | 0.33 | 0.77 (0.61-0.96) | .020 |
| MARTHA | 0.38 | 0.36 | 0.86 (0.68-1.09) | .200 | ||||
| FARIVE | 0.39 | 0.37 | 0.89 (0.74-1.08) | .236 | ||||
| All | 0.84 (0.74-0.95) | .007 | ||||||
| 19, GP6 | 60,228,407 | rs1613662 | T/C | GWAS | 0.15 | 0.13 | 0.76 (0.59-0.99) | .039 |
| MARTHA | 0.15 | 0.14 | 0.86 (0.70-1.06) | .157 | ||||
| FARIVE | 0.16 | 0.13 | 0.76 (0.59-0.99) | .041 | ||||
| All | 0.80 (0.70-0.92) | .002 | ||||||
Common/minor alleles observed in controls.
In the GWAS study, the number of controls and cases were 1228 and 419. In MARTHA and FARIVE, corresponding numbers were 801 and 1150, and 607 and 607, respectively.
OR associated with the minor alleles under the assumption of additive effects for the rs2227589 polymorphism and dominant effects for the rs1613662 and rs13146272 polymorphisms.
Imputed genotype data were used for the rs2227589 in the GWAS study, as this SNP was not represented on the Illumina chip.
Discussion
The GWAS approach is now advocated as a powerful approach to identify new susceptibility loci to common diseases. In the present GWAS, the strongest associations were observed at the FV and ABO loci, 2 known loci for VTE. Heterozygosity for FV Leiden is known to be associated with a 7-fold increased risk in VTE.5 It was then natural to detect the FV locus effect in our GWAS because heterozygote FV Leiden carriers were not excluded from our GWAS samples. It is also well established that, as observed in our study, non-O blood group carriers have a higher risk of VTE than O carriers, this effect on VTE risk being probably mediated by an effect on VWF and FVIII levels.20 This is reinforced by the fact that A2 blood group carriers who have been known to have lower VWF and FVIII levels than other non-O blood group persons21 were at lower risk of VTE in our study. As MARTHA was composed of 2 different control groups to study epistatic effects of FV Leiden and FII 20210A mutations, we investigated whether the effect of the ABO blood groups could be modulated by these mutations. No evidence for heterogeneity in the ABO blood group effect on VTE according to these 2 mutations was observed (Figure S1) in contrast to what was observed in the LETS study.22 It is noteworthy that, although the relative risks associated with the ABO blood groups are lower than that associated with the FV Leiden, its population-attributable fraction is much higher (∼ 30% for the non-O status vs 17% for FV Leiden5 ). Despite its higher population-attributable fraction, ABO determination is not included yet in the routine thrombophilia testing as it has not proved to modify the therapeutic approach in daily practice.
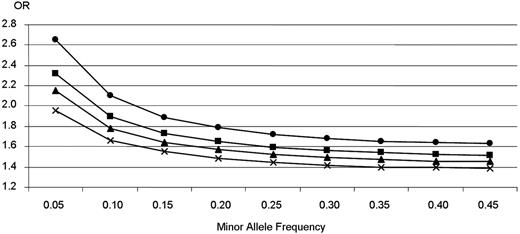
Unlike most previously published GWAS on human complex diseases where the strongest signals were observed either at unexpected known genes or in gene desert regions, the strongest associations observed in the present GWAS were at 2 known candidate loci for VTE. However, as indicated in Figure 3,23 our GWAS study was not well powered to detect genetic effect of modest size, ie, with OR less than 1.4, as generally observed in GWAS, even at a less stringent significance level of 10−3. This, in addition to the stringent statistical criteria we have used, probably explains why we did not identify any other loci despite the use of a selected group of patients with idiopathic VTE at young age aimed at increasing the power of the study. The 2 positive SNPs in the FV locus have an OR of approximately 2.3 and allele frequency of approximately 0.06. Had another SNP of the same MAF and effect size existed, we would have a power of 63% to detect it with our GWAS sample size at the genome-wide significance levels of 1.7 × 10−7. Likewise, the 3 SNPs in the ABO locus have an OR approximately 1.9 and MAF approximately 0.35, an SNP of this level of MAF and OR would have a chance of 100% to be detected in our study. The fact that no other SNP approached the P value of 1.17 × 10−7 suggests that it is doubtful that there is another gene making a contribution comparable with FV and ABO. However, it is entirely possible that other causative variants of lower allele frequencies and/or smaller OR do exist, and further work is thus required to focus on less extreme P values to identify additional disease SNPs.
Genetic effects that could be detected with a power of 80% in a case-control study of 1228 controls and 419 patients. The figure shows the allelic odds ratio (OR) our GWAS study could detect with a power of 80% according to disease allele frequencies and statistical significance levels. ●, ■, ▴, and × represent genome-wide statistical significance level fixed to 10−7, 10−5, 10−4, and 10−3, respectively. Power calculations were performed using the CaTS software. Reprinted from Skol et al23 with permission.
Genetic effects that could be detected with a power of 80% in a case-control study of 1228 controls and 419 patients. The figure shows the allelic odds ratio (OR) our GWAS study could detect with a power of 80% according to disease allele frequencies and statistical significance levels. ●, ■, ▴, and × represent genome-wide statistical significance level fixed to 10−7, 10−5, 10−4, and 10−3, respectively. Power calculations were performed using the CaTS software. Reprinted from Skol et al23 with permission.
Our data also enable us to replicate, for the first time, the findings obtained from a recent scan of nonsynonymous SNPs for VTE based on an initial screening of pooled DNA samples of 443 cases and 453 controls.11 Whereas we did not observe any association between rs2227589 (SERPINC1 gene) and VTE, we confirmed those regarding the rs1613662 in GP6 gene and the rs13146272 in CYP4V2 gene. Besides, these effects were homogeneous according to FV and FII mutations (data not shown). In addition, 172 SNPs available in the array used for the GWAS were located in candidate genes involved in the coagulation process, and 2 of them displayed promising associations (P < 10−3) with VTE, albeit not reaching the a priori threshold (ie, P < 10−7) required for replication. The first one is a nonsynonymous (Ser219Gly) polymorphism located in the EPCR gene. The Gly allele is known to be associated with higher soluble endothelial protein C receptor levels24 ; however, its influence on the risk of VTE is still debated.25,26 The second one, rs867186, is located in the vicinity of the FGB/FGA/FGG cluster and has never been studied in VTE. However, according to HapMap database, rs867186 is in strong LD with both the nonsynonymous rs6050 (Thr312Ala) of the FGA gene (D′ = 1) and the FGG rs2066865 (D′ = 0.96). Interestingly, the rs2066865-T allele belongs to a haplotype that has been recently suspected to be associated with an increased risk of VTE and with reduced fibrinogen γ′ levels.27 Finally, the F2 gene is a known susceptibility locus for VTE. However, only one F2 gene SNP was on the array used for our GWAS scan, and it was not in LD with the FII 20210A mutation. This explains why the F2 gene was not detected in our GWAS.
In conclusion, this report suggests that common susceptibility alleles with rather strong effects, apart from the FV and ABO loci, probably do not contribute to VTE risk in European populations and that joint efforts still have to be done to gather a large collection of VTE patients and to identify VTE-associated SNPs with modest size effect.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Isabelle Martinez, Véronique Remones, Fouad Dali-Ali, Narie Billerey and Gwenaelle Burgos for their excellent technical assistance, and D. Brunet, M.C. Barthet, K. Pouymayou, and C. Frere for the recruitment of MARTHA patients.
The FARIVE study was supported by grants from the Fondation pour la Recherche Médicale, the Program Hospitalier de recherché Clinique, the Fondation de France, and the Leducq Foundation. The MARTHA study was supported by a grant from the Program Hospitalier de la Recherche Clinique.
A complete list of MARTHA and FARIVE investigators appears online.
Authorship
Contribution: D.-A.T. and P.-E.M. designed research, analyzed and interpreted data, and wrote the manuscript; S.H. analyzed and interpreted data and wrote the manuscript; N.S. collected, analyzed, and interpreted data; I.J.-V., L.T., and M.L. designed research and analyzed and interpreted data; C.B.-A., J.-F.S., G.P., L.D., and J.E. collected data; and P.G., D.Z., and M.-C.A. analyzed and interpreted data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre-Emmanuel Morange, Laboratory Hematology Centre Hospitalier Universitaire Timone, 264 rue Saint-Pierre, Marseille, 13385 Cedex 05, France; e-mail: pmorange@ap-hm.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal