Abstract
In the control of T-helper type I (Th-1) polarization, dendritic cells (DCs) must interpret a complex array of stimuli, many of which are poorly understood. Here we demonstrate that Th-1 polarization is heavily influenced by DC-autonomous phenomena triggered by the loading of DCs with antigenically matched major histocompatibility complex (MHC) class I and class II determinants, that is, class I and II peptide epitopes exhibiting significant amino acid sequence overlap (such as would be physiologically present during infectious processes requiring Th-1 immunity for clearance). Data were derived from 13 independent antigenic models including whole-cell systems, single-protein systems, and 3 different pairs of overlapping class I and II binding epitopes. Once loaded with matched class I and II antigens, these “Th-1 DCs” exhibited differential cytokine secretion and surface marker expression, a distinct transcriptional signature, and acquired the ability to enhance generation of CD8+ T lymphocytes. Mechanistically, tRNA-synthetases were implicated as components of a putative sensor complex involved in the comparison of class I and II epitopes. These data provide rigorous conceptual explanations for the process of Th-1 polarization and the antigenic specificity of cognate T-cell help, enhance the understanding of Th-1 responses, and should contribute to the formulation of more effective vaccination strategies.
Introduction
Dendritic cells (DCs) are the master regulators of adaptive immunity.1 Implicit in this regulation is the ability of DCs to prime a polarized T-helper (Th) response. In both mouse and human, a T-helper type I (Th-1)–polarized response is characterized by CD8+ T-cell priming and the release of Th-1 cytokines such as interleukin (IL)–12, IL-2, and interferon-γ (IFN-γ). A Th-2 response is characterized by humoral immunity (eg, immunoglobulin G1 [IgG1]), IgE-mediated allergic-type immunity, and the release of Th-2 cytokines IL-4, IL-5, and IL-10. Furthermore, DCs must also regulate the balance between the production of Th-1–dependent antibody responses (eg, IgG2) and the priming of cellular immune responses within the broader context of Th-1 immunity. While many factors are known to influence the functional development of Th polarization, the principle signals that initiate such development are poorly understood. Indeed, the differentiated Th response to various antigens and epitopes has been vaguely referred to as “antigen-dependent” and “sequence-dependent,” respectively.2
Our previous work demonstrated the surprising result that the tandem loading of DCs with both major histocompatibility complex (MHC) class I and class II antigenic determinants (ie, mRNA preparations and cell lysate preparations) elicited superior CD8+ T-cell responses in comparison to DCs singly loaded with either mRNA or lysate alone. Using a model system of active immunotherapy for the treatment of acute myelogenous leukemia (AML), we demonstrated a strong Th-1 polarization based upon IFN-γ enzyme-linked immunosorbent spot (ELISpot), IL-12 enzyme-linked immunosorbent assay (ELISA), enhanced production of activated CD8+ lymphocytes, and elevated killing of specific targets.3 The data suggested that these “doubly loaded DCs” were acquiring a Th-1–polarizing phenotype based solely upon loading criteria; however, this hypothesis was not directly addressed by the earlier work.
Here, we tested the hypothesis that DCs acquire the ability to prime a Th-1–polarized response when loaded with MHC class I and class II determinants that are antigenically similar or identical, the rationale being that such a scenario would be commonly observed in vivo during an active viremia. Class I determinants would be produced endogenously by infected DCs,4,5 and class II determinants (ie, the infectious particles) would be taken up exogenously by normal DC phagocytic processes.1 A match of class I and class II determinants, likely in conjunction with other inflammatory signals,6 would indicate an intracellular-type infection necessitating clearance by Th-1 mediated immunity and the priming of CD8+ T cells.7 There is much anecdotal evidence in the literature that supports such a hypothesis. Lopez and colleagues demonstrated that the induction of Th-1 type immunity requires actively replicating virus8 and, subsequently, a TLR-independent induction of DC maturation in response to viral infection.9 This result was confirmed by Hornung, who demonstrated that only actively replicating virus can be detected by plasmacytoid DCs and that such detection occurs independent of protein kinase R (PKR), Toll-like receptor (TLR)–7, TLR-8, and TLR-9.10
To answer this question specifically in human DCs, we used a variety of pathogen-associated molecular pattern (PAMP)–independent systems of pooled antigens derived from mammalian sources.3 We also used additional systems consisting of single proteins and/or pairs of overlapping MHC class I and class II binding peptides. The data suggest that DCs can regulate Th-1 polarization and the CD8 response in an autonomous, T cell–independent fashion by comparing the sequence similarity of the MHC class I and class II antigens that have gained access to the antigen-presenting cell (APC).
Methods
Generation of immature DCs, preparation of antigenic materials, and DC loading and maturation
DCs were generated as described previously from healthy donor, granulocyte colony-stimulating factor (G-CSF) mobilized peripheral blood progenitor cells (PBPCs) donated for research under M. D. Anderson Institutional Review Board protocol Lab02-630.3 Cellular tumor antigens were prepared as described previously.3 Human leukocyte antigen (HLA)–restricted peptide sequences derived from the sequence of the influenza A/New Caledonia hemagglutinin (HA) antigen were characterized as described.11 Immature DCs were loaded with either tumor lysate, tumor mRNA, both, or neither, as described previously.3 Conditions for delivering single antigens (ie, single proteins and/or plasmids) were identical unless stated otherwise. DCs were loaded with one of 2 different protein antigens [enhanced green fluorescent protein (EGFP; Invitrogen, Carlsbad, CA) or mIL-4 (eBioscience, San Diego, CA)] by incubation of cells in 100 μg/mL antigen for 3 hours subsequent to the electroporation of 1 μg plasmid DNA/106 cells. When electroporated with proteins, conditions were as described previously.11 When loaded with peptides, immature DCs were incubated for 90 minutes at 10 μg/mL per peptide. DC were then matured for 2 hours as described,3 after which they were washed to remove excess peptide. DCs were then resuspended in AIM-V medium (Invitrogen) supplemented as described3 and matured for an additional 36 hours. If loaded in the presence of ethanolamine, ethanolamine was added in conjunction with the peptide epitopes at concentrations between 0.04% and 0.08% (6.7-13.3 mM). Ethanolamine was washed from mature DCs before T-cell stimulation.
Transcriptome analysis
DC preparations were pooled according to the manner by which they had been loaded. Total RNA was then generated as described3 from unloaded preparations, mRNA-loaded preparations, lysate-loaded preparations, matched, doubly loaded preparations, and mismatched, doubly loaded preparations. The 5 total RNA samples were used to probe the Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA) in duplicate (10 arrays total) in conjunction with Codon Biosciences, LP (Houston, TX). For comparison of transcript expression levels, Cohen d was calculated for each transcript by the method of pooled standard deviation using the root mean square of the standard deviations: [d = (M1 − M2)/√(σ12 + σ22)/2]. Transcripts of matched, doubly loaded DCs were identified as differentially regulated if they conformed to Cohen's d greater than 1.0 (large effect) and q value (Benjamini-Hochberg false discovery) less than 0.01. Functional class analysis of transcripts was performed using the data mining software, Web-Based Gene Set Analysis Toolkit (WEBGESTALT).12 Minimum information about a microarray experiment (MIAME)–compliant microarray data are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) using accession number GSE7247.
Cytotoxic T-lymphocyte antigen-4 (CTLA-4) reverse transcription polymerase chain reaction and IL-12 ELISA
cDNA of differentially loaded DC populations was generated from total RNA pools using the SMART cDNA Synthesis Kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. Exon 2 of CTLA-4 was amplified from DC cDNA with forward primer 5′-TGGCCCAGCCTGCTGTGG-3′ and reverse primer 5′-TCTGGGTTCCGTTGCCTATG-3′ for 25 or 30 cycles, annealing at 58°C. IL-12 ELISA was performed as described previously.3
Statistical analysis
Statistical differences were calculated by Student unpaired 2-tailed t test unless stated otherwise. Significance was defined as P less than or equal to .05. Error bars in all figures are equal to plus or minus standard deviation (SD) unless otherwise indicated.
Results
DC IL-12 secretion and CD83 expression are regulated by loading with matched class I and class II antigens
To confirm cell-autonomous DC phenomena, we demonstrated the absence of accessory T cells in DC cultures. After monocyte isolation and ex vivo tissue culture for 8 days, cell cultures were completely devoid of CD3+ cells as determined by flow cytometry (Figure 1). Subsequently, up to 4 different allogeneic populations of DCs were mixed together in an attempt to detect the presence of contaminating T cells by alloreactive stimulus. 3H-thymidine uptake in mixed DC wells was indistinguishable from that of the assay background (data not shown). We concluded from these data that DC populations were effectively devoid of accessory CD3+ cells. DCs used in subsequent experiments were also verified by flow cytometry to be free of CD3+ cells.
DC cultures are devoid of accessory CD3+ cells. (A) Isotype control. (B) DC culture on day 8 demonstrating no CD3+ events. (C) CD3+ control. x-axis, CD209-PE; y-axis, CD3-PerCP.
DC cultures are devoid of accessory CD3+ cells. (A) Isotype control. (B) DC culture on day 8 demonstrating no CD3+ events. (C) CD3+ control. x-axis, CD209-PE; y-axis, CD3-PerCP.
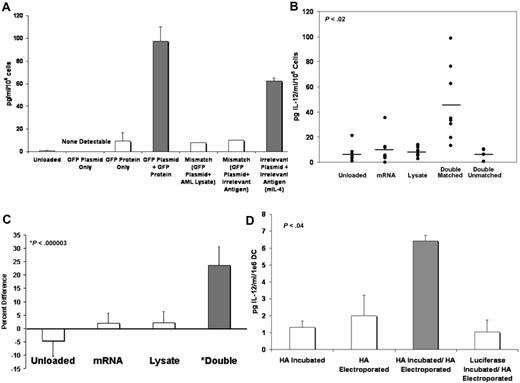
We previously reported that doubly loaded DCs secrete the Th-1 cytokine IL-12 at log-fold higher levels than their unloaded and singly loaded counterparts.3 To demonstrate that up-regulation of IL-12 secretion is specific to the presence of matched MHC class I and class II antigenic determinants, we departed from our well-characterized, multiantigen system,3,6,7,13 developing a simple, single antigen system. Using GFP as a model antigen and the murine isoform of IL-4 (biologically inactive in human systems) as an irrelevant, unmatched antigen, immature DCs were loaded by electroporation of GFP or mIL-4 expression plasmids, incubation with protein antigen, both plasmid and protein, or neither. After maturation, DC IL-12 secretion was determined by ELISA of culture supernatants. Figure 2A demonstrates that only DCs loaded with identical class I (expression plasmid) and class II (purified recombinant protein) antigens exhibited up-regulation of IL-12 secretion over weak baseline. Singly loaded and mismatched doubly loaded DCs did not exhibit significant IL-12 secretion (P < .001, representative experiment shown). This result was reproducible within this particular system as well as within other systems including delivery of the class I antigen (rHA) by electroporation as described previously11 (P < .04, Figure 2D), as well as the system in which the class I antigen (GFP) was delivered by recombinant retroviral vector (P < .03, data not shown).
DCs loaded with matched class I and II antigens exhibit enhanced IL-12 secretion and CD83 expression. (A) Double-loading of DCs with matched MHC class I and II determinants enhances DC IL-12 secretion in a single-antigen system.  equals IL-12 secretion from DCs loaded with matched class I (plasmid) and class II (soluble protein). □ equals IL-12 secretion from other DC loaded by any other method. y-axis: pg IL-12/mL/106 cells (A,B,D). (B) Double-loading of DCs with matched MHC class I and II antigens enhances DC IL-12 secretion. Eight experiments shown (double unmatched, 5 experiments shown). (C) Double-loading of DC with matched class I and II determinants causes enhanced up-regulation of CD83 expression. Average MFI value of unloaded and singly loaded DCs equals 0. □ equals percent by which CD83 MFI of all unloaded or singly loaded DCs differs from the average.
equals IL-12 secretion from DCs loaded with matched class I (plasmid) and class II (soluble protein). □ equals IL-12 secretion from other DC loaded by any other method. y-axis: pg IL-12/mL/106 cells (A,B,D). (B) Double-loading of DCs with matched MHC class I and II antigens enhances DC IL-12 secretion. Eight experiments shown (double unmatched, 5 experiments shown). (C) Double-loading of DC with matched class I and II determinants causes enhanced up-regulation of CD83 expression. Average MFI value of unloaded and singly loaded DCs equals 0. □ equals percent by which CD83 MFI of all unloaded or singly loaded DCs differs from the average.  equals percent by which CD83 MFI of all doubly loaded DCs differs from the average (P < .001). y-axis equals percent difference of CD83 MFI of CD83+ DC from the average of unloaded and singly loaded DCs. (D) Loading of DC MHC class I by electroporation of rHA and class II by incubation of rHA11 enhances DC IL-12 secretion.
equals percent by which CD83 MFI of all doubly loaded DCs differs from the average (P < .001). y-axis equals percent difference of CD83 MFI of CD83+ DC from the average of unloaded and singly loaded DCs. (D) Loading of DC MHC class I by electroporation of rHA and class II by incubation of rHA11 enhances DC IL-12 secretion.  equals IL-12 secretion from DCs loaded with matched class I and class II proteins (rHA). □ equals IL-12 secretion from DC loaded singly or loaded doubly with disparate class I and class II antigens (ie, rHA and luciferase).
equals IL-12 secretion from DCs loaded with matched class I and class II proteins (rHA). □ equals IL-12 secretion from DC loaded singly or loaded doubly with disparate class I and class II antigens (ie, rHA and luciferase).
DCs loaded with matched class I and II antigens exhibit enhanced IL-12 secretion and CD83 expression. (A) Double-loading of DCs with matched MHC class I and II determinants enhances DC IL-12 secretion in a single-antigen system.  equals IL-12 secretion from DCs loaded with matched class I (plasmid) and class II (soluble protein). □ equals IL-12 secretion from other DC loaded by any other method. y-axis: pg IL-12/mL/106 cells (A,B,D). (B) Double-loading of DCs with matched MHC class I and II antigens enhances DC IL-12 secretion. Eight experiments shown (double unmatched, 5 experiments shown). (C) Double-loading of DC with matched class I and II determinants causes enhanced up-regulation of CD83 expression. Average MFI value of unloaded and singly loaded DCs equals 0. □ equals percent by which CD83 MFI of all unloaded or singly loaded DCs differs from the average.
equals IL-12 secretion from DCs loaded with matched class I (plasmid) and class II (soluble protein). □ equals IL-12 secretion from other DC loaded by any other method. y-axis: pg IL-12/mL/106 cells (A,B,D). (B) Double-loading of DCs with matched MHC class I and II antigens enhances DC IL-12 secretion. Eight experiments shown (double unmatched, 5 experiments shown). (C) Double-loading of DC with matched class I and II determinants causes enhanced up-regulation of CD83 expression. Average MFI value of unloaded and singly loaded DCs equals 0. □ equals percent by which CD83 MFI of all unloaded or singly loaded DCs differs from the average.  equals percent by which CD83 MFI of all doubly loaded DCs differs from the average (P < .001). y-axis equals percent difference of CD83 MFI of CD83+ DC from the average of unloaded and singly loaded DCs. (D) Loading of DC MHC class I by electroporation of rHA and class II by incubation of rHA11 enhances DC IL-12 secretion.
equals percent by which CD83 MFI of all doubly loaded DCs differs from the average (P < .001). y-axis equals percent difference of CD83 MFI of CD83+ DC from the average of unloaded and singly loaded DCs. (D) Loading of DC MHC class I by electroporation of rHA and class II by incubation of rHA11 enhances DC IL-12 secretion.  equals IL-12 secretion from DCs loaded with matched class I and class II proteins (rHA). □ equals IL-12 secretion from DC loaded singly or loaded doubly with disparate class I and class II antigens (ie, rHA and luciferase).
equals IL-12 secretion from DCs loaded with matched class I and class II proteins (rHA). □ equals IL-12 secretion from DC loaded singly or loaded doubly with disparate class I and class II antigens (ie, rHA and luciferase).
Returning to our model system of AML immunotherapy, we characterized IL-12 secretion from DCs singly loaded with tumor mRNA, singly loaded with tumor lysate, doubly loaded with mRNA and lysate derived from the same tumor product, or doubly loaded with tumor mRNA and a lysate derived from disparate tissues (human erythroid or xenogenic stromal cell lines). After maturation, culture supernatants were examined for IL-12 (p70) secretion by ELISA. Figure 2B demonstrates that only DCs loaded with mRNA and lysate taken from the same tumor or cell line (P < .02) were able to produce markedly elevated levels of IL-12 secretion. Results were derived from 8 independent experiments using a variety of different healthy donors. Results were in good agreement with similar experiments reported previously.3
CD83 is one of the most consistent indicators of DC maturity. Upon maturation, its expression is up-regulated on the DC surface from undetectable prematuration levels.14 CD83 is a sialic acid–binding Ig-like lectin adhesion receptor15 vitally important in the development of CD4+ T cells16 as well as in the promulgation of productive CD8+ T-cell responses.14,16-21 Soluble CD83 blocks both allogeneic and autologous CD8+ T-cell proliferation in a concentration-dependent manner in vitro, and its administration abolishes experimental autoimmune encephalomyelitis by down-regulating T-cell responses in vivo.17,18 While phenotyping preparations of DCs, we noted that doubly loaded DCs always expressed higher levels of CD83 than singly loaded controls. To characterize CD83 surface expression, we compared CD83 surface expression of doubly loaded versus singly loaded and unloaded DCs after maturation. In 8 independent experiments (Figure 2C), mean fluorescence intensity (MFI) of CD83+ DC was demonstrated to be 24% higher among doubly loaded DCs (P < .001) than among the mean expression level of all singly loaded and unloaded controls. Examination of other surface markers did not demonstrate this pattern of up-regulation (data not shown).
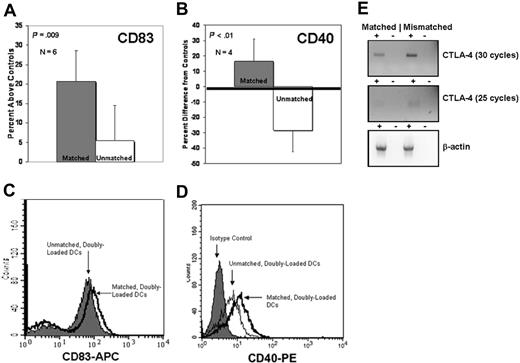
To test the hypothesis that the enhanced up-regulation of DC CD83 was specific to loading with matched class I and class II antigenic determinants, DCs were loaded with matched mRNA and lysate (derived from the same tumor product) or with unmatched determinants derived from disparate combinations of AML tumor products, the human Tri-factor dependent 1a (TF-1a) erythroblast cell line, or the mouse FBMD-1 stromal cell line. Singly loaded controls in each experiment were loaded with the AML-derived antigenic determinant(s). Enhanced up-regulation of CD83 among the matched and unmatched doubly loaded DC populations was then compared with average CD83 expression levels of unloaded and singly loaded controls independently in each of 6 different experiments. As demonstrated in Figure 3A, DCs doubly loaded with matched determinants exhibited a 21% enhancement of CD83 up-regulation, while DCs loaded with unmatched determinants exhibited an enhancement of only 5.5% (P = .009), statistically indistinguishable from the 2% to 3% up-regulation observed among singly loaded controls. A typical flow cytometry overlay of this phenomenon is shown by Figure 3C.
Differential regulation of CD83, CD40, and CTLA-4 is dependent upon the double-loading of DCs with matched class I and II antigens. (A) Percent difference of CD83 expression from average of unloaded and singly loaded controls between matched doubly loaded ( ) and unmatched doubly loaded (□) DCs (P = .009). y-axis equals percent difference from average of unloaded and singly loaded controls. Compilation of 6 independent experiments. (B) Same as panel A but with CD40 staining rather than CD83 (P < .01). Compilation of 4 independent experiments. (C,D) Histograms demonstrating representative results for A (CD83) and B (CD40). (E) Semiquantitative RT-PCR demonstrates differential expression of CTLA-4 between DCs doubly loaded with matched antigens and DCs doubly loaded with mismatched antigens. Matched equals DC loaded with matched class I and II antigens. Mismatched equals DC loaded with mismatched class I and II antigens. +/−, +/− reverse transcriptase.
) and unmatched doubly loaded (□) DCs (P = .009). y-axis equals percent difference from average of unloaded and singly loaded controls. Compilation of 6 independent experiments. (B) Same as panel A but with CD40 staining rather than CD83 (P < .01). Compilation of 4 independent experiments. (C,D) Histograms demonstrating representative results for A (CD83) and B (CD40). (E) Semiquantitative RT-PCR demonstrates differential expression of CTLA-4 between DCs doubly loaded with matched antigens and DCs doubly loaded with mismatched antigens. Matched equals DC loaded with matched class I and II antigens. Mismatched equals DC loaded with mismatched class I and II antigens. +/−, +/− reverse transcriptase.
Differential regulation of CD83, CD40, and CTLA-4 is dependent upon the double-loading of DCs with matched class I and II antigens. (A) Percent difference of CD83 expression from average of unloaded and singly loaded controls between matched doubly loaded ( ) and unmatched doubly loaded (□) DCs (P = .009). y-axis equals percent difference from average of unloaded and singly loaded controls. Compilation of 6 independent experiments. (B) Same as panel A but with CD40 staining rather than CD83 (P < .01). Compilation of 4 independent experiments. (C,D) Histograms demonstrating representative results for A (CD83) and B (CD40). (E) Semiquantitative RT-PCR demonstrates differential expression of CTLA-4 between DCs doubly loaded with matched antigens and DCs doubly loaded with mismatched antigens. Matched equals DC loaded with matched class I and II antigens. Mismatched equals DC loaded with mismatched class I and II antigens. +/−, +/− reverse transcriptase.
) and unmatched doubly loaded (□) DCs (P = .009). y-axis equals percent difference from average of unloaded and singly loaded controls. Compilation of 6 independent experiments. (B) Same as panel A but with CD40 staining rather than CD83 (P < .01). Compilation of 4 independent experiments. (C,D) Histograms demonstrating representative results for A (CD83) and B (CD40). (E) Semiquantitative RT-PCR demonstrates differential expression of CTLA-4 between DCs doubly loaded with matched antigens and DCs doubly loaded with mismatched antigens. Matched equals DC loaded with matched class I and II antigens. Mismatched equals DC loaded with mismatched class I and II antigens. +/−, +/− reverse transcriptase.
DC loaded with overlapping class I and II HLA-binding peptides support enhanced Th-1 lymphocyte responses
Although these data suggested DC autonomous phenomena, it could be argued that, in the absence of specific defined MHC class I and class II binding peptides, the data are difficult to interpret. It might further be argued that, in a total antigen system, the DC-specific phenomenon of cross-presentation22 renders impossible any discernment that important MHC class I epitopes were derived from a pool of mRNA or from an expression plasmid. To address these potential concerns and to remove the phenomenon of cross-presentation as a confounding variable, we developed a model system comprised solely of HLA-restricted binding peptides derived from the hemagglutinin (HA) antigen of influenza A/New Caledonia. The positioning of the peptides along the primary HA sequence is illustrated in Figure 4A. In this new model system, HLA-compatible DCs were loaded phagocytically (ie, the peptides are serving principally as soluble antigens) with class I and II peptides for 90 minutes, after which the peptides were washed away, and the DCs were matured for 30 to 36 hours. Because the half-life of MHC class I is only 4 hours, the majority of any peptide antigen bound by surface MHC class I during the loading process is gone after the 36-hour maturation. Hence, differences in the abilities of differentially loaded DCs to prime Th-1 responses are due to signals that were sent after the initial loading process 36 hours earlier. After maturation, DCs were cocultured with autologous T cells, and the ability of DCs to support Th-1 responses was assayed by IFN-γ release and flow cytometry of proliferating T cells.
Aspects of the HA peptide model system. (A) Positioning of MHC class I and II peptides along the primary sequence of influenza HA antigen. (B) Representation of the class I/II overlapping peptide pair (B8-166/DR3-162), the sequence comparison of which is predicted to be disrupted by ethanolamine. The control class I/II overlapping peptide pair (A2-443/DR3-440) is predicted to be unaffected by the presence of ethanolamine.
Aspects of the HA peptide model system. (A) Positioning of MHC class I and II peptides along the primary sequence of influenza HA antigen. (B) Representation of the class I/II overlapping peptide pair (B8-166/DR3-162), the sequence comparison of which is predicted to be disrupted by ethanolamine. The control class I/II overlapping peptide pair (A2-443/DR3-440) is predicted to be unaffected by the presence of ethanolamine.
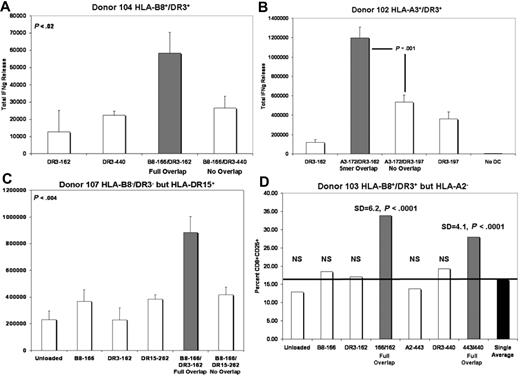
Figure 5A illustrates a representative IFN-γ release experiment, in which DCs loaded with overlapping class I (B8-162) and class II (DR3-166) peptides stimulated the secretion of excess IFN-γ from autologous T cells in comparison to DCs loaded with the same class I peptide (B8-162) and a nonoverlapping class II peptide (DR3-440). Indeed, IFN-γ release stimulated by DC loaded with nonoverlapping peptides was indistinguishable from singly loaded class II controls (P < .02). In a similar series of experiments, we sought to determine whether a partial sequence overlap between class I and II epitopes would be sufficient to enhance the release of IFN-γ. In the representative experiment illustrated by Figure 5B, DC loading with class I and II peptides that overlapped by 5 amino acid residues (A3-172/DR3-162) were still sufficient to promote enhanced IFN-γ secretion. An overlap of 2 residues was insufficient to elicit enhanced IFN-γ production (data not shown). With repeated experiments in both HLA-compatible and HLA-disparate donor backgrounds, we were able to demonstrate that stringent HLA-matching between defined peptide epitope and donor was helpful, but not always an absolute requirement for all peptides (eg, Figure 5C). Such a result is predictable given the high concentration at which the binding peptides were loaded (10 μg/mL) and the likely existence of multiple HLA-binding affinities to undefined haplotypes.
A system of overlapping/nonoverlapping defined class I and II peptides demonstrates that enhanced Th-1 responses mediated by DC are dependent upon sequence overlap of loaded class I and II antigenic epitopes. (A) Full class I/II sequence overlap. (B) Partial (5 residues) class I/II sequence overlap. (C) Peptides defined as B8- and DR3-restricted are somewhat promiscuous and can elicit responses in an HLA-B8−/HLA-DRβ1*03− background (P < .004 in this representative experiment). For panels A, B, and C:  equals IFN-γ release induced by DC loaded with overlapping class I and II peptides, □ equals IFN-γ release induced by DC loaded with nonoverlapping class I and II peptides or singly loaded DC; y-axis equals total IFN-γ release in (in μM2). (D) DCs loaded with overlapping class I and II peptide epitopes support enhanced production of activated CD8+ cells (P < .001).
equals IFN-γ release induced by DC loaded with overlapping class I and II peptides, □ equals IFN-γ release induced by DC loaded with nonoverlapping class I and II peptides or singly loaded DC; y-axis equals total IFN-γ release in (in μM2). (D) DCs loaded with overlapping class I and II peptide epitopes support enhanced production of activated CD8+ cells (P < .001).  equals percent CD8+CD25+ cells induced by DC loaded with overlapping class I and II peptides. □ equals percentage of CD8+CD25+ cells induced by singly loaded DC. ■ equals average percentage of CD8+CD25+ cells induced by all populations of singly loaded DC; y-axis equals percentage of CD8+CD25+ cells.
equals percent CD8+CD25+ cells induced by DC loaded with overlapping class I and II peptides. □ equals percentage of CD8+CD25+ cells induced by singly loaded DC. ■ equals average percentage of CD8+CD25+ cells induced by all populations of singly loaded DC; y-axis equals percentage of CD8+CD25+ cells.
A system of overlapping/nonoverlapping defined class I and II peptides demonstrates that enhanced Th-1 responses mediated by DC are dependent upon sequence overlap of loaded class I and II antigenic epitopes. (A) Full class I/II sequence overlap. (B) Partial (5 residues) class I/II sequence overlap. (C) Peptides defined as B8- and DR3-restricted are somewhat promiscuous and can elicit responses in an HLA-B8−/HLA-DRβ1*03− background (P < .004 in this representative experiment). For panels A, B, and C:  equals IFN-γ release induced by DC loaded with overlapping class I and II peptides, □ equals IFN-γ release induced by DC loaded with nonoverlapping class I and II peptides or singly loaded DC; y-axis equals total IFN-γ release in (in μM2). (D) DCs loaded with overlapping class I and II peptide epitopes support enhanced production of activated CD8+ cells (P < .001).
equals IFN-γ release induced by DC loaded with overlapping class I and II peptides, □ equals IFN-γ release induced by DC loaded with nonoverlapping class I and II peptides or singly loaded DC; y-axis equals total IFN-γ release in (in μM2). (D) DCs loaded with overlapping class I and II peptide epitopes support enhanced production of activated CD8+ cells (P < .001).  equals percent CD8+CD25+ cells induced by DC loaded with overlapping class I and II peptides. □ equals percentage of CD8+CD25+ cells induced by singly loaded DC. ■ equals average percentage of CD8+CD25+ cells induced by all populations of singly loaded DC; y-axis equals percentage of CD8+CD25+ cells.
equals percent CD8+CD25+ cells induced by DC loaded with overlapping class I and II peptides. □ equals percentage of CD8+CD25+ cells induced by singly loaded DC. ■ equals average percentage of CD8+CD25+ cells induced by all populations of singly loaded DC; y-axis equals percentage of CD8+CD25+ cells.
Ultimately, DCs doubly loaded with overlapping class I and II peptides had to demonstrate the ability to expand CD8+ T cells to levels significantly different from unloaded DC, singly loaded DCs, or DCs doubly loaded with nonoverlapping class I and II peptides. In Figure 5D, this reproducible phenomenon is demonstrated. DCs loaded with overlapping class I and II peptides (eg, B8-166/DR3-162 and A2-443/DR3-440) were able to induce and support much higher levels of activated CD8+ T cells than DCs loaded by other methods (P < .001).
DCs loaded with matched class I and II antigenic determinants exhibit a unique transcriptional signature
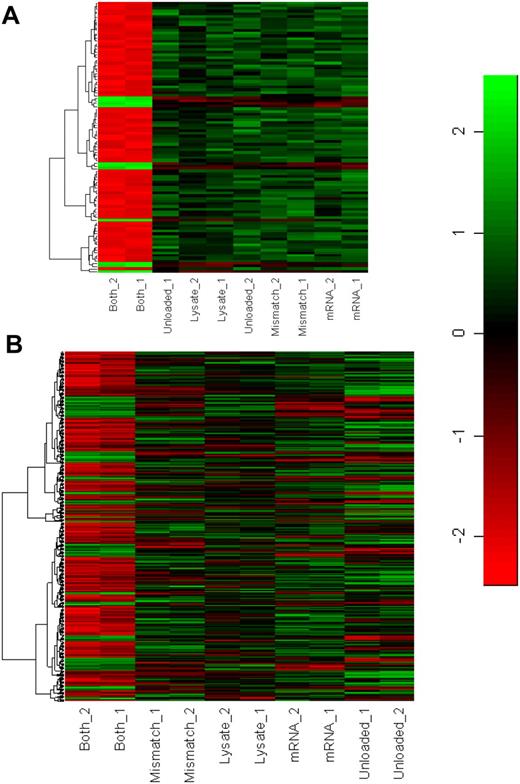
According to our hypothesis, DCs doubly loaded with matched class I and II antigens should exhibit a unique functional phenotype with, presumably, a correspondingly unique transcriptional signature differing from that of DCs loaded by any other method. To investigate this hypothesis, we performed a substantial analysis of the DC transcriptome using extensive controls and stringent statistical inclusion criteria. After loading by one of 5 different methods (unloaded control, mRNA-loaded only control, lysate-loaded only control, mismatched mRNA/lysate doubly loaded control, and the matched mRNA/lysate doubly loaded experimental group), DCs were matured for 48 hours and cryopreserved. Matured DCs from at least 6 distinct individual donors were then pooled together according to the method by which they had been loaded. RNA was isolated from pooled DCs, and samples were analyzed in duplicate using the Affymetrix U133 Plus 2.0 Array, a human genome microarray containing more than 54 000 transcripts (GEO accesion no. GSE7247). Expression concordance among sample duplicates averaged 99.2%. Cohen's d (a measure of effect size) was calculated for each transcript, and transcripts that differed in expression level between matched, doubly loaded DC and all of the other 4 control groups were identified as differentially expressed if they stringently conformed to Cohen's d greater than 1.0 (large effect is > 0.8) and q value (Benjamini-Hochberg false discovery value) less than 0.01. Given the stringent statistical parameters, the likelihood that any given gene has been identified in error is at most 1%.
Figure 6A and B demonstrate that DCs loaded with matched class I and II determinants did indeed exhibit a unique transcriptional signature in comparison to unloaded DCs and DCs loaded by any other method, including double loading with mismatched class I and II determinants. Figure 6A illustrates the top 100 genes that were differentially regulated between DCs doubly loaded with matched class I and II determinants and the 4 control groups. Cohen's d for these genes ranged from 17.1 to 7.7. Figure 6B shows the full genetic signature of 1750 differentially expressed genes as defined by the statistical inclusion criteria. A majority of these differentially expressed genes were down-regulated (red).
Gene expression signatures of DCs as a function of loading methodology. (A) Signatures of the top 100 most differentially expressed genes. Duplicate results shown. Unloaded, unloaded DCs; mRNA, DC loaded by mRNA electroporation; lysate, DC loaded by incubation with cell lysates; mismatch, DC doubly loaded with mRNA/lysate preparations derived from disparate cell types; both, DC doubly loaded with mRNA/lysate preparations derived from same cell type. (B) Signatures of all 1750 differentially regulated genes.
Gene expression signatures of DCs as a function of loading methodology. (A) Signatures of the top 100 most differentially expressed genes. Duplicate results shown. Unloaded, unloaded DCs; mRNA, DC loaded by mRNA electroporation; lysate, DC loaded by incubation with cell lysates; mismatch, DC doubly loaded with mRNA/lysate preparations derived from disparate cell types; both, DC doubly loaded with mRNA/lysate preparations derived from same cell type. (B) Signatures of all 1750 differentially regulated genes.
Differentially regulated genes of DCs doubly loaded with matched class I and II antigens promote Th-1 polarization
According to our hypothesis, known genes identified as differentially regulated among DCs doubly loaded with matched class I and II determinants should be related in some fashion to the promotion of Th-1 type cellular immunity. To examine this issue, differentially expressed genes with immune-related functions were identified by the data-mining software WEBGESTALT, and Cohen's d values for each identified transcript were independently reviewed to verify low standard deviations between experimental and controls groups. This analysis revealed 43 immune-related genes involved with cell survival, antigen presentation, T-cell regulation, antiviral responses, and T-helper polarization (Table 1). Significantly, the results demonstrated both a substantial up-regulation of some class I presentation components and a significant down-regulation of the class II presentation machinery including the MHC class II transcriptional activator (CIITA). Antiviral, Th-1 promoting, and IFN-inducible responses were significantly up-regulated. Genes involved in the generation of humoral immunity were significantly down-regulated.
Immune-related genes (as identified by WEBGESTALT) differentially regulated between matched, doubly loaded DCs and DCs loaded by any other method
| Gene function/gene . | Regulation . | Cohen's d . | q . | Function . |
|---|---|---|---|---|
| Cell survival | ||||
| TNFRSF9 | Down | 8.67 | < 0.002 | Induction of cell death |
| IL-2Rβ | Up | 7.73 | < 0.009 | Inhibition of cell death |
| IL-1α | Down | 4.76 | < 0.002 | Multiple (including survival) |
| CLU (2) | Up | 3.39 | < 0.001 | Multiple (including survival) |
| Antigen presentation | ||||
| IGλ locus | Up | 6.72 | < 0.0007 | MHC I antigen presentation |
| IGHμ | Up | 4.14 | < 0.008 | MHC I antigen presentation |
| HLA-DRβ1/ DRβ3 | Down | 3.80 | < 0.004 | MHC II antigen presentation |
| HLA-DQα1 (2) | Down | 3.33 | < 0.00005 | MHC II antigen presentation |
| CIITA | Down | 3.32 | < 0.001 | MHC II antigen presentation (TF) |
| CD1b | Up | 3.02 | < 0.0003 | Lipid antigen presentation |
| HLA-DQβ1 (2) | Down | 2.99 | < 0.0002 | MHC II antigen presentation |
| G1P2 | Up | 2.71 | < 0.0002 | IFNα-inducible signal transduction |
| CBLB (3) | Down | 2.24 | < 0.002 | Ubiquitination |
| T-cell regulation | ||||
| CTLA-4 (2) | Down | 3.32 | < 0.0003 | Induction of T-cell anergy |
| CD40 (2) | Up | 2.57 | < 0.0006 | CD8+ T-cell priming |
| IL-23α | Down | 1.84 | < 0.00002 | Inhibition of CD8+ responses |
| Antiviral response | ||||
| TRIM22 | Up | 3.74 | < 0.001 | Antiviral response (TF) |
| PRKRA | Down | 3.54 | < 0.0006 | dsRNA antiviral response |
| CTBP1 | Down | 3.04 | < 0.008 | Viral genome replication |
| IRF1 | Down | 2.37 | < 0.004 | Regulation of IFN-inducible response |
| MX2 | Up | 2.32 | < 0.0008 | Antiviral response |
| MX1 | Up | 2.08 | < 0.0004 | Antiviral response |
| CCL4 | Down | 1.74 | < 0.004 | Chemotaxis and viral replication |
| IFI27 | Up | 1.69 | < 0.004 | IFN-inducible immune response |
| GBP2 | Down | 1.53 | < 0.002 | IFN-inducible immune response |
| T-helper polarization | ||||
| TLR-4 | Up | 4.50 | < 0.004 | Pathogen detection and up-regulation of Th-1 responses |
| PTAFR | Down | 4.02 | < 0.002 | Humoral immunity |
| COLEC12 | Down | 3.92 | < 0.003 | Humoral immunity |
| LILR-B2 | Down | 3.82 | < 0.0002 | Humoral immunity |
| LY9 | Down | 3.45 | < 0.002 | Humoral immunity |
| CCL26 | Down | 3.14 | < 0.0000007 | Th-2 response (eosinophil chemotaxis) |
| MYD88 | Up | 3.12 | < 0.002 | TLR signal transduction |
| Other immune-related | ||||
| DCLRE1C | Down | 5.49 | < 0.0008 | DNA damage repair |
| XBP1 | Down | 2.83 | < 0.0005 | Transcription factor |
| IL-1R1 | Down | 2.52 | < 0.0004 | Signal transduction |
| MS4A1 | Up | 2.52 | < 0.01 | Intracellular signal transduction |
| FUS | Down | 2.27 | < 0.002 | Nucleic acid binding |
| OAS2 | Up | 2.14 | < 0.0005 | Microsomal nucleic acid metabolism |
| LST1 (2) | Up | 2.04 | < 0.004 | Dendrite morphogenesis |
| C1Qβ | Up | 1.88 | < 0.0006 | Complement activation |
| CRH | Down | 1.88 | < 0.0006 | Synaptic transmission |
| LIL-RA3 | Up | 1.34 | < 0.001 | Antigen binding |
| C1Qα | Up | 1.32 | < 0.004 | Complement activation |
| Gene function/gene . | Regulation . | Cohen's d . | q . | Function . |
|---|---|---|---|---|
| Cell survival | ||||
| TNFRSF9 | Down | 8.67 | < 0.002 | Induction of cell death |
| IL-2Rβ | Up | 7.73 | < 0.009 | Inhibition of cell death |
| IL-1α | Down | 4.76 | < 0.002 | Multiple (including survival) |
| CLU (2) | Up | 3.39 | < 0.001 | Multiple (including survival) |
| Antigen presentation | ||||
| IGλ locus | Up | 6.72 | < 0.0007 | MHC I antigen presentation |
| IGHμ | Up | 4.14 | < 0.008 | MHC I antigen presentation |
| HLA-DRβ1/ DRβ3 | Down | 3.80 | < 0.004 | MHC II antigen presentation |
| HLA-DQα1 (2) | Down | 3.33 | < 0.00005 | MHC II antigen presentation |
| CIITA | Down | 3.32 | < 0.001 | MHC II antigen presentation (TF) |
| CD1b | Up | 3.02 | < 0.0003 | Lipid antigen presentation |
| HLA-DQβ1 (2) | Down | 2.99 | < 0.0002 | MHC II antigen presentation |
| G1P2 | Up | 2.71 | < 0.0002 | IFNα-inducible signal transduction |
| CBLB (3) | Down | 2.24 | < 0.002 | Ubiquitination |
| T-cell regulation | ||||
| CTLA-4 (2) | Down | 3.32 | < 0.0003 | Induction of T-cell anergy |
| CD40 (2) | Up | 2.57 | < 0.0006 | CD8+ T-cell priming |
| IL-23α | Down | 1.84 | < 0.00002 | Inhibition of CD8+ responses |
| Antiviral response | ||||
| TRIM22 | Up | 3.74 | < 0.001 | Antiviral response (TF) |
| PRKRA | Down | 3.54 | < 0.0006 | dsRNA antiviral response |
| CTBP1 | Down | 3.04 | < 0.008 | Viral genome replication |
| IRF1 | Down | 2.37 | < 0.004 | Regulation of IFN-inducible response |
| MX2 | Up | 2.32 | < 0.0008 | Antiviral response |
| MX1 | Up | 2.08 | < 0.0004 | Antiviral response |
| CCL4 | Down | 1.74 | < 0.004 | Chemotaxis and viral replication |
| IFI27 | Up | 1.69 | < 0.004 | IFN-inducible immune response |
| GBP2 | Down | 1.53 | < 0.002 | IFN-inducible immune response |
| T-helper polarization | ||||
| TLR-4 | Up | 4.50 | < 0.004 | Pathogen detection and up-regulation of Th-1 responses |
| PTAFR | Down | 4.02 | < 0.002 | Humoral immunity |
| COLEC12 | Down | 3.92 | < 0.003 | Humoral immunity |
| LILR-B2 | Down | 3.82 | < 0.0002 | Humoral immunity |
| LY9 | Down | 3.45 | < 0.002 | Humoral immunity |
| CCL26 | Down | 3.14 | < 0.0000007 | Th-2 response (eosinophil chemotaxis) |
| MYD88 | Up | 3.12 | < 0.002 | TLR signal transduction |
| Other immune-related | ||||
| DCLRE1C | Down | 5.49 | < 0.0008 | DNA damage repair |
| XBP1 | Down | 2.83 | < 0.0005 | Transcription factor |
| IL-1R1 | Down | 2.52 | < 0.0004 | Signal transduction |
| MS4A1 | Up | 2.52 | < 0.01 | Intracellular signal transduction |
| FUS | Down | 2.27 | < 0.002 | Nucleic acid binding |
| OAS2 | Up | 2.14 | < 0.0005 | Microsomal nucleic acid metabolism |
| LST1 (2) | Up | 2.04 | < 0.004 | Dendrite morphogenesis |
| C1Qβ | Up | 1.88 | < 0.0006 | Complement activation |
| CRH | Down | 1.88 | < 0.0006 | Synaptic transmission |
| LIL-RA3 | Up | 1.34 | < 0.001 | Antigen binding |
| C1Qα | Up | 1.32 | < 0.004 | Complement activation |
The number of different transcripts detected is listed within parentheses next to each gene.
Of special note, both CD40 and CTLA-4 were shown to be differentially regulated. CD40 is a critical DC surface molecule, the stimulation of which permits “licensing” of CD8+ responses by DC if stimulated by a CD4+ helper T cell expressing CD40L.23-28 In contrast, CTLA-4 suppresses CD8+ T-cell responses, causing T-cell anergy by a variety of hypothesized mechanisms including CD28 antagonism and/or inhibitory signaling. Interestingly, CTLA-4 has not previously been shown to be expressed by DCs, but principally by regulatory CD4+ T cells.29 When class I and II antigens were matched, DCs up-regulated CD40 expression and down-regulated CTLA-4 expression. In addition, WEBGESTALT analysis demonstrated that more than 2% of the differentially regulated genes were directly involved in ubiquitination, a key step in class I antigen processing (Table 2).
Genes related to the ubiquitin cycle as identified by WEBGESTALT (but not specifically identified as immune-related) differentially regulated between matched, doubly loaded DCs and DCs loaded by any other method
| Ubiquitin metabolism . | ||||
|---|---|---|---|---|
| Gene . | Regulation . | Cohen's d . | q . | Function . |
| RFP2 | Down | 8.28 | < 0.0008 | Ubiquitin ligation |
| BIRC4 | Down | 7.74 | < 0.007 | Ubiquitin ligation |
| USP40 | Down | 7.56 | < 0.009 | Ubiquitin-dependent protein catabolism |
| EDEM1 | Down | 7.49 | < 0.00006 | Enhancer of ER protein catabolism |
| MDM4 | Down | 7.26 | < 0.0002 | Ubiquitin ligation |
| UCHL1 | Up | 7.01 | < 0.00002 | Deubiquitination |
| TRIP12 | Down | 6.89 | < 0.0003 | Ubiquitin ligation |
| USP4 | Down | 6.73 | < 0.004 | Ubiquitin-dependent protein catabolism |
| RNF44 | Down | 6.69 | < 0.002 | Transcriptional regulation |
| MAP3K7IP2 (2) | Down | 6.63 | < 0.0005 | Signal transduction |
| MAP3K1 | Down | 6.38 | < 0.0003 | Signal transduction |
| FBXL3 | Down | 5.70 | < 0.002 | Ubiquitin ligation |
| TRIM56 | Down | 5.69 | < 0.007 | Ubiquitin ligation |
| FLJ13855 | Down | 5.62 | < 0.0005 | Ubiquitin ligation |
| TRIM50B | Down | 5.44 | < 0.0002 | Ubiquitin ligation |
| UBE2B (2) | Down | 5.28 | < 0.002 | Ubiquitin ligation |
| MAR7 | Down | 5.16 | < 0.001 | Unknown |
| UBE3C | Down | 5.15 | < 0.01 | Ubiquitin ligation |
| UBE2D3 (3) | Down | 5.13 | < 0.0002 | Ubiquitin ligation |
| SENP6 | Down | 5.00 | < 0.004 | Ubiquitin-dependent protein catabolism |
| FLJ31951 | Down | 4.57 | < 0.0006 | Unknown |
| ANAPC5 | Down | 4.38 | < 0.0003 | Ubiquitin ligation |
| PARK2 | Down | 4.07 | < 0.006 | Ubiquitin ligation |
| VCPIP1 | Up | 3.82 | < 0.008 | Signal transduction |
| TRIM22 | Up | 3.74 | < 0.001 | Transcriptional regulation |
| PSMB2 | Up | 3.61 | < 0.002 | Ubiquitin-dependent protein catabolism |
| SENP8 | Up | 3.59 | < 0.007 | Ubiquitin-dependent protein catabolism |
| BIRC6 | Down | 3.25 | < 0.003 | Ubiquitin ligation |
| RNF34 | Down | 3.03 | < 0.003 | Ubiquitin ligation |
| TBL1X (3) | Down | 2.92 | < 0.0003 | Signal transduction |
| G1P2 | Up | 2.71 | < 0.0002 | IFN-inducible signal transduction |
| PRPF19 | Down | 2.66 | < 0.00003 | Ubiquitin ligation |
| KIAA0317 | Up | 2.64 | < 0.01 | Ubiquitin ligation |
| UBE2I | Down | 2.44 | < 0.004 | Ubiquitin ligation |
| C17orf27 | Down | 2.30 | < 0.003 | Ubiquitin ligation |
| CBLB (3) | Down | 2.24 | < 0.008 | Ubiquitin ligation |
| TSG101 | Up | 2.22 | < 0.002 | Transcriptional regulation |
| FBXO6 | Up | 2.12 | < 0.0006 | Generalized ubiquitination |
| USP18 | Up | 2.10 | < 0.0007 | Ubiquitin-dependent protein catabolism |
| MYLIP (2) | Down | 1.55 | < 0.004 | Ubiquitin ligation |
| Ubiquitin metabolism . | ||||
|---|---|---|---|---|
| Gene . | Regulation . | Cohen's d . | q . | Function . |
| RFP2 | Down | 8.28 | < 0.0008 | Ubiquitin ligation |
| BIRC4 | Down | 7.74 | < 0.007 | Ubiquitin ligation |
| USP40 | Down | 7.56 | < 0.009 | Ubiquitin-dependent protein catabolism |
| EDEM1 | Down | 7.49 | < 0.00006 | Enhancer of ER protein catabolism |
| MDM4 | Down | 7.26 | < 0.0002 | Ubiquitin ligation |
| UCHL1 | Up | 7.01 | < 0.00002 | Deubiquitination |
| TRIP12 | Down | 6.89 | < 0.0003 | Ubiquitin ligation |
| USP4 | Down | 6.73 | < 0.004 | Ubiquitin-dependent protein catabolism |
| RNF44 | Down | 6.69 | < 0.002 | Transcriptional regulation |
| MAP3K7IP2 (2) | Down | 6.63 | < 0.0005 | Signal transduction |
| MAP3K1 | Down | 6.38 | < 0.0003 | Signal transduction |
| FBXL3 | Down | 5.70 | < 0.002 | Ubiquitin ligation |
| TRIM56 | Down | 5.69 | < 0.007 | Ubiquitin ligation |
| FLJ13855 | Down | 5.62 | < 0.0005 | Ubiquitin ligation |
| TRIM50B | Down | 5.44 | < 0.0002 | Ubiquitin ligation |
| UBE2B (2) | Down | 5.28 | < 0.002 | Ubiquitin ligation |
| MAR7 | Down | 5.16 | < 0.001 | Unknown |
| UBE3C | Down | 5.15 | < 0.01 | Ubiquitin ligation |
| UBE2D3 (3) | Down | 5.13 | < 0.0002 | Ubiquitin ligation |
| SENP6 | Down | 5.00 | < 0.004 | Ubiquitin-dependent protein catabolism |
| FLJ31951 | Down | 4.57 | < 0.0006 | Unknown |
| ANAPC5 | Down | 4.38 | < 0.0003 | Ubiquitin ligation |
| PARK2 | Down | 4.07 | < 0.006 | Ubiquitin ligation |
| VCPIP1 | Up | 3.82 | < 0.008 | Signal transduction |
| TRIM22 | Up | 3.74 | < 0.001 | Transcriptional regulation |
| PSMB2 | Up | 3.61 | < 0.002 | Ubiquitin-dependent protein catabolism |
| SENP8 | Up | 3.59 | < 0.007 | Ubiquitin-dependent protein catabolism |
| BIRC6 | Down | 3.25 | < 0.003 | Ubiquitin ligation |
| RNF34 | Down | 3.03 | < 0.003 | Ubiquitin ligation |
| TBL1X (3) | Down | 2.92 | < 0.0003 | Signal transduction |
| G1P2 | Up | 2.71 | < 0.0002 | IFN-inducible signal transduction |
| PRPF19 | Down | 2.66 | < 0.00003 | Ubiquitin ligation |
| KIAA0317 | Up | 2.64 | < 0.01 | Ubiquitin ligation |
| UBE2I | Down | 2.44 | < 0.004 | Ubiquitin ligation |
| C17orf27 | Down | 2.30 | < 0.003 | Ubiquitin ligation |
| CBLB (3) | Down | 2.24 | < 0.008 | Ubiquitin ligation |
| TSG101 | Up | 2.22 | < 0.002 | Transcriptional regulation |
| FBXO6 | Up | 2.12 | < 0.0006 | Generalized ubiquitination |
| USP18 | Up | 2.10 | < 0.0007 | Ubiquitin-dependent protein catabolism |
| MYLIP (2) | Down | 1.55 | < 0.004 | Ubiquitin ligation |
The number of different transcripts detected is listed within parentheses next to each gene.
We verified the differential expression of CD40 with expression data as determined by flow cytometry. In 4 independent experiments (Figure 3B,D), we demonstrated that CD40 surface expression varied significantly between matched doubly loaded DCs (15% above the mean of the unloaded/singly loaded controls) and unmatched, doubly loaded DCs (30% below the mean of unloaded/singly loaded controls; P < .01). Consistent with the literature, we did not detect CTLA-4 expression on the DC surface, nor was it detectable intracellularly. Moreover, the addition of autologous T cells was unable to stimulate expression of CTLA-4 on the DC surface in a manner analogous to B lymphocytes as reported by Kuiper.30 Given these data, we theorized that DCs might be secreting the soluble, secreted isoform of CTLA-4 (sCTLA-4), which lacks the exon 3–encoded transmembrane domain.31 Accordingly, differential CTLA-4 expression in DC RNA pools was confirmed by semiquantitative RT-PCR (Figure 3E), demonstrating high levels of sCTLA-4 expression when class I and II determinants were mismatched and low levels of sCTLA-4 expression when class I and II determinants were matched.
Glycyl-tRNA synthetase inhibitor ethanolamine implicates aminoacyl-tRNA synthetases as components of a “recognisome” sensor complex
Having thoroughly demonstrated that the loading of DCs with “matched” or overlapping MHC class I and II determinants is sufficient to elicit a significant Th-1 polarization, it becomes necessary to develop mechanistic hypotheses that can adequately accommodate this observation. Obviously, such mechanisms must be somewhat speculative at this juncture.
Aminoacyl-tRNA synthetases and their associated tRNA molecules possess the alluring molecular qualities required by a “sensor” system capable of discerning sequence similarities among short peptide epitopes: each tRNA-synthetase is capable of recognizing the amino acid residue for which it is specific, and identical tRNAs are capable of stably recognizing each other via Watson-Crick base-pairing of their extensive stem-loop structures. Again, much anecdotal and experimental evidence exists to support such a hypothesis. It has been reported that the level of extracytoplasmic (microsomal) aminoacyl-tRNA synthetase activity in both human and mouse leukocytes is at least as great as the cytoplasmic tRNA-synthetase activity associated with mRNA translation.32 Furthermore, a substantial body of evidence suggests that tRNA-synthetases and tRNAs very frequently come into close contact with the MHC antigen presentation machinery. Both tRNA-synthetases and tRNAs are the major autoantigens of a wide variety of autoimmune diseases including myositis, systemic lupus erythematosus, interstitial lung disease, and rheumatoid arthritis.33-42 Moreover, our transcriptome analysis revealed that 8 tRNA-aminoacyl synthetases were differentially regulated among DCs doubly loaded with matched class I and II determinants (data not shown). Partly on the basis of these considerations, we generated the hypothesis that a recognisome might form between MHC class I and MHC class II in the postlysosomal microsome and further hypothesized that sequence comparisons within this recognisome could be mediated by extracytoplasmic tRNAs and/or their associated tRNA synthetases reported by Agris.32 To examine this hypothesis, we developed a model system to demonstrate the participation of tRNAs/tRNA synthetases in the process by which the DC compares the sequence similarity of loaded class I and class II epitopes. This model system was able to further emphasize that the generation of Th-1 responses against specific epitopes is highly dependent upon amino acid sequence similarities between class I and class II antigens.
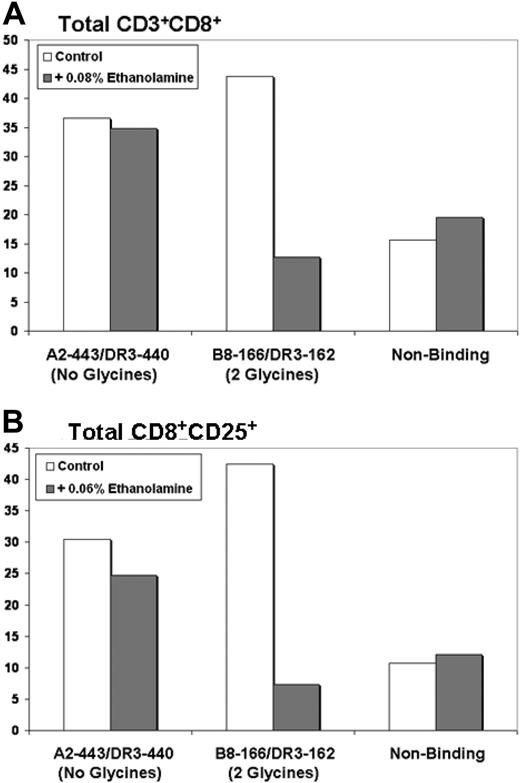
Ethanolamine is the alcohol derivative of the amino acid glycine; its 3′ carboxyl group has been replaced with a hydroxyl moiety. Ethanolamine specifically inhibits glycyl-tRNA synthetase by blocking the first step of glycine aminoacylation, the conversion of glycine and ATP to glycyl-adenylate. Thus, ethanolamine traps glycyl-tRNA synthetase complexed with its substrate molecules and terminates further reaction.43 Because ethanolamine competitively inhibits only glycyl-tRNA synthetase, it is possible to specifically block the generation of Th-1 responses by DCs loaded with glycine-containing class I/class II peptides by the addition of ethanolamine during loading and maturation. Th-1 responses generated by glycine-free overlapping class I and II epitopes should remain unaffected by the presence of ethanolamine. To use such a system, we took advantage of the B8-166/DR3-162 class I/class II peptide pair, possessing 2 glycine residues in the sequence overlap region, and the A2-443/DR3-440 peptide pair that possesses no glycine residues in the overlap region (as outlined in Figure 4B). Because ethanolamine can also inhibit protein synthesis, independent experiments were necessary to establish the proper concentration of ethanolamine for experimentation (range, 0.1%-0.01%; data not shown). Experiments were performed identically to the previous peptide experiments with the exception that 0.04% to 0.08% ethanolamine was added during loading and maturation. No ethanolamine was present after mature DC harvest and during T-cell stimulation. After stimulation, T cells were phenotyped by flow cytometry to determine the ability of peptide-loaded DCs to support the proliferation of CD8 cells.
As hypothesized, ethanolamine had no effect upon the ability of DCs doubly loaded with nonglycine-containing peptides to support the proliferation of CD8+ T cells. In contrast, DCs loaded with peptides containing glycine residues in class I/II sequence overlap region were profoundly affected by the presence of 0.04% to 0.08% ethanolamine. The representative experiment depicted by Figure 7A demonstrates that the presence of ethanolamine during DC loading and maturation resulted in a 71% reduction in the number of CD3+CD8+ T cells (43.7%-12.7% total CD3+CD8+) generated by the overlapping class I/II peptide pair B8-166/DR3-162 (2 glycine residues in the overlap region). In contrast, DCs loaded with the A2-443/DR3-440 peptide pair (no glycine residues in the overlap region) maintained high production levels of CD3+CD8+ cells in either the presence (34.9%) or absence (36.6%) of ethanolamine. DCs loaded with nonbinding control peptides also produced CD3+CD8+ cells at equivalent levels in either the presence or absence of ethanolamine and allowed discernment of background CD3+CD8+ levels. A second representative experiment demonstrates equivalent results in the production of activated CD8+ cells (CD8+CD25+). As demonstrated by Figure 7B, the presence of ethanolamine during DC loading and maturation resulted in an 83% reduction in the number of CD8+CD25+ T cells (from 42.4% to 7.3% total CD8+CD25+) generated by the overlapping class I/II peptide pair B8-166/DR3-162. Ethanolamine treatment did not induce a similar reduction in CD8+CD25+ generation by A2-443/DR3-440–loaded DCs (30.4% vs 24.8%) nor DCs loaded with nonbinding peptides (10.7% vs 12.1%).
Treatment of DC with the glycyl-tRNA synthetase inhibitor ethanolamine inhibits Th-1 responses induced by DC loaded with overlapping class I and II MHC binding peptides only when glycine residues are present in the class I/II sequence overlap region. (A) Addition of ethanolamine reduced CD8+ production by more than 70% when DC were loaded with the glycine-containing B8-166/DR3-162 peptide pair. CD8+ production induced by the non–glycine-containing peptide pair A2-443/DR3-440 was unaffected; y-axis: percentage of CD3+CD8+ cells. (B) Addition of ethanolamine reduced CD8+CD25+ production by more than 80% when DC were loaded with the glycine-containing B8-166/DR3-162 peptide pair. CD8+CD25+ production induced by the nonglycine-containing peptide pair A2-443/DR3-440 was largely unaffected; y-axis: percentage of CD8+CD25+ cells. For panels A and B, 1 representative experiment of 3 independent experiments is shown.
Treatment of DC with the glycyl-tRNA synthetase inhibitor ethanolamine inhibits Th-1 responses induced by DC loaded with overlapping class I and II MHC binding peptides only when glycine residues are present in the class I/II sequence overlap region. (A) Addition of ethanolamine reduced CD8+ production by more than 70% when DC were loaded with the glycine-containing B8-166/DR3-162 peptide pair. CD8+ production induced by the non–glycine-containing peptide pair A2-443/DR3-440 was unaffected; y-axis: percentage of CD3+CD8+ cells. (B) Addition of ethanolamine reduced CD8+CD25+ production by more than 80% when DC were loaded with the glycine-containing B8-166/DR3-162 peptide pair. CD8+CD25+ production induced by the nonglycine-containing peptide pair A2-443/DR3-440 was largely unaffected; y-axis: percentage of CD8+CD25+ cells. For panels A and B, 1 representative experiment of 3 independent experiments is shown.
Discussion
Here, we demonstrate autonomous DC involvement in a novel mechanism by which Th-1 polarization is governed and present evidence that this mechanism is dependent upon DC detection of sequence similarity between MHC class I and MHC class II antigenic epitopes. Such antigenic similarity would be expected to occur in vivo during active viremia, provided that the sentinel DCs themselves were also infected, as is often the case.4,5 We present additional evidence implicating aminoacyl-tRNA synthetases as possible components of the molecular machinery that enables amino acid sequence comparison of MHC class I and II peptide epitopes. Although antigenic similarity between class I and class II epitopes might be necessary for the promulgation of the Th-1 response, clearly it is not sufficient. We have previously demonstrated that potent inflammatory signals are also necessary to observe significant enhancement to Th-1 polarization.6 It is not hypothesized that this newly discovered mechanism operates independently. On the contrary, this mechanism is likely well-integrated amid the vast array of other signaling components that regulate adaptive immunity.
We first demonstrated that DC IL-12 secretion and CD83 surface expression were differentially up-regulated after loading of DCs with matched MHC class I and II antigenic determinants. A second antigenic system, based upon defined, overlapping MHC class I and II peptide epitopes demonstrated DC support of Th-1 responses only when the loaded class I and II peptides possessed significant sequence overlap. This additional system also served to remove cross-presentation22 as a confounding experimental variable. We subsequently demonstrated a unique transcriptional signature among populations of DCs loaded with matched class I and II determinants. Many of the genes and gene pathways that comprised this unique signature, including CD40 and CTLA-4, were known to be important regulatory components of Th-1 responses.
At the cellular level, how might DCs loaded with matched class I and II antigenic determinants (Th-1 DCs) promote Th-1 polarization and enhanced CD8+ T-cell responses? The identification of the differentially regulated genes and gene pathways among this population of DCs could be instrumental toward the elucidation of this question. Th-1 DCs might be longer-lived in vivo, demonstrating up-regulation of antiapoptotic genes as well as concomitant down-regulation of proapoptotic genes. Th-1 DCs might present class I antigens more efficiently, exhibiting up-regulation of the some MHC class I presentation components. Interestingly, Th-1 DCs also down-regulate their MHC class II presentation machinery, including the transcriptional activator, CIITA, suggesting a more specialized function in the priming of CD8+ effectors. Th-1 DCs have up-regulated multiple genes involved with antiviral defenses as well as IFN-inducible genes, despite the absence of exogenous IFNs. Th-1 DCs appear to support a highly polarized Th-1 cellular immune response, up-regulating Th-1 promoting genes and down-regulating genes involved with Th-2 responses and/or humoral immunity. Further, Th-1 DCs exhibit broad, genome-wide alterations of the ubiquitination machinery, particularly the E2 and E3 ligases crucial in determining the specificity of presented class I antigens.44,45 Significantly, Th-1 DCs up-regulate CD40 expression while down-regulating sCTLA-4 transcriptional activity. Such a profile would be predicted to promote the licensing of CD8+ T-cell responses23-28 while preventing the formation of anergic T cells,29-31 a scenario supported by previous work.3
A similar question can be asked at the molecular level: What kind of molecular system might account for the observation that DCs possess the ability to compare MHC class I and MHC class II antigenic sequences? Although aminoacyl-tRNA synthetases are typically identified as parts of the cellular translational machinery, aminoacyl-tRNA synthetase activity is also known to localize heavily to the microsomal subfraction in both human and mouse cells of hematopoietic origin.32 In addition, there is extensive information in the literature implicating tRNA synthetases and their associated tRNAs as the autoreactive antigens in a vast array of autoimmune diseases,33-42 suggesting preferential access of these molecules to MHC under conditions in which proteolytic damage might occur (ie, in the lysosomally derived compartments of DCs), thereby allowing the presentation of antigenic aminoacyl-tRNA synthetase peptides. Although it is beyond the scope of this manuscript to model the manner by which tRNA synthetases may participate in the comparison of class I and II antigenic peptide sequences, we implicated them mechanistically in this process by experiments using the glycyl-tRNA synthetase inhibitor ethanolamine.
The specificity of T-cell help in the control of humoral responses is well-documented; however, unambiguous demonstration of antigen-specific T-cell help in the control of the cellular immune response is lacking. Through the use of peptide antigens and other experimental systems, investigators have previously demonstrated that priming of an active CD8+ T-cell response in vivo requires covalent linkage between the MHC class II epitope that binds the CD4+ T-cell receptor (TCR) and the MHC class I epitope that attracts the cognate CD8+ effector cell; the implication being that such epitopes must be present in the same APC for the induction of a productive immune response.46-49 Clearly, however, other mechanisms governing specificity must regulate CD8 responses, otherwise almost any soluble antigen might provide help for CD8+ T cells recognizing almost any presented MHC class I peptide. Our observations detail a mechanism by which such specificity might be imparted. The data could be suggestive of a cross-licensing model by which DCs loaded with identical MHC class I and MHC class II antigenic determinants become receptive to subsequent licensing by CD40L expression of helper CD4+ T cells.23-28
In summary, we present data describing a novel mechanism by which DC control of Th-1 polarization appears to be regulated. The presence of antigenically matched MHC class I and class II determinants within the DCs results in a phenotypic transition, endowing the DCs with an enhanced capability to mediate Th-1 immune responses and generate CD8+ effectors. These data allow a potential explanation for the epitopic specificity of Th-1/Th-2 polarization and elucidate an additional level of regulation governing the cellular immune response. The characterization of this previously unrecognized regulatory mechanism could have significant implications for the future of clinical immunology and might substantially impact the design and implementation of next-generation vaccination strategies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Malcolm K. Brenner, MD, PhD, Alan Munro, PhD, John R. Rodgers, PhD, and Jose Engelmayer, PhD, MBA, for their support and insight. The authors also acknowledge the technical support of Suzanne Colicos, Wei Yu, PhD, and John W. Belmont, MD, PhD (all formerly of Codon Biosciences, LP); Farhang Farhangfar, formerly of MithraGen, Inc; and Michael W. Thomas, MS, of the M. D. Anderson Cancer Center. The authors graciously acknowledge Amar Safdar, MD, for providing the influenza HA peptides.
This work was supported in part by National Institutes of Health (NIH, Bethesda, MD) grant no. 5-R01 CA061508-13 (to E.J.S.).
National Institutes of Health
Authorship
Contribution: W.K.D. designed the research, performed research, analyzed data, and wrote the paper; S.L. performed research; D.X., S.N.R., H.Y., D.S., and K.V.K. analyzed data; and E.J.S. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William K. Decker, The University of Texas M.D. Anderson Cancer Center, Department of Stem Cell Transplantation and Cellular Therapy, Unit 065, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: wkdecker@mdanderson.org.