Resistance to chemotherapy is an unsolved medical problem and no clear solutions have been identified despite decades of research. In this issue of Blood, Zenz and colleagues identify miR-34a as a component of the chemotherapy resistance network in CLL. This is a remarkable finding with possible diagnostic and therapeutic consequences.
One of the most unexpected and fascinating discoveries of the past few years in molecular oncology is that abnormalities in both protein coding genes (PCGs) and non-coding RNAs (ncRNAs; RNAs that do not codify for a protein) can be identified in tumors, and the interplay between them is causally involved in cancer initiation, progression, and dissemination. The most studied ncRNAs are microRNAs (miRNAs), small 19 to 25 nt transcripts involved in gene regulation of the majority of PCGs.1 Initially identified in chronic lymphocytic leukemia (CLL),2 alterations of miRNAs have since been detected in every type of human tumor analyzed to date (both benign and malignant, solid, and liquid), incriminating miRNA in tumorigenesis. Furthermore, miRNA expression profiling has identified signatures associated with diagnosis, staging, progression, prognosis, and response to treatment.3
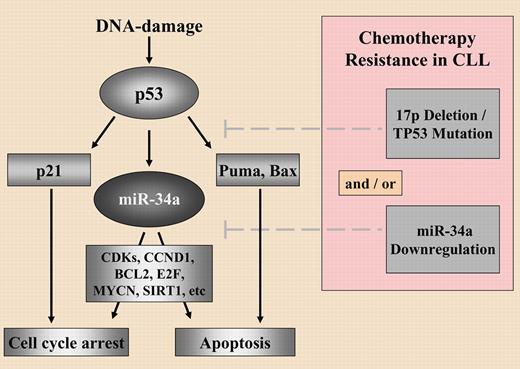
Why is the study by Zenz and colleagues4 important? The first reason is due to the novelty of the findings. The authors were able to elegantly prove in a relatively large set of cytogenetically well-defined CLL samples a link between the expression levels of miR-34a, a member of a miRNA family already known to be positively regulated by TP53,5 and response to DNA damage, TP53 status and, most importantly, response to fludarabine treatment. A picture of the complex interplay between PCGs and miRNAs in determining response of CLL cells to therapy is clearly emerging (see figure): in the presence of functional TP53, miR-34a levels are up-regulated by DNA damage, and low miR-34 levels instead are associated with impaired DNA damage response, TP53 mutations, and fludarabine-refractory disease, in either the presence or the absence of TP53 deletions. These are straightforward findings that yield an additional piece to the newly commenced puzzle of miRNAs and drug resistance/sensitivity. Other pieces in this puzzle include the observation that an 829C>T polymorphism in a dihydrofolate reductase (DHFR) gene binding site for miR-24 leads to loss of miR-24 function and results in DHFR overexpression and methotrexate resistance.6 In addition, changing the cellular levels of let-7i, miR-16, and miR-21 in cancer cells affects the potencies of a number of anticancer agents by up to 4-fold.7 Furthermore, miR-15b and miR-16, through regulation of BCL2 expression, can modulate the sensitivity of gastric cancer cells to specific anticancer drugs.8 The latter finding is of special interest in light of the data found by Zenz et al, as miR-16 was proved to be located in the most frequently deleted region in CLL (13q14), down-regulated in the majority of CLL patients, and acts as a tumor suppressor by targeting BCL2 and MCL1 oncogenes.9
Resistance to chemotherapy in CLL as an interplay between protein coding genes and miR-34a. See the complete figure in the article beginning on page 3801.
Resistance to chemotherapy in CLL as an interplay between protein coding genes and miR-34a. See the complete figure in the article beginning on page 3801.
Second, the study of Zenz and colleagues opens intriguing avenues of research that can bring mir-34a investigations closer to CLL patients' bedsides. It can be speculated that plasma levels of miR-34a at diagnosis are different between patients who will respond and patients who will be resistant to fludarabine therapy. The premise of such exciting findings already exists—high miR-21 expression in colon adenocarcinomas is associated with poor response to adjuvant chemotherapy, so miR-21 levels can predict the way in which these patients respond to therapy.10
Third, Zenz et al's results highlight miRNA (ie, miR-34a) as either new targets or agents for alternative cancer therapies. miRNAs participating in CLL pathogenesis can be exploited as therapeutic drugs against messenger RNAs of PCGs involved in CLL, or can be directly targeted by synthetic RNAs (such as antagomirs) or other compounds. Zenz et al found that cells with reduced miR-34a expression show increased viability after DNA damage independently of 17p status. Therefore, restoration of miR-34a levels, either by reactivation or by reinsertion in CLL cells, can be employed as a therapeutic intervention to restore sensitivity to therapy.
As with all landmark studies, the contribution by Zenz and colleagues raises intriguing questions: Is miR-34a involved in CLL resistance to therapies other than fludarabine? What are the TP53-independent mechanisms of miR-34 regulation in CLL andcan these be exploited to the patient's advantage? What are the other miRNAs involved in chemotherapy resistance in CLL? Are there other ncRNAs involved in refractory/resistant disease? Certainly, these questions and many more will not be left without answers for long. The time when the ncRNA revolution in CLL society will shift paradigms in chemotherapy is already here!
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal