Abstract
By presenting antigenic peptides on the cell surface, human leukocyte antigen (HLA) class I molecules are critical for immune defense. Their surface density determines, to a large extent, the level of CD8+ T cell–dependent immune reactions; their loss is a major mechanism of immune escape. Therefore, powerful processes should regulate their surface expression. Here we document the mechanisms used by CD99 to mediate HLA class I modulation. Up-regulation of HLA class I by IFN-γ requires CD99. In the trans Golgi network (TGN), and up to the cell surface, CD99 and HLA class I are physically associated via their transmembrane domain. CD99 also binds p230/golgin-245, a coiled-coil protein that recycles between the cytosol and buds/vesicles of the TGN and which plays a fundamental role in trafficking transport vesicles. p230/golgin-245 is anchored within TGN membranes via its Golgin-97, RanBP1, IMh1p, P230 (GRIP) domain and the overexpression of which leads to surface and intracellular down-modulation of HLA class I molecules.
Introduction
CD99 is a ubiquitous 32-kDa integral transmembrane protein encoded by the pseudoautosomal MIC2 gene, which shares no homology with any known protein family.1-4 However, a gene called XG, situated in the pseudoautosomal region (PAR) immediately proximal (centromeric) to MIC2, that encodes the protein termed Xg, shares 48% homology with CD99 but its functions are unknown.5 To date, no CD99 counterpart has been identified with certainty in mice, when considering the lack of similar functional commitment and the distant homologies of the CD99-like molecules described in both species.6 CD99 is heavily O-glycosylated and, as a result of alternative splicing that introduces a stop codon in the intracytoplasmic region, is made of a long- (185 residues) and a short-deleted (161 residues) isoform.7
From a functional viewpoint, CD99 has been described as a T-cell costimulator, and as a strong activator of the actin cytoskeleton and of α4β1 integrin, promoting cell adhesion and homotypic aggregation, immediate arrest on an inflamed vascular endothelium, and cell migration through the vascular endothelium.8-11 In addition, ligation of CD99 promotes apoptosis that is limited to particular cell types, including immature T cells.12 The function of CD99 is dictated by the CD99 isoforms expressed by the cells, the long or short, or both isoforms. For instance, activation of actin cytoskeleton (CSK) requires only the expression of the long isoform.13
HLA class I molecules are ubiquitously expressed by almost all cell types. They are decisive elements in major defense mechanisms against intracellular infectious agents, such as viruses, and against nascent tumor cells; they capture intracellular antigens and display them on the cell surface, inducing specific cytotoxic CD8+ cells to destroy potentially harmful cells. In turn, to escape destruction, infected or tumor cells have developed mechanisms that downregulate HLA class I expression. Viruses have been particularly efficient in this way and numerous proteins of viral origin have been described that have the capacity to turn down class I expression. How tumor cells, particularly in the early phases of transformation, turn down their class I expression remains largely unknown.14 Yet the host immune system includes natural killer cells, which have the capacity to recognize cells deprived of HLA class I expression and therefore to counterattack the viral or tumor escape mechanisms.15
It has been observed that decreasing CD99 expression in a B-cell line with an antisense RNA promotes the generation of cells with a Hodgkin-Reed Sternberg phenotype, a morphologic hallmark of Hodgkin disease. Interestingly, in the B cells rendered CD99 negative, HLA class I expression was also decreased and it is known that HLA class I molecules are usually poorly expressed on Reed-Sternberg cells.16 Using this B-cell line, it was shown that decreased expression of CD99 resulted in HLA class I retention within the Golgi.17
The Golgi complex plays a central role in glycoprotein processing and in quality control of secreted cargo in eukaryotic cells. This complex is strongly linked to the cytoskeleton, which facilitates concentration of cargo during transport from the endoplasmic reticulum (ER) through the Golgi stacks, up to the cell surface or other final destinations.18 Typical of Golgi proteins, the golgins form a family of large proteins characterized by extensive predictive coiled-coil structures and are associated with the TGN via their C-terminal domain GRIP.19,20 The approximately 50-residue GRIP domain is both necessary and sufficient for targeting to the Golgi apparatus and is conserved in animals, fungi, plants, and protozoa.21,22 GRIP golgins are required for both anterograde or retrograde transport processes and are devoted to the transport of given cargoes.23-25
We have observed that CD99 surface expression is required for the action of IFN-γ on HLA class I expression. Without CD99, upon IFN-γ stimulation HLA class I molecules accumulate within the Golgi. Based on the molecular interactions we have observed, we present here a model based on molecular scaffolds that could drive HLA class I to the cell surface in a quantitatively regulated manner.
Methods
Cell lines
Jurkat cells (clone E6-1) from ATCC (Molsheim, France) were grown as reported. BLA2 cell line was a gift from Dr F. Chouaib (Inserm U487, Villejuif, France). CD99-deficient Jurkat cells and CD99pos transfectants were obtained by chemical mutation as previously described.13 Where indicated, 200 U/mL recombinant human IFN-γ (R&D Systems, Lille, France), 6 μg/mL−1 Nocodazole (Sigma-Aldrich, Lisle d' Abeau, France) or 1 μg/mL−1 cytochalasin B (Sigma-Aldrich).
Constructs
Plasmid constructions, cloning, and DNA sequencing were carried out according to standard protocols. The full-length of CD99 cDNA was previously subcloned into pcDNA3 and pcDNA4 expression vectors (Invitrogen, Cergy-Pontoise, France).13 For pcDNA4-CD99 long- and short-form constructs, the sense primer for polymerase chain reaction (PCR) was 5′-ATGGCCCGCGGGGCTGCGCTG-3′ and the antisense primers were, respectively, 5′-CTATTTCTCTAAAAGAGTACG-3′and 5′-TCAGCCATCATTTTCTTTGAAGC-3′, using the pcDNA3-CD99 Long or Short Form plasmids. Both amplified fragments were introduced into the pcDNA4/HisMax TOPO TA vector, according to the manufacturer's instructions (Invitrogen).
CD99 ShRNA targeting 2 sequences shared by both isoforms of CD99 were designed using software provided by the Invitrogen website and converted into 2 pairs of complementary oligonucleotides encoding a hairpin structure (F: GTCCCTGTAACTCAAATGTCA; R: TGAATTTGAGTTACAGGGAC). Oligonucleotides were annealed separately and cloned into pENTER/H1TO (Invitrogen). pGEX4T2-p230(1-270aa) was prepared by PCR using the primers 5′-TCCGGATCCACCATGGAACAGAAACTGATCTCT-3′ and 5′-CGGCCTCGAGTC ATGGCTCTCCATCA C-3′. For pGEX4T2-p230(2000-2230aa), the primers were 5′-GGCAGGATCCATGAGGGAGTTTATACACAGC-3′ and 5′-GCCTCGAGTCAGAAG ATACCACTGCGAGG-3′. The amplified DNA was then cut with BamHI and XhoI and inserted into the BamHI and XhoI sites of pGEX4T2.
For pEGFP-p230(2000-2230aa) and pcDNA4 p230(2000-2230aa), the primers were 5′-GGCACTCGAGATGAGG GAGTTTAATACACAGC-3′ and 5′-TTGGATCCTCAAGATGAAGATCGGAGCC-3′. The amplified DNA was then cut with XhoI and BamHI and inserted into the XhoI and BamHI sites of pEGFP-C3.
For pGEX4T2-CD99 (1-186aa), (1-161aa), (1-121aa), (1-147aa), (1-148aa) constructs, the sense primer 5′-GCGGATCCGCTTACCAGAAAAAGAAGCTATGC-3′ was used with the antisense primers 5′-GGCCGCTCGAGGCTATTTCTCTAAAAGAG-3′, 5′-TATAGCTC GAGGTCAGCCATCATTTTCTTT-3′, 5′-CTTCTCGAGTCAGTCGGCCTCTTCC-3′, 5′-GCGCTCGAGTCAAGCAATGAAGCTAG-3′, and 5′-GCGCTCGAGTCAGTAAGCAATGA AGC-3′, respectively. The amplified fragment was digested with BamHI and XhoI and exchanged for the BamHI/XhoI fragment of pGEX4T2.
For pGEX4T2-IC long-form CD99 (147-185aa) and -IC short-form CD99 (147-161aa), the sense primer was 5′-GCGGATCCGCTTACCAGAAAAAGAAGCTATGC-3′ and the antisense was 5′-GCGCAACGTTCTTGCCATTGCTGCAG-3′, using the pGEX4T2-CD99 long form and pcDNA3-CD99 short form, respectively. The amplified fragment was digested with BamHI and PstI and exchanged for the BamHI/PstI fragment of pGEX4T2.
For pGEX4T2-TM140-147 + IC CD99 short form (140-161aa) and pGEX4T2-TM130-147 + IC CD99 short form (130-161aa), the sense primers were 5′-GCGGATCCGGAG CCATCTCTAGCTTCATTGC-3′ and 5′-GCGGATCCATTGTGGGGGCTGTCGTG-3′, respectively, with the antisense primer 5′-TATAGCTCGAGGTCAGCCATCATTTTCTTT-3′, using the pGEX4T2-CD99 short form (1-161aa) as template. The amplified fragment was digested with BamHI and XhoI and exchanged for the BamHI/XhoI fragment of pGEX4T2.
pGEX4T2-CD99 TM mutant 1 (130-161aa) was prepared using the primers 5′-CC ATTGTGGGGGCTCTCCTGATCGCCGTGGCTGGAGC-3′ and 5′-GCTCCAGCCAC GGCGATCAGGAGAGCCCCCACAATGG-3′; pGEX4T2-CD99 TM mutant 2 (130-161aa) was prepared using the primers 5′-CCATTCTGGGGGCTCTCCTGATCGCCCTGG CTGGAGCC-3′ and 5′-GGCTCCAGCCAGGGCGATCAGGAGAGCCCCCAGAATGG-3′. pGEX4T2-CD99 short form was used as template for mutagenesis, according to the manufacturer's instructions (Stratagene, Amsterdam, The Netherlands). All constructs were confirmed by sequencing.
Transfections
Jurkat cells were electroporated or transfected with lipofectamin as previously described and BLA 2 cells transfected with lipofectamine 2000 (Invitrogen), as described.13 Selection was carried out 72 hours later with 200 μg/mL Zeocin (Invitrogen) and stable transfectant cells were analyzed for CD99 or His-Tagged p230 GRIP expression 3 weeks later.
Site-directed mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene).
Kinetics of HLA I expression by flow cytometry
For the kinetics of HLA I expression under IFN-γ stimulation, after 2 washings, cells (5 × 105/mL) were treated with 200 IU/mL IFN-γ (R&D Systems) and incubated at 37°C. At the appropriate time points, cells were stained with FITC-HLA class I “W6/32” mAb (10 μg/mL), and then analyzed by flow cytometry.
For the kinetics of HLA I molecule internalization, experiments were performed as previously described. Cells were incubated with the anti–HLA I “W6/32” mAb (10 μg/mL) in phosphate-buffered saline (PBS) at 4°C for 60 minutes, washed, and grown at 37°C. At different time points, W6/32 mAb-bound HLA I surface molecules were stained with rabbit anti–mouse-FITC before analysis by flow cytometry. Data are the ratio of the mean fluorescence at various time points to the values obtained at time 0. For externalization experiments, cell surfaces were saturated with unconjugated W6/32 mAbs (250 μg/mL) for 60 minutes at 4°C, washed, and transferred to 37°C in culture medium. Cells were removed at various time points for staining with FITC-HLA I “W6/32” mAb (10 μg/mL) and analyzed by flow cytometry. The relative Ag intensity (RAI) was calculated by subtracting mean fluorescence values of W6/32 mAb-HLA class I complex obtained at time 0 from all values at different time points, and then dividing the mean fluorescence values obtained at time 0. All values are means plus or minus the standard deviation (SD) of 3 independent experiments.
Antibodies and flow cytometry
CD99 mAbs (0662 and 12E7 clones), antihuman HLA I mAb (W6/32), and beta 1 integrin/CD29 (K20) were produced in our laboratory; antihuman HLA I (H300) comes from Santa Cruz Biotechnology (Santa Cruz, CA); anti–His G (6x Histidine tag) was from Invitrogen; PE-conjugated CD99 mAb (3B2/TA8 clone) and FITC-conjugated antihuman LAMP1/CD107a (eBioRDR5) were from e-Bioscience (CliniScience, Montrouge, France); FITC-conjugated W6/32, R-PE–conjugated W6/32, biotin-conjugated W6/32 mAbs, and sheep anti-TGN46 were from AbD Serotec (Dusseldorf, Germany); control Ig, FITC-conjugated CD44 (NKI-P2), CD45 (MEM-28), and alpha 4 integrin/CD49d (BU49) mAbs were from Immunotools (Friesoythe, Germany); TR-SAV was from Perbio Science (Brebieres, France); CD99 (TÜ12), CD47 (B6H12), R-PE–conjugated mouse anti–human IFN-γ receptor α-chain monoclonal antibody (CD119), anti-GRP78, anti-LAMP1/CD107a, anti-GS27, anti-Syntaxin11, anti-Rab8, anti-p230, anti-GM130, and FITC-conjugated GM130 mAbs were from BD Biosciences (Le Pont de Claix, France); anti–p-STAT1 (A-2), anti-STAT1 (9H2) mAbs and rabbit anti-TAP1 (CSA-620), goat anti-LMP2 (C-20), and goat anti-Actin (I-19) antibodies were from Tebu-bio (Le Perray en Yvelines, France).
Secondary FITC-conjugated rabbit anti–mouse Ig comes from Dako Denmark A/S (Trappes, France); TXRD-conjugated horse anti–mouse or rabbit anti–sheep Ig was from Vector Laboratories (CliniSciences); horseradish peroxidase–conjugated anti–mouse and anti–rabbit secondary antibodies were from Cell Signaling Technology (Ozyme, St Quentin Yvelines, France).
Microscopic analysis
Cells were plated on polylysine (PLL)–coated slides (Kindler, Biovalley Marne la Vallée, France) and fixed with 3.7% paraformaldehyde. Cells were neutralized with 50 mM NH4Cl, permeabilized with 0.2% Triton X-100, and incubated with 0.05% saponin plus 1% bovine serum albumin (BSA). Staining was then performed with indicated antibodies for 30 minutes and analyzed with the Hamamatsu Orca CCD camera using a ×63 objective on a Zeiss Axiovert 200 M inverted microscope (Zeiss, Le Pecq, France). Images were taken and deconvoluted if necessary using the Volocity software package (Improvision, Coventry, United Kingdom).
Capping assays
Cells were incubated with saturating amounts of biotinylated anti–HLA class I (W6/32) mAb. After 3 washings with ice-cold PBS, cells were incubated with saturating amounts of Texas-Red-Streptaudin. Cells were induced to cap at 37°C for 60 minutes. The reaction was stopped by adding ice-cold PBS plus 0.1% sodium azide. After saturation with Balb/c mouse serum, labeling of CD99 was realized with CD99 mAb (0662) supplemented with TR-conjugated antibody, for 30 minutes at 4°C. Cells were next plated onto PLL-coated coverslips, fixed and visualized by microscopy as described above.
Electron microscopy, ultracryomicrotomy and immunogold labeling
Cells were fixed with 4% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer (PB). Cell pellets were washed with PB, embedded in 10% (wt/vol) gelatin and infused in 2.3 M sucrose. Mounted gelatin blocks were frozen in liquid nitrogen and ultrathin sections were prepared with an Ultracut FCS ultracryomicrotome (Leica, Wien, Austria). Ultrathin cryosections were collected with 2.3 M sucrose and single or double immunogold labeled with antibodies and protein A coupled to 10 and 15 nm gold (protein A–gold [PAG] obtained from Cell Microscopy Center, Utrecht, The Netherlands). When mouse monoclonal antibodies were used for labeling, sections were incubated with a polyclonal rabbit anti-mouse IgG (Dako, Denmark) before submitted to protein A–gold. This step is omitted when rabbit polyclonal antibodies were used for labeling. Thin sections were analyzed on a Philips CM12 TEM (Eindhoven, The Netherlands) and acquisitions were made with a Morada digital camera (Olympus SIS, Germany).
GST pull-down assays and coimmunoprecipiation
GST fusion proteins were expressed in OneSHOTBL21(DE3)pLysS bacterial cells (Invitrogen) with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3 hours at 30°C.
GST proteins were purified and stored at −80°C on glutathione Sepharose 4B beads (GE Healthcare, Orsay, France) as a 50% slurry in PBS 1 × 50% glycerol and protease inhibitors (10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM PMSF). The amount of fusion protein was estimated by Coomassie blue staining of gels after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
For GST precipitation, Jurkat cells were lysed on ice in TTX lysis buffer (1% Triton X-100, protease inhibitors, in TA1X). Lysates were centrifuged at 16000g at 4°C. Precleared proteins levels were quantified using the Bradford assay (Biorad, Schiltigheim, France). Protein (1 mg) was incubated with the indicated GST fusion proteins on glutathione Sepharose 4B beads at 4°C with rotating for 15 hours. After extensive washing bound proteins were analyzed by immunoblotting.
Coimmunoprecipitation experiments were performed as previously described.13 Jurkat cell lysates were obtained with TBS containing 0.5% TX-100 and protease inhibitors. Normalized supernatants were incubated for 16 hours at 4°C with 5 μg CD99 “O662.” Immunoprecipitation was performed with 70 μL protein A Sepharose (GE Healthcare) for 4 hours at 4°C.
Samples resolved by SDS-PAGE were transferred to nitrocellulose membranes (GE Healthcare), and probed with primary and horseradish peroxidase (HRP)–conjugated antibodies followed by detection by enhanced chemiluminescence.
Results
Fine-tuning between surface expression of CD99 and HLA class I molecules; requirement for CD99 in up-regulation of HLA class I expression by IFN-γ
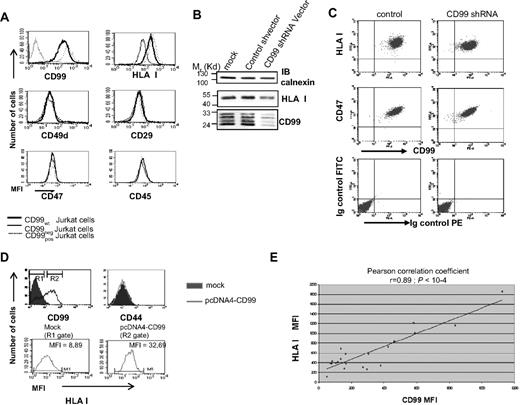
CD99 and HLA class I expressions are correlatively linked in the Jurkat T-cell line as shown by chemical mutation (CD99neg), subsequent re-expression after transfection of CD99neg cells (CD99pos; Figure 1A), and use of a CD99-shRNA (Figure 1B,C), overexpression of CD99 in naturally negative lung adenocarcinoma (Figure 1D) and breast cancer cell lines (not shown). Continuous correlation could be seen in a series of stable CD99 transfection experiments resulting in a strong linear correlation between the surface densities of CD99 and of HLA class I (Pearson correlation coefficient r = 0.89; P < 10−4) (Figure 1E).
Quantitative expression of CD99 and HLA class I molecules is closely related. (A) CD99 and HLA class I surface expression was measured by flow cytometry for wild-type Jurkat T cells, CD99neg and CD99pos cells. CD49d (alpha 4 integrin), CD29 (beta 1 integrin), CD47, and CD45 were measured as controls using “BU49,” “K20,” “B6H12,” and “MEM-28” mAbs, respectively. (B) Effects of a CD99 shRNA on HLA class I expression measured by Western blot analysis with the anti–HLA class I “W6/32” mAb; CD99 expression was measured using the “12E7” mAb. (C) Flow cytometric analysis was performed with a PE-CD99 “3B2/TA8” mAb and with an FITC-HLA class I “W6/32” mAb; analysis was performed 72 hours after transfection with CD99-shRNA or control vector. Results are reported as dot-plot profiles and are representative of at least 3 independent experiments. For controls we included staining with an Ig control isotype and FITC-CD47 (B6H12). (D) Surface expression of HLA class I on BLA2 cells, a lung adenocarcinoma cell line, forced to express CD99. Two gates (R1 and R2) were defined on CD99 histograms (top left panel) and HLA class I expression was measured on cells from each gate (bottom panel). CD44 expression was used as control. Results are representative of at least 3 independent experiments. (E) Correlation between CD99 and HLA class I molecules at the surface of Jurkat CD99neg cells stably transfected with various amounts of CD99 cDNA. Cells were costained with the FITC-CD99 “3B2/TA8” mAb and a PE-HLA class I “W6/32” mAb (Pearson correlation coefficient r = 0.89; P < 10−4).
Quantitative expression of CD99 and HLA class I molecules is closely related. (A) CD99 and HLA class I surface expression was measured by flow cytometry for wild-type Jurkat T cells, CD99neg and CD99pos cells. CD49d (alpha 4 integrin), CD29 (beta 1 integrin), CD47, and CD45 were measured as controls using “BU49,” “K20,” “B6H12,” and “MEM-28” mAbs, respectively. (B) Effects of a CD99 shRNA on HLA class I expression measured by Western blot analysis with the anti–HLA class I “W6/32” mAb; CD99 expression was measured using the “12E7” mAb. (C) Flow cytometric analysis was performed with a PE-CD99 “3B2/TA8” mAb and with an FITC-HLA class I “W6/32” mAb; analysis was performed 72 hours after transfection with CD99-shRNA or control vector. Results are reported as dot-plot profiles and are representative of at least 3 independent experiments. For controls we included staining with an Ig control isotype and FITC-CD47 (B6H12). (D) Surface expression of HLA class I on BLA2 cells, a lung adenocarcinoma cell line, forced to express CD99. Two gates (R1 and R2) were defined on CD99 histograms (top left panel) and HLA class I expression was measured on cells from each gate (bottom panel). CD44 expression was used as control. Results are representative of at least 3 independent experiments. (E) Correlation between CD99 and HLA class I molecules at the surface of Jurkat CD99neg cells stably transfected with various amounts of CD99 cDNA. Cells were costained with the FITC-CD99 “3B2/TA8” mAb and a PE-HLA class I “W6/32” mAb (Pearson correlation coefficient r = 0.89; P < 10−4).
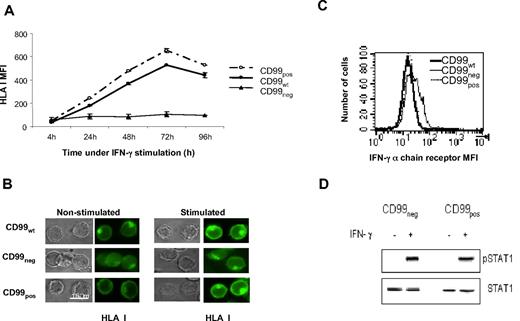
IFN-γ powerfully up-regulates class I expression. Therefore, we examined whether this effect could require CD99 expression. Wild-type Jurkat T cells (CD99WT), CD99neg or CD99 reconstituted (CD99pos) Jurkat T cells, were treated with IFN-γ for various periods of time, and their levels of HLA class I surface expression were measured by flow cytometry (Figure 2A). While no increase in class I expression was observed on CD99neg cells, a strong increase, peaking at 72 hours, occurred on CD99-positive cells, either natural (CD99WT) or reconstituted (CD99pos).
IFN-γ requires CD99 to up-regulate HLA class I expression. (A) Kinetics of HLA class I surface expression in IFN-γ–stimulated cells. After treatment with IFN-γ, cells were incubated at 37°C for the indicated time and were then stained with the FITC-HLA class I “W6/32” mAb. The levels of HLA class I molecules in CD99wt, CD99neg, and CD99pos Jurkat T cells were then measured by flow cytometry. (B) CD99wt, CD99neg, and CD99pos Jurkat cells were grown in medium alone or stimulated with IFN-γ. Cells were permeabilized and HLA class I molecules were detected using the FITC-HLA class I “W6/32” mAb, and analyzed by deconvolution fluorescence microscopy. (C) Surface expression of the IFN-γ receptor α-chain in CD99wt, CD99neg, and CD99pos Jurkat cells were analyzed by flow cytometry using a PE-STAT1 mAb. (D) STAT1 phosphorylation was tested by Western blotting. Cell lysates were prepared from untreated (−) or 20-minute IFN-γ–treated (+) CD99neg andCD99pos Jurkat cells. Cell lysates were immunoblotted for phospho-Tyr 701 STAT1 (top panel). The membrane was stripped and reprobed with an anti-STAT1 Ab (bottom panel).
IFN-γ requires CD99 to up-regulate HLA class I expression. (A) Kinetics of HLA class I surface expression in IFN-γ–stimulated cells. After treatment with IFN-γ, cells were incubated at 37°C for the indicated time and were then stained with the FITC-HLA class I “W6/32” mAb. The levels of HLA class I molecules in CD99wt, CD99neg, and CD99pos Jurkat T cells were then measured by flow cytometry. (B) CD99wt, CD99neg, and CD99pos Jurkat cells were grown in medium alone or stimulated with IFN-γ. Cells were permeabilized and HLA class I molecules were detected using the FITC-HLA class I “W6/32” mAb, and analyzed by deconvolution fluorescence microscopy. (C) Surface expression of the IFN-γ receptor α-chain in CD99wt, CD99neg, and CD99pos Jurkat cells were analyzed by flow cytometry using a PE-STAT1 mAb. (D) STAT1 phosphorylation was tested by Western blotting. Cell lysates were prepared from untreated (−) or 20-minute IFN-γ–treated (+) CD99neg andCD99pos Jurkat cells. Cell lysates were immunoblotted for phospho-Tyr 701 STAT1 (top panel). The membrane was stripped and reprobed with an anti-STAT1 Ab (bottom panel).
Experiments using permeabilized cells where the readout was performed by deconvolution fluorescence microscopy of IFN-γ–stimulated CD99pos Jurkat T cells, showed that a considerable up-regulation of HLA class I surface expression occurred (Figure 2B). In contrast, under the same IFN-γ treatment, CD99neg cells exhibited a very faint surface expression of HLA class I. Moreover, in CD99neg cells treated with IFN-γ, large quantities of class I accumulated within the cells (Figure 2B).
Controls indicated that IFN-γ had no effect on CD99 expression by itself and that the IFN-γ pathway was not altered in CD99neg cells. These controls included α chain expression of IFN-γ receptors (Figure 2C), STAT1 expression, and phosphorylation, which occurred to the same extent in both CD99neg and CD99pos IFN-γ–treated cells (Figure 2D). Thus, CD99 is required for up-regulation of class I by IFN-γ. This cytokine is known to increase the HLA class I externalization process at the cell surface.26,27 As measured by reverse transcription (RT)–PCR, we found that all the genes of the antigen processing machinery were expressed at the same levels in CD99neg and CD99pos cells (not shown). This is particularly striking for the proteasome components that characteristically appear on stimulation with IFN-γ. However, consistent with other observations we found the LMP2 protein to be an exception (not shown).17 It is expressed in higher amounts in CD99neg cells, but we have no explanation for this as, on the contrary, it has been described that forced overexpression of LMP2 leads to enhanced HLA class I expression at the cell surface.28
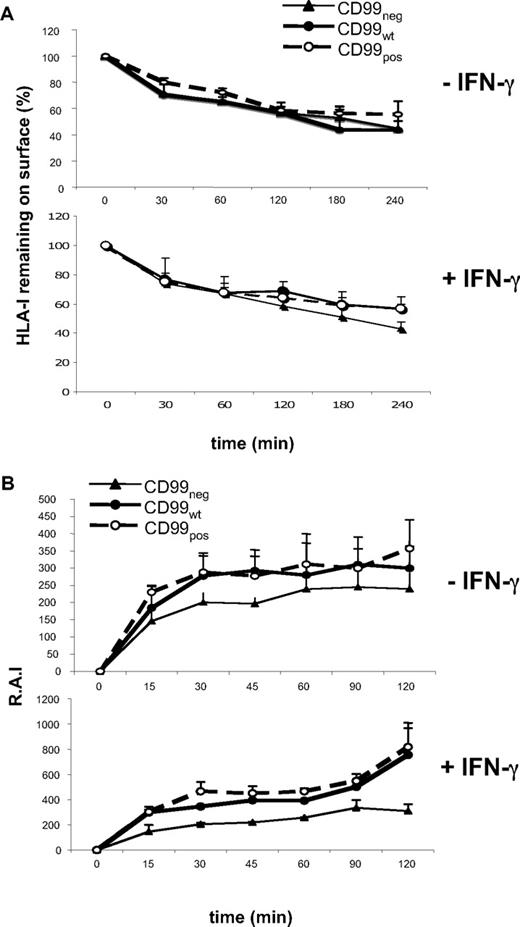
Internalization and externalization of HLA class I were investigated on the 3 Jurkat cell lines, CD99pos, CD99wt, and CD99neg. From Figure 3A, it can be seen that CD99 expression did not modify class I internalization. In contrast, class I externalization was markedly accelerated with IFN-γ treatment of CD99pos but not CD99neg cells (Figure 3B).
CD99 regulates HLA class I externalization. (A) Internalization kinetics of surface-bound anti–HLA class I mAb on untreated (−) or 72-hour IFN-γ–treated (+) CD99neg, CD99pos, and CD99wt Jurkat T cells, as described in “Methods.” Results are expressed as a percentage of remaining HLA class I molecules at the cell surface. All values are means plus or minus SD of 3 independent experiments. (B) Kinetics of cell-surface expression of HLA class I molecules of untreated (−) or 72-hour IFN-γ–treated (+) CD99neg, CD99pos, and CD99wt Jurkat T cells. Surface levels of HLA class I molecules labeled with an FITC-conjugated “W6/32” mAb were analyzed by flow cytometry and reported as relative Ag intensity (RAI), as described in “Kinetics of HLA I expression by flow cytometry.” All values are means plus or minus SD of 3 independent experiments.
CD99 regulates HLA class I externalization. (A) Internalization kinetics of surface-bound anti–HLA class I mAb on untreated (−) or 72-hour IFN-γ–treated (+) CD99neg, CD99pos, and CD99wt Jurkat T cells, as described in “Methods.” Results are expressed as a percentage of remaining HLA class I molecules at the cell surface. All values are means plus or minus SD of 3 independent experiments. (B) Kinetics of cell-surface expression of HLA class I molecules of untreated (−) or 72-hour IFN-γ–treated (+) CD99neg, CD99pos, and CD99wt Jurkat T cells. Surface levels of HLA class I molecules labeled with an FITC-conjugated “W6/32” mAb were analyzed by flow cytometry and reported as relative Ag intensity (RAI), as described in “Kinetics of HLA I expression by flow cytometry.” All values are means plus or minus SD of 3 independent experiments.
Imaging an association between CD99 and HLA class I
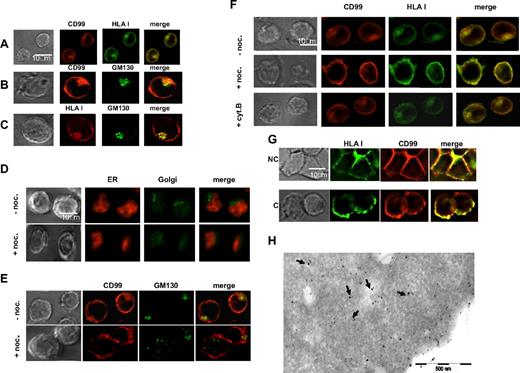
Immunofluorescence analysis of CD99neg cells treated with IFN-γ showed, as indicated above, that large quantities of class I accumulate within the cells, whereas much lesser material was seen in untreated cells (Figure 2B). Double labeling experiments showed colocalization of CD99 and HLA class I (Figure 4A), both at the cell surface and within the cells. Labeling for the Golgi-matrix protein GM130 (Figure 4B,C) revealed that CD99 and HLA class I both localized to the Golgi apparatus. In contrast, no colocalization was detected within the ER using the BIP/GRP 78 marker, a major chaperone of the ER lumen, or with lysosomes using the LAMP1 marker (not shown).
Colocalization of CD99 and HLA class I. Wild-type Jurkat T cells were fixed, permeabilized, and stained for immunofluorescence microscopy (IFM). (A) CD99 molecules were visualized by indirect fluorescence using the CD99 “O662” mAb followed by a Texas Red–conjugated horse anti–mouse IgG. HLA class I molecules were detected with an FITC-conjugated “W6/32” mAb. (B,C) “O662” or “W6/32” mAbs were used to visualize CD99 or HLA class I molecules, respectively; specific binding was revealed with a Texas Red–conjugated horse anti–mouse IgG. The Golgi apparatus was stained with a FITC-GM130 mAb. (D-F) Intracellular colocalization of CD99 and HLA class I molecules is Golgi dependent. Jurkat cells treated with or without nocodazole (noc.) were fixed, permeabilized, and stained for deconvolution fluorescence microscopy. Nocodazole disturbs the Golgi integrity while the ER remains intact. ER and Golgi compartments were costained either with an anti-Bip/GRP78 mAb followed by a Texas Red–conjugated horse anti–mouse IgG and a FITC-GM130 mAb, respectively. Nocodazole affects the intracellular localization of CD99. The “0662” mAb was used for CD99 labeling, which was detected with a Texas Red–conjugated horse anti–mouse IgG; Golgi was detected with an FITC-conjugated GM130 mAb. CD99 and HLA class I labeling was performed using the “O662” mAb with a Texas Red–conjugated horse anti–mouse IgG and the FITC-“W6/32” mAb, respectively. As a control, cells were treated with cytochalasin B (cyt. B), which disturbs the actin cytoskeleton. (G) Cocapping of CD99 and HLA class I molecules. Jurkat T cells were treated as described in “Methods.” Briefly, cells were incubated for 30 minutes with biotinyled anti–HLA class I “W6/32” mAb followed by a 30-minute incubation with Texas Red–streptavidin. Cells were incubated at 37°C for 60 minutes to allow capping. Labeling for CD99 was then performed by incubation with the “0662” mAb followed by incubation for 30 minutes at 4°C with a Texas Red–conjugated rabbit antimouse antibody. Finally, cells were plated onto PLL-coated slides and fixed before microscopic analysis. Cells in noncapping (NC) and capping (C) conditions are shown in the upper panel and in the lower panel, respectively. No cross-reactivity between the anti–Ig conjugates and the primary antibodies was observed in control cells. (H) Immuno-electron microscopy identifies colocalization of HLA class I and CD99. Consecutive immuno-gold double labeling of Jurkat cells: detection of HLA class I by W632 mAb and protein A coupled to 15-nm gold particles followed by labeling with CD99 (O662 mAb) and protein A coupled to 10-nm gold particles. The arrows point to HLA class I and CD99 colocalization areas.
Colocalization of CD99 and HLA class I. Wild-type Jurkat T cells were fixed, permeabilized, and stained for immunofluorescence microscopy (IFM). (A) CD99 molecules were visualized by indirect fluorescence using the CD99 “O662” mAb followed by a Texas Red–conjugated horse anti–mouse IgG. HLA class I molecules were detected with an FITC-conjugated “W6/32” mAb. (B,C) “O662” or “W6/32” mAbs were used to visualize CD99 or HLA class I molecules, respectively; specific binding was revealed with a Texas Red–conjugated horse anti–mouse IgG. The Golgi apparatus was stained with a FITC-GM130 mAb. (D-F) Intracellular colocalization of CD99 and HLA class I molecules is Golgi dependent. Jurkat cells treated with or without nocodazole (noc.) were fixed, permeabilized, and stained for deconvolution fluorescence microscopy. Nocodazole disturbs the Golgi integrity while the ER remains intact. ER and Golgi compartments were costained either with an anti-Bip/GRP78 mAb followed by a Texas Red–conjugated horse anti–mouse IgG and a FITC-GM130 mAb, respectively. Nocodazole affects the intracellular localization of CD99. The “0662” mAb was used for CD99 labeling, which was detected with a Texas Red–conjugated horse anti–mouse IgG; Golgi was detected with an FITC-conjugated GM130 mAb. CD99 and HLA class I labeling was performed using the “O662” mAb with a Texas Red–conjugated horse anti–mouse IgG and the FITC-“W6/32” mAb, respectively. As a control, cells were treated with cytochalasin B (cyt. B), which disturbs the actin cytoskeleton. (G) Cocapping of CD99 and HLA class I molecules. Jurkat T cells were treated as described in “Methods.” Briefly, cells were incubated for 30 minutes with biotinyled anti–HLA class I “W6/32” mAb followed by a 30-minute incubation with Texas Red–streptavidin. Cells were incubated at 37°C for 60 minutes to allow capping. Labeling for CD99 was then performed by incubation with the “0662” mAb followed by incubation for 30 minutes at 4°C with a Texas Red–conjugated rabbit antimouse antibody. Finally, cells were plated onto PLL-coated slides and fixed before microscopic analysis. Cells in noncapping (NC) and capping (C) conditions are shown in the upper panel and in the lower panel, respectively. No cross-reactivity between the anti–Ig conjugates and the primary antibodies was observed in control cells. (H) Immuno-electron microscopy identifies colocalization of HLA class I and CD99. Consecutive immuno-gold double labeling of Jurkat cells: detection of HLA class I by W632 mAb and protein A coupled to 15-nm gold particles followed by labeling with CD99 (O662 mAb) and protein A coupled to 10-nm gold particles. The arrows point to HLA class I and CD99 colocalization areas.
To confirm that colocalization of CD99 and HLA class I occurs in the Golgi, we used nocodazole, a drug known to bind tubulin and, consequently, to disrupt the Golgi apparatus but not the ER (Figure 4D). Such treatment of Jurkat cells resulted in dispersion of detectable intracellular CD99 (Figure 4E), and in a loss of detectable intracellular labeling of both CD99 and class I, while their colocalization at the cell periphery was intact (Figure 4F). These results also show that treatment with cytochalasin B, which specifically disrupts the actin CSK, had no effect on the localization of both molecular species.
Physical link between CD99 and HLA class I molecules
A series of experiments was next performed to determine whether a physical association could link CD99 to HLA class I molecules: 2-hybrid analysis in yeast using CD99 as bait allowed recovery of class I molecules in our hands (not shown) and in similar investigations performed by others (H. Kovar; personal communication). To determine whether direct binding occurs between CD99 and HLA class I, we prepared appropriate fusion proteins (Figure 5A).
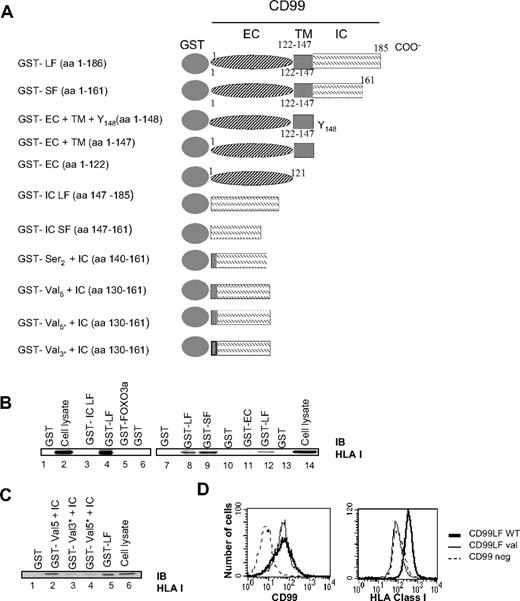
HLA class I molecules are physically linked to CD99 molecules in Jurkat cells. (A) Schematic diagram of GST-CD99 fusion proteins prepared for this study. LF indicates long form; SF, short form; IC, CD99 intracytoplasmic region; EC, CD99 extracellular region; TM, CD99 transmembrane region. For GST-val 3* val 134,135,136 were mutated in Leu, Leu, Ile, and for GST-val 5* mutations of val 131 and 138 to Leu were added. (B) The short form of CD99 can bind HLA class I molecules. GST pull-down assays were performed on Jurkat cell lysates using the indicated GST-CD99 fusion proteins (above lanes 1-6 and 7-14). After extensive washings, bound proteins were separated by SDS-PAGE and immunoblotted (IB) with the “H300” Ab directed against HLA class I molecules. The whole-cell lysates used for HLA class I binding analysis are shown on the immunoblots in lanes 2 and 14. (C) Val 134-136 residues of the transmembrane and HLA class I binding domains. GST pull-down assays were performed using GST-CD99 fusion proteins extending into the transmembrane region GST-Val 5 + IC). Mutations to Leu or Ile were introduced at Val 131, 134-136, and 138 (GST-Val 3* + IC and GST-Val 5* + IC). (D) Expression of HLA class I by transfectants showing CD99 long form mutated on the valines 134-136 (CD99 LF Val) compared with CD99 long form WT (CD99LFwt) transfectants and CD99 negative cells (CD99neg).
HLA class I molecules are physically linked to CD99 molecules in Jurkat cells. (A) Schematic diagram of GST-CD99 fusion proteins prepared for this study. LF indicates long form; SF, short form; IC, CD99 intracytoplasmic region; EC, CD99 extracellular region; TM, CD99 transmembrane region. For GST-val 3* val 134,135,136 were mutated in Leu, Leu, Ile, and for GST-val 5* mutations of val 131 and 138 to Leu were added. (B) The short form of CD99 can bind HLA class I molecules. GST pull-down assays were performed on Jurkat cell lysates using the indicated GST-CD99 fusion proteins (above lanes 1-6 and 7-14). After extensive washings, bound proteins were separated by SDS-PAGE and immunoblotted (IB) with the “H300” Ab directed against HLA class I molecules. The whole-cell lysates used for HLA class I binding analysis are shown on the immunoblots in lanes 2 and 14. (C) Val 134-136 residues of the transmembrane and HLA class I binding domains. GST pull-down assays were performed using GST-CD99 fusion proteins extending into the transmembrane region GST-Val 5 + IC). Mutations to Leu or Ile were introduced at Val 131, 134-136, and 138 (GST-Val 3* + IC and GST-Val 5* + IC). (D) Expression of HLA class I by transfectants showing CD99 long form mutated on the valines 134-136 (CD99 LF Val) compared with CD99 long form WT (CD99LFwt) transfectants and CD99 negative cells (CD99neg).
Pull-down experiments with GST-CD99 fusion protein recovered specifically HLA class I molecules (Figure 5B). To map the binding site of CD99 to HLA class I, pull-down assays with various GST-CD99 constructs were performed (Figure 5B). Exclusive expression of the intracellular (GST-IC LF) or extracellular (GST-EC) CD99 regions did not lead to any class I binding. Unexpectedly, we found that the binding region of CD99 to HLA class I appears to extend from residues 130 to 147, which include part of the transmembrane region (Figure 5C, and Figures S1,S2, available on the Blood website; see the Supplemental Materials link at the top of the online article). This segment is characterized by a valine-rich region. Mutation of the 3 valines 134-136 (GST-Val 3* + IC) was sufficient to prevent any binding between CD99 and class I (Figure 5C) and as expected mutation of the 5 valines resulted in the same effect (GST-Val 5* + IC). GST pull-down results not shown in Figure 5 are provided in Figure S1. Transfection of the CD99 cDNA mutated for the 3 valines did not allow restoration of the expression of HLA class I obtained with the wild-type CD99 cDNA (Figure 5D). Thus, and puzzlingly, CD99 could hold HLA class I via a horseback-like region requiring 3 valines (134-136).
Physical association between CD99 and p230/golgin-245
Among the specific markers of protein maturation and vesicular transport that we have explored, the p230/golgin-245 protein, which is restricted to the trans- and post-Golgi network, strongly colocalized with HLA class I molecules (Figure 6A).29 Moreover, double-label immuno-electron microscopy for p230/golgin and CD99 confirmed that they colocalized (Figure 6B). It must be noted that GS27, another TGN resident protein,30 and HLA class I also colocalize. Yet, no direct association of CD99 with GS27 was observed probably because GS27 is not a GRIP domain protein (not shown). Thus, we looked at whether CD99 and p230/golgin-245 could physically associate. A series of CD99-GST fusion proteins used in GST pull-down assays, as depicted in Figure 5A, were immunoblotted with an anti-p230 antibody. This revealed that the whole transmembrane domain of CD99 is required for binding to p230 (Figure 6C). In a second set of experiments we prepared 2 p230-GST fusion proteins either including only their proline-rich N-terminal domain or their C-terminal region (2000-2230 amino acids) encompassing the GRIP domain.25 Clearly, only the p230 GRIP domain region physically associates with CD99 (Figure 6D).
The transmembrane region of CD99 associates with the p230 GRIP domain region. (A) Colocalization of HLA class I molecules and p230/golgin-245. Jurkat T cells were fixed, permeabilized, and stained for IFM. Trafficking markers were visualized by indirect fluorescence using specific mAbs, detected with a Texas Red–conjugated horse anti–mouse IgG. HLA class I molecules were detected with the FITC-“W6/32” mAb. Bip/GRP78: a major chaperone of the ER lumen; LAMP1: a glycoprotein of lysosome membranes; GM130: a Golgi matrix protein, also associated with the cis-Golgi; GS27/membrin: a Golgi-associated SNARE which acts in medial- to trans-Golgi protein movement; Syntaxin 11: a t-SNARE from the post-Golgi; Rab8: involved in the regulation of vesicular transport from the TGN to the plasma membrane; p230/golgin-245: a protein of the TGN structure. (B) Immuno-electron microscopy identifies colocalization of p230 and CD99. Consecutive immuno-gold double labeling of Jurkat cells: detection of p230 (anti-p230 mAb) and protein A coupled to 15-nm gold particles followed by labeling with CD99 (O662 mAb) and protein A coupled to 10-nm gold particles. The arrows point to p230 and CD99 colocalization areas. (C) GST-CD99 fusion proteins were incubated with Jurkat cell extracts. Bound proteins were detected with a p230-specific antibody. The whole-cell lysates used for the analysis of p230 binding are shown in lanes 4 and 9 of the immunoblot. LAMP1 staining was also used as a control. (D) GST-p230 fusion proteins expressing only the proline-rich domain (1-270aa) or the C-terminal region encompassing the GRIP domain of p230 (2000-2230aa) were incubated with Jurkat cell extracts. The eluted material was analyzed by Western blotting for CD99 using the “12E7” mAb. The whole-cell lysate is shown in lane 4. GM130 staining was also used as a control. (E) Jurkat cells were lysed in 0.5%TTX lysis buffer. Samples were immunoprecipitated with the “O662” CD99 mAb. Immunoprecipitates were analyzed by Western blot against HLA class I (H300 mAb) and anti-p230.
The transmembrane region of CD99 associates with the p230 GRIP domain region. (A) Colocalization of HLA class I molecules and p230/golgin-245. Jurkat T cells were fixed, permeabilized, and stained for IFM. Trafficking markers were visualized by indirect fluorescence using specific mAbs, detected with a Texas Red–conjugated horse anti–mouse IgG. HLA class I molecules were detected with the FITC-“W6/32” mAb. Bip/GRP78: a major chaperone of the ER lumen; LAMP1: a glycoprotein of lysosome membranes; GM130: a Golgi matrix protein, also associated with the cis-Golgi; GS27/membrin: a Golgi-associated SNARE which acts in medial- to trans-Golgi protein movement; Syntaxin 11: a t-SNARE from the post-Golgi; Rab8: involved in the regulation of vesicular transport from the TGN to the plasma membrane; p230/golgin-245: a protein of the TGN structure. (B) Immuno-electron microscopy identifies colocalization of p230 and CD99. Consecutive immuno-gold double labeling of Jurkat cells: detection of p230 (anti-p230 mAb) and protein A coupled to 15-nm gold particles followed by labeling with CD99 (O662 mAb) and protein A coupled to 10-nm gold particles. The arrows point to p230 and CD99 colocalization areas. (C) GST-CD99 fusion proteins were incubated with Jurkat cell extracts. Bound proteins were detected with a p230-specific antibody. The whole-cell lysates used for the analysis of p230 binding are shown in lanes 4 and 9 of the immunoblot. LAMP1 staining was also used as a control. (D) GST-p230 fusion proteins expressing only the proline-rich domain (1-270aa) or the C-terminal region encompassing the GRIP domain of p230 (2000-2230aa) were incubated with Jurkat cell extracts. The eluted material was analyzed by Western blotting for CD99 using the “12E7” mAb. The whole-cell lysate is shown in lane 4. GM130 staining was also used as a control. (E) Jurkat cells were lysed in 0.5%TTX lysis buffer. Samples were immunoprecipitated with the “O662” CD99 mAb. Immunoprecipitates were analyzed by Western blot against HLA class I (H300 mAb) and anti-p230.
To definitely conclude on the physical association of CD99 with both HLA class I and p230 we performed coimmunoprecipitation experiments. In accordance with colocalization and GST pull-down results HLA class I and p230 were detected in CD99 immunoprecipitates (Figure 6E).
Effects of overexpression of p230/golgin-245
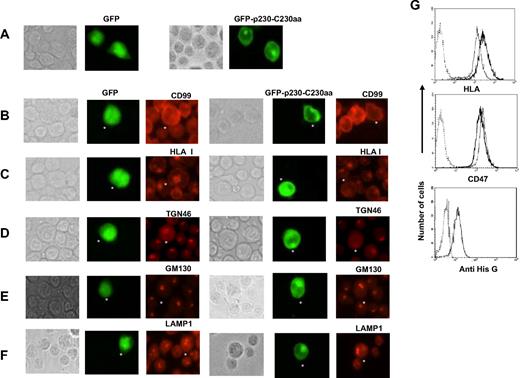
To examine whether the p230 GRIP domain would contribute to orchestrate CD99 and HLA class I targeting to the Golgi, we prepared a GFP construct with the p230/C230aa fragment (GFP-p230-C230aa). After transfection, the GFP-p230-C230aa fusion protein efficiently localized to the Golgi, this is in contrast with wild-type GFP alone, which gave both cytoplasmic and nuclear staining (Figure 7A). These observations are consistent with previous reports.21,22,31,32 Most importantly, we saw that overexpression of the p230-C230aa segment affected the intracellular distribution of CD99 and HLA class I molecules, which lost their Golgi localization and became dispersed throughout the cytoplasm (Figure 7B,C). Similarly, overexpression of the p230-C230aa segment affected the distribution of TGN46, a TGN resident protein (Figure 7D) which showed faint and diffused staining throughout the cytoplasm. In contrast, GM130 Golgi distribution was preserved in spite of p230-C230aa overexpression (Figure 7E); finally, the characteristic distribution of lysosomes also was not affected (Figure 7F). From the above, it may be concluded that the overexpression of a region of the p230/golgin-245 encompassing the GRIP domain prevented the localization of the CD99-HLA class I complex to the Golgi, an effect that could be localized to the trans-Golgi network, while the cis-Golgi network appeared not to be affected. To better conclude on the effect of p230 GRIP on HLA class I trafficking, we selected Jurkat cells stably transfected with the His-Tagged p230 GRIP domain and their HLA class I expression was measured using flow cytometry. We observed significant down-regulation of HLA class I expression in Jurkat cells overexpressing the p230 GRIP domain while Jurkat cells transfected with the empty vector show no modification of HLA class I expression. The expression level of CD47, used as a control, was not modified by overexpression of the p230 GRIP (Figure 7G).
Overexpression of the C-terminal p230 GRIP domain disrupts the intracellular localization of both CD99 and HLA class I and induces down regulation of HLA class I at the cell surface. (A) Wild-type Jurkat T cells that were transiently transfected with plasmid expressing GFP-p230-C230aa were analyzed 72 hours later by IFM. GFP-p230-C230aa fusion protein is efficiently Golgi localized. (B-F) Cells were stained for endogenous CD99, HLA class I, TGN46, GM130, and LAMP1 with, respectively, the CD99 “O662” and the “W6/32” mAbs, a sheep anti-TGN46 antibody, the “35” anti-GM130 and anti-LAMP1“25CD107a” mAbs followed by appropriate secondary Texas Red–conjugated antibodies. (G) Wild-type Jurkat T cells that were stably transfected with a His-tagged plasmid expressing p230-C230aa were analyzed for HLA class I expression using the “W632” mAb; CD47 “B6H12” mAb was used as control. The bottom panel shows the expression level of His-tagged p230-C230aa protein compared with control cells using the anti–His G mAb after 20 minutes permeabilization in methanol.
Overexpression of the C-terminal p230 GRIP domain disrupts the intracellular localization of both CD99 and HLA class I and induces down regulation of HLA class I at the cell surface. (A) Wild-type Jurkat T cells that were transiently transfected with plasmid expressing GFP-p230-C230aa were analyzed 72 hours later by IFM. GFP-p230-C230aa fusion protein is efficiently Golgi localized. (B-F) Cells were stained for endogenous CD99, HLA class I, TGN46, GM130, and LAMP1 with, respectively, the CD99 “O662” and the “W6/32” mAbs, a sheep anti-TGN46 antibody, the “35” anti-GM130 and anti-LAMP1“25CD107a” mAbs followed by appropriate secondary Texas Red–conjugated antibodies. (G) Wild-type Jurkat T cells that were stably transfected with a His-tagged plasmid expressing p230-C230aa were analyzed for HLA class I expression using the “W632” mAb; CD47 “B6H12” mAb was used as control. The bottom panel shows the expression level of His-tagged p230-C230aa protein compared with control cells using the anti–His G mAb after 20 minutes permeabilization in methanol.
Discussion
The requirement for CD99 to bring into play the effects of IFN-γ on HLA class I pinpoints the importance of this molecule in the mechanism(s) that control, overall, the levels of class I expression. In cells rendered CD99 negative, HLA class I molecules were produced but retained in large amounts in the Golgi apparatus, particularly upon IFN-γ stimulation. Studies into the internalization and externalization kinetics of class I indicated that CD99 acts at a late stage in the transport process. As compared with CD99pos cells, CD99neg cells present a delay in HLA class I externalization, which is particularly marked upon IFN-γ stimulation.
Maybe CD99 controls class I expression through multiple mechanisms. We found a higher amount of HLA class I transcripts in CD99pos cells, as compared with CD99neg cells, which may participate in the higher cell-surface expression of class I. While CD99pos cells express a normal amount of HLA class I, this amount is considerably decreased in CD99neg cells. This raises the question of the reciprocal contribution of the 2 mechanisms, Golgi retention and reduced transcription, in the overall dampening of class I expression by CD99. Given the high level of class I retained within the Golgi apparatus, particularly in the presence of INF-γ in CD99neg cells, it is likely that Golgi retention could play a major role.
The papilloma virus E5 protein was also shown to act on HLA expression both in inhibiting transcription and in inducing Golgi retention.33 E5 was shown to be localized in transmembrane compartments of the ER and of the Golgi apparatus. Moreover, a physical association was shown to occur between the E5 protein and HLA class I molecules, and it was assumed that such an association and location are responsible for the effects on transport of HLA class I, retained in an irreversible manner before reaching the cell surface.34
To determine the molecular mechanisms by which CD99 exerts tight regulation of HLA class I, we first looked for a physical linkage between these 2 proteins. We found that CD99 and HLA class I molecules clearly colocalize both in the Golgi apparatus and at the cell surface.
Through coimmunoprecipitation and GST pull-down experiments, we found that both CD99 and HLA class I molecules physically interact with each other. Strikingly, this association occurs at the transmembrane level. In addition, we found that 3 valines (134-136), located in the transmembrane region, are required for the binding to HLA class I molecules, likely in relation with their hydrophobicity. In contrast, we found no binding with a GST construct linked to the intracytoplasmic domain of CD99. In mice, a transmembrane protein (BAP31) was found to bind MHC class I, while playing a role in the ER-to-Golgi step of the biosynthetic pathway.35,36 Moreover BAP31 also associates with 2 surface molecules, by associating their transmembrane domains.
Little is known concerning the molecular basis of HLA class I transport from the ER to the cell surface. It has been assumed that the exit of HLA class I molecules from the ER is guided by a selective mechanism of export.36,37 Several mechanisms could explain the regulated export of HLA class I. First, HLA class I may contain, in their sequence, specific motifs involved in the export process, although none of them have been fully identified and characterized so far. Second, HLA class I could associate with a transport receptor, which would exhibit an export motif.38,39 Although intracytoplasmic tail residues have been identified to promote internalization, degradation and ER retrieval, HLA class I molecules do not appear to possess any identifiable export signal.40,41
Consistent with receptor-mediated exit, ER-to-Golgi transport of HLA class I molecules was shown not to require their cytoplasmic tails, which themselves lack ER export motifs.36,42,43 Truncation of the intracytoplasmic fragment of the HLA-A2 molecules does not prevent their export to the cell surface.42 Furthermore, mouse MHC class I molecules deleted of their intracytoplasmic region were still able to localize to the cell surface.43 Finally, tail swapping experiments also support a selective export mechanism.36 Collectively, these data suggest the involvement of an unknown HLA transport receptor.
Moreover, our results suggest that CD99 is located in close proximity to a golgin, the coiled-coil protein p230/golgin-245.29,44 We confirmed, by coimmunoprecipitation and GST pull-down assays, physical linkage between both molecules. This interaction mapped between the transmembrane domain of CD99 and the golgin GRIP domain. Although we found a clear association between CD99 and HLA class I on the one hand, and CD99 with p230 on the other hand, we could not directly demonstrate a physical link between HLA class I and p230.
Critically, overexpression of the C-terminal region encompassing the p230 GRIP domain affects the intracellular localization of CD99 and HLA class I molecules. It is well established that a high level of expression of GRIP domain–containing fragments disrupts TGN resident protein distribution such as for p230, likely reflecting competition for a limited GRIP domain binding site at the TGN.21,22 Taken together, our data suggest that saturation of GRIP domain binding site on TGN membranes results in mislocalization accompanied by down-modulation of CD99 and HLA class I, confirming the tight functional link occurring between these molecules and p230/golgin-245. GRIP domains form independent dimers which anchor to the Golgi membrane upon binding to a small G protein of the Arf family, Arl1.19,20,45 When bound to GDP, Arl1 is detached in the cytoplasm; when bound to GTP, Arl1 associates with the golgin GRIP domain and anchors into the Golgi outer membrane via its myristoylated N-terminus. It can be assumed that CD99 binds the GRIP domain through a different region from the GRIP binding site for Arl1. Interaction of CD99 with p230/golgin-245 could participate in the stabilization of the Golgi membrane. The p230/golgin-245 protein acts as a membrane matrix tethering protein within the Golgi stack that recycles between the cytosol and the buds/vesicles of TGN membranes under the regulation of G proteins.45 Its C-terminal GRIP domain links p230/golgin-245 to the TGN outer membrane, while the coiled-coil segment extends far into the cytoplasm. In addition to Arl-GTP, p230 was described to bind to MACF1 and to cross-link microtubules to the actin cytoskeleton.25 This interaction provides a molecular link for protein transport along the microtubule and actin cytoskeleton. Other golgins were also shown to specifically conduct export traffic of other cargo proteins.19,29,46
Endogenous p230/golgin-245 depletion resulted in Golgi dispersion to the cell periphery, forming “mini stacks” with failure to migrate along microtubules toward the microtubules organizing center.24 Thus, our observations fit the present knowledge of Golgi molecular organization. They provide a model of molecular interactions involving CD99 and HLA class I transport whereby CD99 binds class I molecules via their transmembrane domains, on the one hand, and CD99 binds p230/golgin-245, on the other hand, still at the transmembrane level. Arl-GTP could provide a decisive link by appropriately positioning the GRIP domain, and the coiled-coil domain, extending into the cytoplasm, which would connect the vesicular complex to tether actin and tubulin networks allowing the transport structure to be pulled out along these organelles toward the cell surface.
Fitting into this model, we found that CD99 can associate with ezrin and moesin in Jurkat cells (data not shown). The actin binding complex is known to act as a linker between actin CSK and transmembrane proteins.47 CD99 is known for its capacity to activate the actin CSK and to bind zyxin, a protein taking part in actin polymerization.9-11,13,48 It is tempting to assume that a connection occurs between the pathway of class I transport and the actin CSK, to ensure proper regulation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Inserm, Association pour la Recherche sur le Cancer (ARC), La Ligue contre le cancer, European project Prognosis and Therapeutic Targets in the “Ewing” Family of Tumors (PROTHETS), Institut National du Cancer (INCA) Cancéropôle PACA, Ministère de la Recherche et des Technologies, Fondation de France, Fondation de la Recherche Médicale (FRM), and Agence Nationale de Biomédecine.
Authorship
Contribution: A. Bremond and O.M. performed research and analyzed data; K.M., S.C., and M.T. performed research; K.S. and P.G. analyzed data; J.P.B. performed flow cytometry analysis; A.B. designed research, provided financial support, analyzed data, and wrote the paper; and G.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ghislaine Bernard, Inserm UMR 576, Hôpital de l'Archet BP 3079, 06202 Nice, France; e-mail: bernardg@unice.fr.
References
Author notes
*A.B. and O.M. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal