Abstract
Anaplastic large cell lymphoma represents a subset of neoplasms caused by translocations that juxtapose the anaplastic lymphoma kinase (ALK) to dimerization partners. The constitutive activation of ALK fusion proteins leads to cellular transformation through a complex signaling network. To elucidate the ALK pathways sustaining lymphomagenesis and tumor maintenance, we analyzed the tyrosine-kinase protein profiles of ALK-positive cell lines using 2 complementary proteomic-based approaches, taking advantage of a specific ALK RNA interference (RNAi) or cell-permeable inhibitors. A well-defined set of ALK-associated tyrosine phosphopeptides, including metabolic enzymes, kinases, ribosomal and cytoskeletal proteins, was identified. Validation studies confirmed that vasodilator-stimulated phosphoprotein and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/inosine monophosphate cyclohydrolase (ATIC) associated with nucleophosmin (NPM)–ALK, and their phosphorylation required ALK activity. ATIC phosphorylation was documented in cell lines and primary tumors carrying ALK proteins and other tyrosine kinases, including TPR-Met and wild type c-Met. Functional analyses revealed that ALK-mediated ATIC phosphorylation enhanced its enzymatic activity, dampening the methotrexate-mediated transformylase activity inhibition. These findings demonstrate that proteomic approaches in well-controlled experimental settings allow the definition of informative proteomic profiles and the discovery of novel ALK downstream players that contribute to the maintenance of the neoplastic phenotype. Prediction of tumor responses to methotrexate may justify specific molecular-based chemotherapy.
Introduction
Cell transformation is the result of the sequential acquisition of multiple genetic defects, which provide a growth and survival advantage to the cancerous cells and the acquisition of metastatic potential.1 The activation of oncogenes and the loss of tumor suppressor genes are pivotal in cancer development, as they deregulate multiple metabolic pathways and contribute to the neoplastic phenotype. Better understanding of key metabolic checkpoints in cancer cells would allow the design of novel therapeutic strategies. Dividing cells heavily rely on de novo purine synthesis, whereas normal cells prefer the salvage pathway.2 Glycinamide ribonucleotide formyltransferase and the bifunctional 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) formyltransferase/inosine monophosphate (IMP) cyclohydrolase (AICAR-FT/IMP-CHase, named ATIC) have raised considerable attention because of their role in cancer. Both enzymes are folate-dependent and have become exquisite targets of chemotherapeutic intervention.2-4
ATIC is a bifunctional enzyme that catalyzes the final 2 steps of de novo purine biosynthesis pathway.3-5 The AICAR formyltransferase (AICAR-FT) domain (residues 199-592) catalyzes the transfer of the one-carbon formyl group from the cofactor N10-formyl-tetrahydrofolate (10-f-THF) to the substrate AICAR to produce N-formyl-5-aminoimidazole-4-carboxamide ribonucleotide (F-AICAR) and tetrahydrofolate. The IMP cyclohydrolase domain (IMP-Chase; residues 1-198) then enhances the intramolecular cyclization of N-formyl-AICAR to the final product of the pathway, IMP.6
The ATIC gene is fused, as result of cryptic inversion [inv(2) (9p23q35)], to the anaplastic lymphoma kinase (ALK) in a subset of anaplastic large cell lymphoma (ALCL). ALCL, a distinct entity among T-cell non-Hodgkin lymphoma (NHL), is a hematologic disorder that accounts for approximately 30% of all pediatric NHLs. Many ALCLs carry translocations that involve ALK and variable partner genes (mainly nucleophosmin [NPM1]). In ATIC-ALK, the N-terminus of ATIC fuses to the intracytoplasmic region of ALK and encodes a novel oncogenic chimeric protein.7-9
ALK chimeras have constitutive tyrosine kinase activity with oncogenic potential. In vitro and in vivo studies have demonstrated that ALK signaling induces cell transformation by modulating many adaptor proteins involved in cell-cycle progression, survival, cytoskeletal rearrangement, and cell migration.10 ALK signaling is required and necessary to maintain the neoplastic phenotype because the loss of ALK activity causes cell-cycle arrest and cell death in vitro, and tumor regression in vivo.11,12 These findings have fostered the discovery of ALK small-molecule inhibitors that are now in early clinical trials or on the verge of entering the clinical arena. The discovery that deregulated expression of ALK can be seen in a subset of nonhematologic tumors, including inflammatory myofibroblastic tumors, non–small cell lung cancer, sarcoma, and neuroblastoma,12 has increased the interest on ALK, as a promising target for specific therapies.
Because some signaling molecules essential for ALK-mediated transformation10 display a key function in other ALK− tumors, several groups have undertaken high throughput (HTP) analyses, including gene expression profiling assays13,14 and proteomic-based approaches,15,16 to discover selective ALK targets. Liquid chromatography–tandem mass spectrometry (LC-MS/MS) and HTP proteomics focusing on tyrosine phosphopeptides provide a fast and reliable method for large-scale analysis of cellular proteins differentially expressed in normal and tumor samples, and it is a powerful tool to identify selective signatures in kinase-driven hematologic and nonhematologic malignancies.15,17-20
Here we used 2 complementary proteomic-based approaches to dissect the ALK signaling. Taking advantage of shRNA and ALK kinase inhibitors, we compared the differential ALK tyrosine-phosphorylation profiling in different settings. We found that ALK activity is associated with a defined set of phosphorylated proteins regulating key cellular functions. Among novel ALK-associated proteins, we have shown that vasodilator-stimulated phosphoprotein (VASP) and ATIC are directly phosphorylated by ALK. The enzymatic activity of ATIC was enhanced after tyrosine phosphorylation via several oncogenes and phospho-ATIC was less efficiently inhibited by the methotrexate. These findings provide novel insights into ALK-mediated transformation and support the selection of tailored chemotherapeutic protocols.
Methods
Cell lines and reagents
Human ALCL cell lines TS (a subclone of Sup-M2), Sup-M2, JB-6, SU-DHL1, and Karpas-299 were previously described.11-21 T-cell leukemic cell lines CCRF-CEM and Jurkat were obtained from ATCC (Manassas, VA); Mac-1 was kindly provided by Dr M. Kadin (Harvard University, Boston, MA). Cell lines were grown at 37°C in 5% CO2 humidified air in RPMI 1640 medium (Lonza Verviers SPRL, Verviers, Belgium).
HEK-293T and HEK-293T-Rex Tet-on NPM-ALK cells22 were grown at 37°C in 5% CO2 humidified air in Iscove modified Dulbecco medium, supplemented with 10% fetal calf serum. For antiphosphotyrosine immunoprecipitation, HEK-293T-Rex Tet-on NPM-ALK cells were grown in nonadherent conditions on poly (2-hydroxyethylmethacrylate; Sigma-Aldrich, St Louis, MO)–coated plates, starved for 12 hours, and then induced with 1 μg/mL of tetracycline for 24 hours.
Self-inactivating retroviral particles for NPM-ALK and the kinase dead mutant NPM-ALKK210R were produced as described previously.11 Aliquots of virus, plus 8 μg/mL of polybrene, were used to infect exponentially growing cells (CCRF-CEM and Mac-1, 105/mL). Fresh medium was supplemented 24 hours after infection. The infectivity was determined after 72 hours of infection by fluorescence-activated cell sorting analysis of green fluorescent protein (GFP)–positive cells. GFP+ cells were sorted by MoFlo High-Performance fluorescence-activated cell sorting (Dako North America, Carpinteria, CA) and expanded.
For the kinase inhibition experiments, NPM-ALK–positive and –negative cells were treated with 300 nM of CEP11988 or CEP1408323 for 6 hours.
Phosphopeptide immunoprecipitation and LC-MS/MS mass spectrometry
Phosphopeptide immunoprecipitation from cell lines was performed as described previously15 using the PhosphoScan Kit (P-Tyr-100) from Cell Signaling Technology (Danvers, MA; Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
All spectra and all sequence assignments obtained using Sequest (Thermo Scientific, Waltham, MA) were then imported into a relational database based on FileMaker Pro (FileMaker, Santa Clara, CA) and MySQL (Sun Microsystems, Santa Clara, CA), as described.15 Comparison between large datasets was performed using a custom-made Perl script (www.perl.com), to find overlapping and/or recurrent sequences (see Document S1).
Anti-phosphotyrosine protein immunoprecipitation and LC/MS-MS analysis
Anti-phosphotyrosine immunoprecipitation on HEK-293T-Rex Tet-on cells (Invitrogen, Carlsbad, CA), transfected with a wild-type NPM-ALK or a kinase-dead mutant control NPM-ALKK210R, and LC/MS-MS analyses were performed as previously described.22 Protein identification via peptide MS/MS spectra was achieved using the Mascot software (http://matrixscience.com) for searching the National Center for Biotechnology Information nonredundant human protein database (released April 29, 2003; containing 37 490 protein sequences).
Stable isotope labeling of amino acid in cell culture analysis of shALK cells
Cells were grown in RPMI medium lacking arginine and lysine, supplemented with 10% dialyzed fetal bovine serum, penicillin/streptomycin, and L-lysine/HCl and L-arginine/HCl (Sigma-Aldrich) for light cultures or L-arginine/HCL (U-13C6, 98%) and L-lysine/2HCl (U-13C6, 98%; Cambridge Isotope Laboratories, Andover, MA) for heavy cultures as described.24,25 Cells were grown to a density of approximately 106 cells/mL for a total of 108 cells per each cell culture type (2 × 108 cells total). After lysis, heavy and light cultures were combined and carried through the phosphopeptide immunoprecipitation protocol.
Protein immunoprecipitation and Western blotting
Immunoprecipitation and Western blot analysis were performed as described previously26 (Document S1). The protein content of cell suspensions was assessed with the Lowry kit from Bio-Rad (Hercules, CA).
The following primary antibodies were used: mouse anti-ALK (1:4000, 4C5B8) and anti-STAT3 (5G7) from Zymed Laboratories (South San Francisco, CA); mouse anti–phospho-tyrosine (PY100; 1:2000), rabbit anti–phospho STAT3 (Tyr 705, 1:1000), rabbit antiphospho-ERK1/2 (Thr202/Thy204) (#9101 1:1000), rabbit anti–phospho-SHP2 (Tyr 542) (1:1000), rabbit anti–phospho-SHC (Tyr 317) (1:1000), rabbit anti–phospho-ALK (Tyr 1604) (1:1000), rabbit anti–SHC (1:1000), rabbit anti–p44/42 MAPK (1:1000), anti–VASP (1:1000), and rabbit anti–SHP2 (1:1000) from Cell Signaling Technology; mouse anti-actin (1:2000) from Millipore (Billerica, MA); mouse anti-ATIC (1:1000) from Abcam (Cambridge, United Kingdom) and agarose-conjugated 4G10 from Upstate Biotechnology (Charlottesville, VA).
Enzyme-kinase assay
ATIC enzyme and NPM-ALK kinase were immunoprecipitated from 2 mg of HEK-293T and HEK-293T NPM-ALK total cell lysate, respectively. The immunoprecipitated proteins were resuspended in kinase buffer (25 mM Tris-HCl, pH 7.5, 2 mM dithiothreitol (DTT), 0.1 mM Na3VO4, 10 mM MgCl2, 5 mM b-glycerophosphate) and then combined at an equal ratio. The reaction was started by adding 0.2 mM adenosine triphosphate (ATP) to the mixture and incubating for 30 minutes at 37°C and then stopped by freezing the samples at −80°C.
Where indicated, CEP14083 (300 nM) was added to the reaction mixture before adding ATP, then incubated for 30 minutes at 37°C. Methotrexate (MTX) was added at the indicated concentrations, and the reaction mixture was incubated again at 37°C for 2 hours.
Measurement of AICAR-FT/IMP-CHase activity
The synthesis of N10-formyl-tetrahydrofolate from (6R, 6S)-5-formyltetrahydropteroil-L-glutamate was performed according to Uyeda and Rabinowitz.27 The amount of N10-formyl-tetrahydrofolate was assessed by measuring the absorbance at 298 nm (ϵ = 9.54 cm−1·M−1). AICAR-FT activity was evaluated by coupling it with the reactions of serine-hydroxy-methyltransferase and methylene-tetrahydrofolate reductase28 : 50 μg of total cellular lysates or immunoprecipitated proteins was resuspended in the following 1-mL reaction mix: 66 mM Tris (pH 7.4), 20 mM K3PO4, 5 mM DTT, 2 mM N10-formyl-tetrahydrofolate, 1 mM AICAR. After 5 minutes, samples were incubated at 37°C in the presence of 1 mM L-serine, 0.2 μg serine-hydroxy-methyltransferase, and 0.2 μg of methylene-tetrahydrofolate reductase. Then (after 2 minutes), 0.05 mM nicotinamide adenine dinucleotide phosphate was added and the absorbance at 340 nm was measured for 10 minutes using a Lambda 3 spectrophotometer (PerkinElmer Life and Analytical Sciences, Waltham, MA). Preliminary experiments showed that, in these experimental conditions, the oxidation rate of nicotinamide adenine dinucleotide phosphate was linear throughout the observation time and stoichiometrically equivalent to the rate of AICAR disappearance by AICAR-FT (data not shown). Results were expressed as nmol NADP+/minute per mg cell proteins.
The activities of IMP-CHase or AICAR-FT plus IMP-CHase were measured by a coupling assay with inosine 5′-monophosphate dehydrogenase29 ; 50 μg of total cellular lysates or immunoprecipitated proteins were incubated 30 minutes at 37°C in a reaction mix containing 66 mM Tris (pH 7.4), 5 mM DTT, 50 mM KCl (final volume, 1 mL). To detect the activity of IMP-Chase, 0.1 mM N-formylaminoimidazole-4-carboxamide ribonucleotide (F-AICAR), synthesized as previously described30 and quantified by measuring the absorbance at 268 nm (ϵ = 10 900 cm−1·M−1), was added. For the total activity of AICAR-FT plus IMP-CHase, samples were incubated with 2 mM N10-formyl-tetrahydrofolate and 1 mM AICAR instead of F-AICAR. Then 0.1 μg inosine 5′-monophosphate dehydrogenase and 0.1 mM NAD+ were added. The reduction rate of NAD+, stoichiometrically equivalent to the rate of IMP synthesis under these experimental conditions (data not shown), was evaluated by measuring the absorbance at 340 nm. The reaction was linear throughout a 10-minute observation time and the results were expressed as nmol NADH/min/mg cell proteins.
Measurement of 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, lactic dehydrogenase, and ornithine decarboxilase activities
Enzymatic activity for glucose 6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (6PGD), and lactic dehydrogenase (LDH) was measured as previously described.31 Ornithine decarboxilase activity was determined as the amount of 14CO2 released from 0.5 μCi DL-[1-14C]ornithine, as described.32
Results
LC-MS/MS identifies a set of phosphotyrosine peptides in NPM-ALK+ cells
To determine a global profile of tyrosine (Tyr)–phosphorylated proteins in NPM-ALK cells, we used 2 different LC-MS/MS-based proteomic approaches. Both methods implied immunoaffinity precipitation of Tyr-phosphorylated proteins by specific anti–phosphotyrosine antibodies, but whereas the first approach required the excision of bands of interest from sodium dodecyl sulfate–polyacrylamide gel electrophoresis (15-20 bands differentially expressed, compared with control), the second approach allowed a more global mapping of Tyr-phosphorylated peptides. We have first assessed the feasibility of both strategies using a tetracycline-inducible NPM-ALK HEK-293T-Rex cell line (Figure 1A). A list of 40 common proteins detected by both approaches is provided in Table 1. These studies confirmed several known interactors of NPM-ALK (STAT3, SHC, and PTPN11)21,26,33 and discovered novel Tyr-phosphorylated proteins. The immunoaffinity profiling of phosphopeptides (PhosphoScan)15 identified a total of 167 peptides (corresponding to 137 phosphorylated proteins), granting a better specificity compared with conventional approaches (Table S1). This technique excludes all proteins that are not phosphorylated but bind specifically to a phosphorylated peptide and identifies multiple Tyr-phosphorylated peptides corresponding to the same protein, providing a higher degree of confidence.
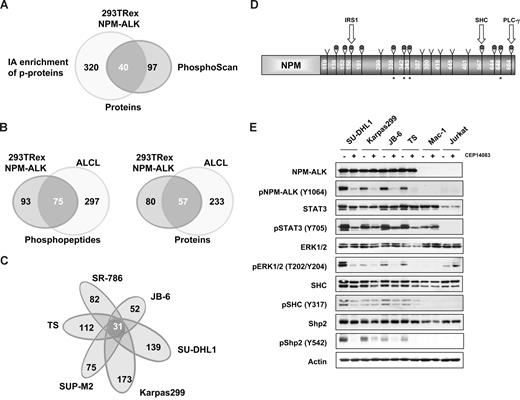
LC-MS/MS profiling of NPM-ALK+ cells identifies a common set of Tyr-phosphorylated proteins. (A) Comparison of the datasets derived from the immunoaffinity (PhosphoScan) and conventional LC-MS/MS-based proteomic approaches (Table 1). (B) Immunoaffinity profiling of NPM-ALK+ ALCL cell lines compared with NPM-ALK-positive HEK-293T-Rex cells. (C) Comparative PhosphoScan performed on 6 different ALCL cell lines. (D) Tyrosine phosphorylated sites on the NPM-ALK protein are represented with a gray dot (9 of these were identified in all ALK+ cell lines); the star represents newly discovered sites. Tyrosine sites known to interact with IRS1, SHC, and PLC-g are indicated. (E) The phosphorylation status of selected proteins was assessed in ALCL cell lines after treatment with CEP14083 and detected by phospho-specific antibodies as indicated.
LC-MS/MS profiling of NPM-ALK+ cells identifies a common set of Tyr-phosphorylated proteins. (A) Comparison of the datasets derived from the immunoaffinity (PhosphoScan) and conventional LC-MS/MS-based proteomic approaches (Table 1). (B) Immunoaffinity profiling of NPM-ALK+ ALCL cell lines compared with NPM-ALK-positive HEK-293T-Rex cells. (C) Comparative PhosphoScan performed on 6 different ALCL cell lines. (D) Tyrosine phosphorylated sites on the NPM-ALK protein are represented with a gray dot (9 of these were identified in all ALK+ cell lines); the star represents newly discovered sites. Tyrosine sites known to interact with IRS1, SHC, and PLC-g are indicated. (E) The phosphorylation status of selected proteins was assessed in ALCL cell lines after treatment with CEP14083 and detected by phospho-specific antibodies as indicated.
Proteins identified in HEK 293T NPM-ALK cells by 2 complementary proteomic approaches
| Gene symbol . | NCBI access no. . | Protein . | No. of peptides* . | Sequence coverage,* % . |
|---|---|---|---|---|
| Kinases and phosphatases | ||||
| ALK | NP_004295 | Anaplastic lymphoma kinase Ki-1 | 30 | 38 |
| GSK3A | NP_063937 | Glycogen synthase kinase 3 α | 1 | 2 |
| PFKP | NP_002618 | Phosphofructokinase, platelet | 3 | 3 |
| PKM2 | NP_872270 | Pyruvate kinase 3 isoform 2 | 8 | 20 |
| PTPN11 | NP_002825 | Shp2 | 4 | 6 |
| Metabolism | ||||
| ACLY | NP_001087 | ATP citrate lyase | 6 | 6 |
| CUL2 | NP_003582 | Cullin 2 | 5 | 9 |
| DDX3X | NP_004651 | DEAD/H box-3 | 5 | 7 |
| DNAJC7 | NP_003306 | DnaJ (Hsp40) homolog | 7 | 13 |
| ENO1 | NP_001419 | Enolase 1 | 48 | 57 |
| HNRPA1 | NP_112420 | Heterogeneous nuclear ribonucleoprotein A1 | 5 | 22 |
| HNRPA2B1 | NP_112533 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 11 | 30 |
| HNRPF | NP_004957 | Heterogeneous nuclear ribonucleoprotein F | 3 | 8 |
| HNRPH1 | NP_005511 | Heterogeneous nuclear ribonucleoprotein H1 | 2 | 3 |
| HNRPR | NP_005817 | Heterogeneous nuclear ribonucleoprotein R | 2 | 3 |
| HNRPU | NP_004492 | Heterogeneous nuclear ribonucleoprotein U | 11 | 12 |
| SFPQ | NP_005057 | Splicing factor proline/glutamine rich | 5 | 5 |
| TARS | NP_689508 | Threonyl-tRNA synthetase | 3 | 4 |
| ZNF598 | NP_835461 | Zinc finger protein 598 | ||
| Adaptor proteins | ||||
| HGS | NP_004703 | Hepatocyte growth factor-regulated tyrosine kinase substrate | 3 | 4 |
| HSP90AB1 | NP_031381 | Heat shock 90-kDa protein 1, β | 25 | 25 |
| HSPA1B | NP_005337 | Heat shock 70-kDa protein 1B | 16 | 28 |
| HSPA2 | NP_068814 | Heat shock 70-kDa protein 2 | 1 | 1 |
| HSPA4 | NP_002145 | Heat shock 70-kDa protein 4 | 1 | 1 |
| HSPA8 | NP_006588 | Heat shock 70-kDa protein 8 | 1 | 3 |
| IRS4 | NP_003595 | Insulin receptor substrate 4 | 48 | 27 |
| RPLP0 | NP_444505 | Ribosomal protein P0 | 3 | 10 |
| SHC | NP_003020 | SHC | 2 | 2 |
| TRAP1 | NP_057376 | Tumor necrosis factor type 1 receptor-associated protein | 1 | 2 |
| Signal transduction | ||||
| ARHGEF2 | NP_004714 | rho/rac guanine nucleotide exchange factor 2 | 1 | 1 |
| CNOT1 | NP_057368 | KIAA1007 protein; CCR4-NOT transcription complex, subunit 1 | 1 | 1 |
| LMO7 | NP_005349 | LIM domain only 7 | 1 | 1 |
| STAT3 | NP_644805 | Signal transducer and activator of transcription 3 | 2 | 2 |
| YWHAG | NP_036611 | 14-3–3 γ | 7 | 24 |
| Cytoskeleton | ||||
| ACTB | NP_001092 | β actin | 8 | 20 |
| BICD2 | NP_056065 | Bicaudal D homolog 2 | 7 | 11 |
| VIM | NP_003371 | Vimentin | 18 | 38 |
| Unknown function | ||||
| FHL1 | NP_001440 | Four and a half LIM domains 1 | 2 | 6 |
| NCAPH | NP_056156 | Barren | 7 | 13 |
| UBAP2L | NP_055662 | NICE-4 protein | 4 | 6 |
| Gene symbol . | NCBI access no. . | Protein . | No. of peptides* . | Sequence coverage,* % . |
|---|---|---|---|---|
| Kinases and phosphatases | ||||
| ALK | NP_004295 | Anaplastic lymphoma kinase Ki-1 | 30 | 38 |
| GSK3A | NP_063937 | Glycogen synthase kinase 3 α | 1 | 2 |
| PFKP | NP_002618 | Phosphofructokinase, platelet | 3 | 3 |
| PKM2 | NP_872270 | Pyruvate kinase 3 isoform 2 | 8 | 20 |
| PTPN11 | NP_002825 | Shp2 | 4 | 6 |
| Metabolism | ||||
| ACLY | NP_001087 | ATP citrate lyase | 6 | 6 |
| CUL2 | NP_003582 | Cullin 2 | 5 | 9 |
| DDX3X | NP_004651 | DEAD/H box-3 | 5 | 7 |
| DNAJC7 | NP_003306 | DnaJ (Hsp40) homolog | 7 | 13 |
| ENO1 | NP_001419 | Enolase 1 | 48 | 57 |
| HNRPA1 | NP_112420 | Heterogeneous nuclear ribonucleoprotein A1 | 5 | 22 |
| HNRPA2B1 | NP_112533 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 11 | 30 |
| HNRPF | NP_004957 | Heterogeneous nuclear ribonucleoprotein F | 3 | 8 |
| HNRPH1 | NP_005511 | Heterogeneous nuclear ribonucleoprotein H1 | 2 | 3 |
| HNRPR | NP_005817 | Heterogeneous nuclear ribonucleoprotein R | 2 | 3 |
| HNRPU | NP_004492 | Heterogeneous nuclear ribonucleoprotein U | 11 | 12 |
| SFPQ | NP_005057 | Splicing factor proline/glutamine rich | 5 | 5 |
| TARS | NP_689508 | Threonyl-tRNA synthetase | 3 | 4 |
| ZNF598 | NP_835461 | Zinc finger protein 598 | ||
| Adaptor proteins | ||||
| HGS | NP_004703 | Hepatocyte growth factor-regulated tyrosine kinase substrate | 3 | 4 |
| HSP90AB1 | NP_031381 | Heat shock 90-kDa protein 1, β | 25 | 25 |
| HSPA1B | NP_005337 | Heat shock 70-kDa protein 1B | 16 | 28 |
| HSPA2 | NP_068814 | Heat shock 70-kDa protein 2 | 1 | 1 |
| HSPA4 | NP_002145 | Heat shock 70-kDa protein 4 | 1 | 1 |
| HSPA8 | NP_006588 | Heat shock 70-kDa protein 8 | 1 | 3 |
| IRS4 | NP_003595 | Insulin receptor substrate 4 | 48 | 27 |
| RPLP0 | NP_444505 | Ribosomal protein P0 | 3 | 10 |
| SHC | NP_003020 | SHC | 2 | 2 |
| TRAP1 | NP_057376 | Tumor necrosis factor type 1 receptor-associated protein | 1 | 2 |
| Signal transduction | ||||
| ARHGEF2 | NP_004714 | rho/rac guanine nucleotide exchange factor 2 | 1 | 1 |
| CNOT1 | NP_057368 | KIAA1007 protein; CCR4-NOT transcription complex, subunit 1 | 1 | 1 |
| LMO7 | NP_005349 | LIM domain only 7 | 1 | 1 |
| STAT3 | NP_644805 | Signal transducer and activator of transcription 3 | 2 | 2 |
| YWHAG | NP_036611 | 14-3–3 γ | 7 | 24 |
| Cytoskeleton | ||||
| ACTB | NP_001092 | β actin | 8 | 20 |
| BICD2 | NP_056065 | Bicaudal D homolog 2 | 7 | 11 |
| VIM | NP_003371 | Vimentin | 18 | 38 |
| Unknown function | ||||
| FHL1 | NP_001440 | Four and a half LIM domains 1 | 2 | 6 |
| NCAPH | NP_056156 | Barren | 7 | 13 |
| UBAP2L | NP_055662 | NICE-4 protein | 4 | 6 |
Number of peptides and sequence coverage refer to IA enrichment method only.
Given that the HEK-293T–inducible system uses a highly controlled but ectopic NPM-ALK expression in a nonlymphoid cells, we applied the PhosphoScan technology (Cell Signaling Technology) to a panel of NPM-ALK–positive ALCL lines, a model that better reproduces the ALK lymphoproliferative processes. This analysis revealed a total of 372 Tyr-phosphorylated peptides within 290 proteins (Figure 1B), which included approximately half of the peptides previously identified in NPM-ALK HEK-293T cells. A further analysis, performed to find the proteins shared by all cell lines and applying both techniques (data not shown), led to the discovery of a small cluster of 26 Tyr-phosphorylated proteins (Table S2), which were also largely represented in previous analyses. We then compared the data obtained from the phospho-tyrosine immunoprecipitation of 6 ALK+ ALCL lines and found a panel of 372 peptides (within 290 different proteins), 31 of which were shared by all 6 ALCL cell lines (Figure 1C).
Of these 31 Tyr-phosphorylated peptides, 12 were also shared by the ALK− control T cell lymphoblastoid line Mac-1 (CD30/STAT3/STAT5-positive). These phosphopeptides belonged to proteins with kinase activity (GSK3B (Y279), CDK3 (Y15), DYRK1A (Y321), PRPF4B (Y849]) or metabolic enzymes (ENO-1 [Y43], LDH-A [Y238], LDH-B [Y239], PKM [Y104]) frequently found in proliferating cells. Notably, some well-documented ALK interactors, such as SHC-1 (Y427) and STAT3 (iso1 Y704, and iso2 Y705), were also found. Similar findings were obtained by comparison of ALK+ cell lines to other 2 ALK− lymphoid cell lines (CCRF-CEM and Jurkat, data not shown).
Nineteen peptides were exclusively found in ALK+ cells, and 9 belonged to the ALK protein itself. Noteworthy, 4 of these were novel ALK phosphorylation sites (Figure 1D). The remaining 10 peptides identified proteins linked to cell metabolism, including ATIC (Y104) and ATP-citrate lyase (ACLY, Y682), G-proteins (GTPase-activating protein RasA1, Y615), and ribosomal proteins (RPs) L31 (Y103) and L18a (Y63). Using a lower cutoff level (peptides identified in > 4 of 6 cell lines), we could define a larger protein set with several novel entities (Table 2); among them, ribosomal (L7 [Y139], S13 [Y37], and P0 [Y24]) and structural proteins, including VASP (Y38), a cytoskeletal-regulating protein recently identified as part of BCR-ABL phospho-tyrosine signature.17
Tyrosine phosphorylated peptides identified by PhosphoScan in 6 ALK+ ALCL cell lines (cutoff level, 67%) versus the ALK− cell line Mac-1
| Gene symbol . | pTyr site . | Peptide . | Karpas299 . | SU-DHL1 . | JB-6 . | TS . | SR-786 . | SUP-M2 . | Mac-1 . |
|---|---|---|---|---|---|---|---|---|---|
| Kinases | |||||||||
| ALK | 1078* | HQELQAMQMELQSPEyK | • | • | • | • | • | • | |
| ALK | 1092* | TSTIMTDyNPNYCFAGK | • | • | • | • | • | • | |
| ALK | 1096* | TSTIMTDYNPNyCFAGK | • | • | • | • | • | • | |
| ALK | 1131* | GLGHGAFGEVyEGQVSGMPNDPSPLQVAVK | • | • | • | • | |||
| ALK | 1278* | DIyRASYYR | • | • | • | • | • | • | |
| ALK | 1282* | DIyRASyYR | • | • | • | • | • | • | |
| ALK | 1283* | DIyRASyyR | • | • | • | • | • | • | |
| ALK | 1507* | NKPTSLWNPTyGSWFTEKPTK | • | • | • | • | • | • | |
| ALK | 1584* | HFPCGNVNyGYQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | |||
| ALK | 1586* | HFPCGNVNYGyQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | • | • | |
| ALK | 1604* | HFPCGNVNYGyQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | • | • | |
| CDC2 | 15 | IEKIGEGTyGVVYK | • | • | • | • | • | • | |
| CDK3 | 15 | VEKIGEGTyGVVYK | • | • | • | • | • | • | • |
| DYRK1A | 321 | IYQyIQSR | • | • | • | • | • | • | • |
| GSK3B | 279 | GEPNVSyICSR | • | • | • | • | • | • | • |
| HIPK1 | 352 | AVCSTyLQSR | • | • | • | • | • | • | |
| MAPK1 | 187 | VADPDHDHTGFLTEyVATR | • | • | • | • | • | • | |
| PGK1 | 195 | ELNyFAK, KELNyFAK | • | • | • | • | • | ||
| PIP5K2B | 93 | FKEyCPMVFR | • | • | • | • | |||
| PRPF4B | 849 | LCDFGSASHVADNDITPyLVSR | • | • | • | • | • | • | • |
| Metabolism | |||||||||
| ACLY | 682 | TTDGVyEGVAIGGDRYPGSTFMDHVLR | • | • | • | • | • | • | |
| ATIC | 104 | VVACNLyPFVK | • | • | • | • | • | • | |
| DCP1B | 110 | LSIyGIWFYDKEECQR | • | • | • | • | |||
| EIF3S7 | 318 | NLAMEATyINHNFSQQCLR | • | • | • | • | |||
| EIF3S9 | 525 | NGDyLCVK | • | • | • | • | • | ||
| ELP3 | 202 | NLHDALSGHTSNNIyEAVK | • | • | • | • | • | ||
| ENO1 | 43 | AAVPSGASTGIyEALELR | • | • | • | • | • | • | • |
| ENO1 | 286 | SFIKDyPVVSIEDPFDQDDWGAWQK | • | • | • | • | |||
| HIST1H4I | 51 | ISGLIyEETR | • | • | • | • | |||
| HNRPF | 306 | ATENDIyNFFSPLNPVR | • | • | • | • | |||
| HSPCB | 483 | SIyYITGESKEQVANSAFVER | • | • | • | • | • | • | |
| LDHA | 238 | QVVESAyEVIK | • | • | • | • | • | • | • |
| LDHB | 239 | MVVESAyEVIK | • | • | • | • | • | • | • |
| MDH2 | 56 | LTLyDIAHTPGVAADLSHIETK | • | • | • | • | |||
| NIT2 | 145 | TLSPGDSFSTFDTPyCR | • | • | • | • | • | • | |
| NYREN18 | 126 | SKIAETFGLQENyIK | • | • | • | • | |||
| PABPC1 | 54 | SLGyAYVNFQQPADAER | • | • | • | • | • | • | |
| PBEF1 | 188 | YLLETSGNLDGLEyKLHDFGYR | • | • | • | • | • | ||
| PKM2 | 104 | TATESFASDPILyRPVAVALDTKGPEIR | • | • | • | • | • | • | • |
| PRIM2A | 381 | IILSNPPSQGDyHGCPFR | • | • | • | • | • | ||
| PSMA2 | 23 | LVQIEyALAAVAGGAPSVGIK | • | • | • | • | • | ||
| PSMA2 | 97 | KLAQQYyLVYQEPIPTAQLVQR | • | • | • | • | • | ||
| PSMB4 | 102 | VNNSTMLGASGDyADFQYLK | • | • | • | • | • | ||
| RNPS1 | 205 | GyAYVEFENPDEAEK | • | • | • | • | |||
| RPL18A | 63 | SSGEIVyCGQVFEK | • | • | • | • | • | • | |
| RPL31 | 103 | NEDEDSPNKLyTLVTYVPVTTFK | • | • | • | • | • | • | |
| RPL7 | 139 | IVEPyIAWGYPNLK | • | • | • | • | • | ||
| RPLP0 | 24 | IIQLLDDyPKCFIVGADNVGSK | • | • | • | • | • | • | • |
| RPS13 | 37 | LTSDDVKEQIyK | • | • | • | • | |||
| TXNRD1 | 11 | SyDYDLIIIGGGSGGLAAAK | • | • | • | • | |||
| TXNRD1 | 13 | SYDyDLIIIGGGSGGLAAAK | • | • | • | • | • | • | |
| WDR54 | 33 | NLTyFGVVHGPSAQLLSAAPEGVPLAQR | • | • | • | • | |||
| G-proteins | |||||||||
| GDI2 | 203 | TDDYLDQPCyETINR | • | • | • | • | • | ||
| RASA1 | 615 | HFTNPyCNIYLNSVQVAK | • | • | • | • | • | • | |
| Adaptor proteins | |||||||||
| GRLF1 | 1105 | NEEENIySVPHDSTQGK | • | • | • | ||||
| HSPA4 | 336 | LKKEDIyAVEIVGGATR | • | • | • | • | • | ||
| HSPD1 | 227 | GYISPyFINTSK | • | • | • | • | |||
| IRS1 | 46 | AASEAGGPARLEyYENEKK | • | • | • | ||||
| SHC1 | 427 | ELFDDPSyVNVQNLDK | • | • | • | • | • | • | • |
| TRAP1 | 498 | NIyYLCAPNR | • | • | • | • | • | • | |
| Transcription factors | |||||||||
| EEF1A1 | 141 | EHALLAyTLGVK | • | • | • | • | • | • | |
| STAT3 | 704 | YCRPESQEHPEADPGAAPyLK | • | • | • | • | • | • | • |
| STAT3 | 705 | YCRPESQEHPEADPGSAAPyLK | • | • | • | • | • | • | • |
| Cytoskeleton | |||||||||
| ACTB | 218 | DIKEKLCyVALDFEQEMATAASSSSLEK | • | • | • | • | |||
| DNCH1 | 3379 | NYMSNPSYNyEIVNR | • | • | • | • | • | ||
| STOML2 | 124 | ASYGVEDPEyAVTQLAQTTMR | • | • | • | • | |||
| VASP | 38 | VQIyHNPTANSFR | • | • | • | • | • | ||
| VIM | 116 | TNEKVELQELNDRFANyIDKVR, FANyIDKVR | • | • | • | • | |||
| WDR1 | 238 | AHDGGIyAISWSPDSTHLLSASGDKTSK | • | • | • | • | • |
| Gene symbol . | pTyr site . | Peptide . | Karpas299 . | SU-DHL1 . | JB-6 . | TS . | SR-786 . | SUP-M2 . | Mac-1 . |
|---|---|---|---|---|---|---|---|---|---|
| Kinases | |||||||||
| ALK | 1078* | HQELQAMQMELQSPEyK | • | • | • | • | • | • | |
| ALK | 1092* | TSTIMTDyNPNYCFAGK | • | • | • | • | • | • | |
| ALK | 1096* | TSTIMTDYNPNyCFAGK | • | • | • | • | • | • | |
| ALK | 1131* | GLGHGAFGEVyEGQVSGMPNDPSPLQVAVK | • | • | • | • | |||
| ALK | 1278* | DIyRASYYR | • | • | • | • | • | • | |
| ALK | 1282* | DIyRASyYR | • | • | • | • | • | • | |
| ALK | 1283* | DIyRASyyR | • | • | • | • | • | • | |
| ALK | 1507* | NKPTSLWNPTyGSWFTEKPTK | • | • | • | • | • | • | |
| ALK | 1584* | HFPCGNVNyGYQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | |||
| ALK | 1586* | HFPCGNVNYGyQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | • | • | |
| ALK | 1604* | HFPCGNVNYGyQQQGLPLEAATAPGAGHyEDTILK | • | • | • | • | • | • | |
| CDC2 | 15 | IEKIGEGTyGVVYK | • | • | • | • | • | • | |
| CDK3 | 15 | VEKIGEGTyGVVYK | • | • | • | • | • | • | • |
| DYRK1A | 321 | IYQyIQSR | • | • | • | • | • | • | • |
| GSK3B | 279 | GEPNVSyICSR | • | • | • | • | • | • | • |
| HIPK1 | 352 | AVCSTyLQSR | • | • | • | • | • | • | |
| MAPK1 | 187 | VADPDHDHTGFLTEyVATR | • | • | • | • | • | • | |
| PGK1 | 195 | ELNyFAK, KELNyFAK | • | • | • | • | • | ||
| PIP5K2B | 93 | FKEyCPMVFR | • | • | • | • | |||
| PRPF4B | 849 | LCDFGSASHVADNDITPyLVSR | • | • | • | • | • | • | • |
| Metabolism | |||||||||
| ACLY | 682 | TTDGVyEGVAIGGDRYPGSTFMDHVLR | • | • | • | • | • | • | |
| ATIC | 104 | VVACNLyPFVK | • | • | • | • | • | • | |
| DCP1B | 110 | LSIyGIWFYDKEECQR | • | • | • | • | |||
| EIF3S7 | 318 | NLAMEATyINHNFSQQCLR | • | • | • | • | |||
| EIF3S9 | 525 | NGDyLCVK | • | • | • | • | • | ||
| ELP3 | 202 | NLHDALSGHTSNNIyEAVK | • | • | • | • | • | ||
| ENO1 | 43 | AAVPSGASTGIyEALELR | • | • | • | • | • | • | • |
| ENO1 | 286 | SFIKDyPVVSIEDPFDQDDWGAWQK | • | • | • | • | |||
| HIST1H4I | 51 | ISGLIyEETR | • | • | • | • | |||
| HNRPF | 306 | ATENDIyNFFSPLNPVR | • | • | • | • | |||
| HSPCB | 483 | SIyYITGESKEQVANSAFVER | • | • | • | • | • | • | |
| LDHA | 238 | QVVESAyEVIK | • | • | • | • | • | • | • |
| LDHB | 239 | MVVESAyEVIK | • | • | • | • | • | • | • |
| MDH2 | 56 | LTLyDIAHTPGVAADLSHIETK | • | • | • | • | |||
| NIT2 | 145 | TLSPGDSFSTFDTPyCR | • | • | • | • | • | • | |
| NYREN18 | 126 | SKIAETFGLQENyIK | • | • | • | • | |||
| PABPC1 | 54 | SLGyAYVNFQQPADAER | • | • | • | • | • | • | |
| PBEF1 | 188 | YLLETSGNLDGLEyKLHDFGYR | • | • | • | • | • | ||
| PKM2 | 104 | TATESFASDPILyRPVAVALDTKGPEIR | • | • | • | • | • | • | • |
| PRIM2A | 381 | IILSNPPSQGDyHGCPFR | • | • | • | • | • | ||
| PSMA2 | 23 | LVQIEyALAAVAGGAPSVGIK | • | • | • | • | • | ||
| PSMA2 | 97 | KLAQQYyLVYQEPIPTAQLVQR | • | • | • | • | • | ||
| PSMB4 | 102 | VNNSTMLGASGDyADFQYLK | • | • | • | • | • | ||
| RNPS1 | 205 | GyAYVEFENPDEAEK | • | • | • | • | |||
| RPL18A | 63 | SSGEIVyCGQVFEK | • | • | • | • | • | • | |
| RPL31 | 103 | NEDEDSPNKLyTLVTYVPVTTFK | • | • | • | • | • | • | |
| RPL7 | 139 | IVEPyIAWGYPNLK | • | • | • | • | • | ||
| RPLP0 | 24 | IIQLLDDyPKCFIVGADNVGSK | • | • | • | • | • | • | • |
| RPS13 | 37 | LTSDDVKEQIyK | • | • | • | • | |||
| TXNRD1 | 11 | SyDYDLIIIGGGSGGLAAAK | • | • | • | • | |||
| TXNRD1 | 13 | SYDyDLIIIGGGSGGLAAAK | • | • | • | • | • | • | |
| WDR54 | 33 | NLTyFGVVHGPSAQLLSAAPEGVPLAQR | • | • | • | • | |||
| G-proteins | |||||||||
| GDI2 | 203 | TDDYLDQPCyETINR | • | • | • | • | • | ||
| RASA1 | 615 | HFTNPyCNIYLNSVQVAK | • | • | • | • | • | • | |
| Adaptor proteins | |||||||||
| GRLF1 | 1105 | NEEENIySVPHDSTQGK | • | • | • | ||||
| HSPA4 | 336 | LKKEDIyAVEIVGGATR | • | • | • | • | • | ||
| HSPD1 | 227 | GYISPyFINTSK | • | • | • | • | |||
| IRS1 | 46 | AASEAGGPARLEyYENEKK | • | • | • | ||||
| SHC1 | 427 | ELFDDPSyVNVQNLDK | • | • | • | • | • | • | • |
| TRAP1 | 498 | NIyYLCAPNR | • | • | • | • | • | • | |
| Transcription factors | |||||||||
| EEF1A1 | 141 | EHALLAyTLGVK | • | • | • | • | • | • | |
| STAT3 | 704 | YCRPESQEHPEADPGAAPyLK | • | • | • | • | • | • | • |
| STAT3 | 705 | YCRPESQEHPEADPGSAAPyLK | • | • | • | • | • | • | • |
| Cytoskeleton | |||||||||
| ACTB | 218 | DIKEKLCyVALDFEQEMATAASSSSLEK | • | • | • | • | |||
| DNCH1 | 3379 | NYMSNPSYNyEIVNR | • | • | • | • | • | ||
| STOML2 | 124 | ASYGVEDPEyAVTQLAQTTMR | • | • | • | • | |||
| VASP | 38 | VQIyHNPTANSFR | • | • | • | • | • | ||
| VIM | 116 | TNEKVELQELNDRFANyIDKVR, FANyIDKVR | • | • | • | • | |||
| WDR1 | 238 | AHDGGIyAISWSPDSTHLLSASGDKTSK | • | • | • | • | • |
The indicated ALK pTyr sites refer to the full-length ALK receptor, and they correspond to NPM-ALK pTyr sites 138, 152, 156, 191, 338, 342, 343, 567, 644, 646, and 664, respectively
ALK inhibition defines a common phospho-tyrosine signature
To identify phosphorylated proteins playing a pathogenic role in NPM-ALK-mediated transformation, we analyzed the Tyr-phosphorylation signature of ALK+ ALCL lines after abrogating the expression or the activity of NPM-ALK by RNA interference (RNAi)11 or by a small molecule ALK inhibitor,23 respectively. Using an inducible shRNA and quantitative proteomic approach, we demonstrated that a total of 101 peptides were no longer phosphorylated after ALK silencing, whereas 24 became phosphorylated and 35 were apparently unaffected by ALK RNAi (Table S3).
To exclude off-target effects by RNAi, ALCL cells were treated with a small-molecule ALK inhibitor (CEP14083) or by a control compound (CEP11988).23 These studies revealed a total of 138 phosphopeptides lost after ALK inhibition, whereas 109 new peptides became phosphorylated and 226 were unchanged after ALK inhibition (Table S4), with a good overlap between the RNAi and ALK inhibitor datasets. Nevertheless, small or no variations of well-known ALK-related Tyr-phosphorylated proteins were seen by the single approach or the combined methods (ie, STAT3). Thus, we used a Stable Isotope Labeling of Amino acid in Cell culture approach on TS cells after induction of a specific ALK shRNA. The PhosphoScan technology allowed the identification of variations in the phosphorylation levels of many proteins, including known NPM-ALK downstream targets and novel proteins, undetectable using previous analyses (Table 3). For instance, a significant down-modulation of p-STAT3 signal (13-fold inhibition) was observed after ALK silencing. Moreover, Western blot validation confirmed the abrogation of NPM-ALK–dependent phosphorylation of several downstream targets (Figure 1E). A combined approach may identify a common set of tyrosine-phosphorylated proteins whose status correlates more precisely to ALK activity.
Tyrosine phosphorylated peptides that changed their phosphorylation status while ALK was inhibited by shRNA or a pharmacologic inhibitor
| Gene symbol . | Peptide . | PTyr site . | Protein . | *Average H:L . | NCBI access no. . | shALK . | ALK inhibitor . |
|---|---|---|---|---|---|---|---|
| Peptides that decrease when ALK is reduced | |||||||
| Kinases | |||||||
| ALK | R.TSTIMTDYNPNY*CFAGK | 1096 | Anaplastic lymphoma kinase | 1:6.20 | NP_004295 | 1 | 1 |
| ALK | R.NKPTSLWNPTY*GSWFTEKPTK | 1507 | Anaplastic lymphoma kinase | 1:3.72 | NP_004295 | 2 | 1 |
| CDC2 | K.IEKIGEGTY*GVVYK | 15 | Cell division cycle 2 protein | 1:2.7 | NP_203698 | 2 | 1 |
| CDK2 | K.VEKIGEGTY*GVVYK | 15 | Cyclin dependent kinase 2 | >1:6 | NP_001789 | 2 | 1 |
| ERK1/MAPK3 | R.IADPEHDHTGFLTEY*VATR | 187 | Mitogen-activated protein kinase 3 | 1:2.07 | NP_002737 | Not found | 2 |
| p38/MAPK14 | R.HTDDEMTGY*VATR | 182 | Mitogen-activated protein kinase 14 | 1:3.22 | NP_620581 | 2 | 1 |
| Adaptors | |||||||
| HGS | R.VCEPCY*EQLNR | 216 | HGF regulated tyrosine kinase substrate | >1:4 | NP_004703 | Not found | 1 |
| PAG | K.SGQSLTVPESTY*TSIQGDPQR | 341 | Phosphoprotein associated with glycosphingo | 1.47:1 | NP_060910 | Not found | 3 |
| PARD3 | R.DVTIGGSAPIY*VK | 489 | par-3 partitioning defective 3 homolog | >1:4 | NP_062565 | Not found | 2 |
| Cytoskeletal proteins | |||||||
| ACTB | R.KDLY*ANTVLSGGTTMYPGIADR.M | 294 | Actin, β | 1:5.71 | NP_001092 | 1 | Not found |
| ACTB | K.EKLCY*VALDFEQEMATAASSSSLEK.S | 218 | Actin, β | >1:3 | NP_001092 | 1 | 1 |
| CORO1C | R.YFEITDESPY*VHYLNTFSSK.E | 301 | Coronin, actin binding protein, 1C | 1:2.47 | NP_055140 | 3 | 1 |
| VASP | R.VQIY*HNPTANSFR.V | 39 | Vasodilator-stimulated phosphoprotein | >1:9 | NP_003361 | 1 | 1 |
| VIM | R.FANY*IDKVR.F | 117 | Vimentin | >1:6 | NP_003371 | 2 | 1 |
| Chaperones | |||||||
| HSP90AB1 | K.SIY*YITGESK.E | 484 | Heat shock protein 90kDa α | 1:2.74 | NP_031381 | 4 | 1 |
| HSPA2 | R.TTPSY*VAFTDTER.L | 43 | Heat shock 70kDa protein 2 | >1:2 | NP_068814 | Not found | 1 |
| HSPA4 | K.LKKEDIY*AVEIVGGATR.I | 336 | Heat shock 70kDa protein 4 | 1:2.66 | NP_002145 | 2 | 1 |
| TRAP1 | R.NIY*YLCAPNR.H | 498 | TNF receptor-associated protein 1 | 1:2.9 | NP_057376 | 1 | 1 |
| Cytokines | |||||||
| PBEF | K.YLLETSGNLDGLEY*KLHDFGYR.G | 188 | Pre-B-cell colony enhancing factor 1 | 1:2.04 | NP_005737 | 1 | 1 |
| Energy metabolism | |||||||
| RASA1 | K.HFTNPY*CNIYLNSVQVAK.T | 615 | RAS p21 protein activator 1 | 1:3.25 | NP_072179 | ||
| Metabolic enzymes | |||||||
| ACLY | R.TTDGVY*EGVAIGGDRYPGSTFMDHVLR.Y | 682 | ATP cytrate lyase | 1:4.27 | NP_001087 | 1 | 1 |
| ACP1 | K.QLIIEDPYYGNDSDFETVY*QQCVR.C | 143 | Acidic phosphatase 1 | 1:5.2 | NP_004291 | 2 | 1 |
| ATIC | VVACNLyPFVK | 104 | AICAR formyltransferase/IMP Chase | 1:3.70 | NP_004035 | 1 | 1 |
| ELP3 | R.NLHDALSGHTSNNIY*EAVK.Y | 202 | Elongation protein 3 | 1:5.57 | NP_060561 | 2 | 1 |
| ENO1 | K.SFIKDY*PVVSIEDPFDQDDWGAWQK.F | 287 | Enolase 1 | 1:2.91 | NP_001419 | 1 | 1 |
| GBE1 | R.EGDNVNY*DWIHWDPEHSYEFK.H | 173 | Glucan (1,4-α-), branching enzyme 1 | 1:1.86 | NP_000149 | 3 | 1 |
| LDHA | K.QVVESAY*EVIK.L | 239 | Lactate dehydrogenase A | 1:3.03 | NP_005557 | 2 | 1 |
| LDHB | K.MVVESAY*EVIK.L | 240 | Lactate dehydrogenase B | 1:4.00 | NP_002291 | 1 | 1 |
| NIT2 | K.TLSPGDSFSTFDTPY*CR.V | 145 | Nitrilase family, member 2 | 1:4.14 | NP_064587 | 1 | 1 |
| NUDT5 | R.TLHY*ECIVLVK.Q | 74 | Nudix-type motif 5 | 1:10.3 | NP_054861 | 2 | 1 |
| PFKFB3 | R.ISCY*EASYQPLDPDKCDR.D | 194 | 6-Phosphofructo-2-kinase | >1:8 | NP_004557 | Not found | 3 |
| PKM2 | R.TATESFASDPILY*RPVAVALDTKGPEIR.T | 105 | Pyruvate kinase 3 | 1:4.56 | NP_002645 | 1 | 1 |
| SUCLA2 | K.SPDEAY*AIAK.K | 84 | succinate-CoA ligase, ADP-forming, β sub | >1:2.64 | NP_003841 | Not found | 1 |
| Metabolism of nucleic acids | |||||||
| HIST1H4I | R.ISGLIY*EETR.G | 52 | H4 histone family | 1:2.45 | NP_068803 | 4 | 1 |
| HNRPF | K.ATENDIY*NFFSPLNPVR.V | 306 | Heterogeneous nuclear ribonucleoprotein F | >1:3 | NP_004957 | Not found | 2 |
| MKI67IP | R.TGNSKGY*AFVEFESEDVAK.I | 88 | FHA domain interacting nucleolar phosphopro | 1:2.8 | NP_115766 | Not found | 1 |
| PABP 1 | R.SLGY*AYVNFQQPADAER.A | 56 | Poly A binding protein, cytoplasmic 4 | >1:8 | NP_003810 | 2 | 1 |
| POLR2A | R.LTHVY*DLCK.G | 145 | DNA directed RNA pol. II polyp. A | 1:3.58 | NP_000928 | 2 | 1 |
| RNPS1 | K.GY*AYVEFENPDEAEK.A | 205 | RNA binding prot S1, serine-rich domain | >1:4 | NP_542161 | 4 | 1 |
| SFPQ | R.FAQHGTFEY*EYSQR.W | 488 | Splicing factor proline/glutamine rich (polypyri | 1:3.63 | NP_005057 | 2 | 3 |
| SYNCRIP | K.LKDY*AFIHFDERDGAVK.A | 373 | Synaptotagmin binding, cytoplasmic RNA inte | 1:3.88 | NP_006363 | Not found | 2 |
| Regulation of translation | |||||||
| PHF19 | K.LTEGQY*VLCR.W | 45 | PHD finger protein 19 | 1:3.5 | NP_082992 | Not found | 2 |
| PHB2 | R.IPWFQY*PIIYDIR.A | 77 | Prohibitin 2 | 1:3.43 | NP_009204 | Not found | 1 |
| PHB2 | R.LGLDY*EER.V | 128 | Prohibitin 2 | 1:1.93 | NP_009204 | Not found | 3 |
| EEF1A1 | K.STTTGHLIY*K.C | 29 | Eukaryotic elongation factor 1 α 1 | >1:3 | NP_001393 | Not found | 3 |
| EIF3S2 | K.SYSSGGEDGY*VR.I | 308 | Eukaryotic translation initiation factor 3 | >1:13 | NP_003748 | 2 | 1 |
| EIF3S9 | K.NGDY*LCVK.V | 525 | Eukaryotic translation initiation factor 3, subun | 1:3.75 | NP_003742 | 2 | 1 |
| EIF3S7 | R.NLAMEATY*INHNFSQQCLR.M | 318 | Eukaryotic translation initiation factor 3,subun | 1:2.68 | NP_003744 | 4 | 1 |
| KNTC2 | R.AQVY*VPLKELLNETEEEINK.A | 458 | Kinetochore associated 2 | >1:5 | NP_006092 | Not found | 1 |
| Ribosomal proteins | |||||||
| RPL18A | K.SSGEIVY*CGQVFEK.S | 63 | Ribosomal protein L18a | 1:3.16 | NP_000971 | 1 | 1 |
| RPL31 | R.NEDEDSPNKLY*TLVTYVPVTTFK.N | 103 | Ribosomal protein L31 | 1:2.87 | NP_000984 | 1 | 1 |
| RPL7 | R.IVEPY*IAWGYPNLK.S | 139 | Ribosomal protein L7 | 1:2.10 | NP_000962 | 2 | 1 |
| RPL8 | R.ASGNY*ATVISHNPETK.K | 133 | Ribosomal protein L8 | 1:3.15 | NP_000964 | 1 | 1 |
| RPLP0 | K.IIQLLDDY*PK.C | 24 | Ribosomal protein P0 | 1:5.23 | NP_000993 | 4 | 1 |
| RPS10 | R.IAIY*ELLFK.E | 12 | Ribosomal protein S10 | 1:2.52 | NP_001005 | Not found | 1 |
| RPS13 | K.LTSDDVKEQIY*K.L | 38 | Ribosomal protein S13 | 1:1.8 | NP_001008 | Not found | 1 |
| Transcription factors | |||||||
| DCP1A | R.SASPY*HGFTIVNR.L | 64 | DCP1 decapping enzyme homolog A | 1:2.24 | NP_060873 | Not found | 1 |
| STAT3 iso1 | K.YCRPESQEHPEADPGSAAPY*LK.T | 705 | Signal transducer and activator of transcriptio | 1:13.23 | NP_644805 | 1 | 1 |
| STAT3 iso2 | K.YCRPESQEHPEADPGAAPY*LK.T | 704 | Signal transducer and activator of transcriptio | 1:13.57 | NP_003141 | 4 | 1 |
| Ubiquitin-proteasome system | |||||||
| PSMA2 | K.LVQIEY*ALAAVAGGAPSVGIK.A | 98 | Proteasome subunit α type 2 | 1:4.37 | NP_002778 | 2 | 1 |
| PSMA2 | R.KLAQQYY*LVYQEPIPTAQLVQR.V | 24 | Proteasome subunit α type 2 | 1:3.18 | NP_002778 | 2 | 1 |
| Others | |||||||
| ANXA2 | K.SLY*YYIQQDTK.G | 316 | Annexin A2 | 1:1.91 | NP_004030 | 2 | 1 |
| GSDMDC1 | R.SRGDNVY*VVTEVLQTQK.E | 71 | Gasdermin domain containing 1 | 1:3.27 | NP_079012 | Not found | 1 |
| FAM62B | R.NLIAFSEDGSDPY*VR.M | 796 | Family with sequence similarity 62 member B | >1:3 | NP_065779 | 2 | 3 |
| WDR1 | K.AHDGGIY*AISWSPDSTHLLSASGDK.T | 98 | WD repeat-containing protein 1 | 1:3.58 | NP_005103 | 2 | 1 |
| Peptides with no significant change | |||||||
| Kinases | |||||||
| DYRK1A | R.KVYNDGYDDDNY*DYIVK.N | 145 | Dual-specificity tyr. phosphorylation regulated | 1:1.15 | NP_001387 | 2 | 1 |
| DYRK1A | R.IYQY*IQSR.F | 321 | Dual-specificity tyr. phosphorylation regulated | 1:1.36 | NP_001387 | 1 | 1 |
| GSK3 | R.GEPNVSY*ICSR.Y | 216 | Glycogen synthase kinase 3 α | 1:1.15 | NP_063937 | 1 | 1 |
| HIPK1 | K.AVCSTY*LQSR.Y | 352 | Homeodomain interacting protein kinase 1 | 1:1.15 | NP_689909 | 2 | 1 |
| HIPK3 | K.TVCSTY*LQSR.Y | 359 | Homeodomain interacting protein kinase 3 | 1:0.99 | NP_005725 | 3 | 1 |
| PRPF4B | K.LCDFGSASHVADNDITPY*LVSR.F | 849 | Serine/threonine-protein kinase PRP4K | 1:1.02 | NP_003904 | 1 | 1 |
| SRC | R.LIEDNEY*TAR.Q | 419 (416) | v-src sarcoma viral Oncogene homolog | 1:1.10 | NP_005408 | Not found | Not found |
| Adaptors | |||||||
| SHC | R.ELFDDPSY*VNVQNLDK.A | 318 | SHC 1 | 1:1.44 | NP_003020 | 1 | 1 |
| Chaperones | |||||||
| HSPD1 | R.GYISPY*FINTSK.G | 227 | Heat shock 60 kDa protein 1 (chaperonin) | 1:1.35 | NP_955472 | Not found | 1 |
| HSP90AA1 | K.HIY*YITGETK.D | 492 | Heat shock 90 kDa protein | 1:1.18 | NP_005339 | Not found | 1 |
| Cytokines | |||||||
| PBEF | K.VY*SYFECR.E | 34 | Pre-B-cell colony enhancing factor 1 | 1:1.02 | NP_005737 | 2 | 1 |
| Energy metabolism | |||||||
| GDI2 | R.TDDYLDQPCY*ETINR.I | 203 | GDP dissociation inhibitor 2 | 1:1.46 | NP_001485 | 2 | 1 |
| RAN | K.SNY*NFEKPFLWLAR.K | 155 | ras-related nuclear protein | 1:1.12 | NP_006316 | Not found | 1 |
| WRNIP1 | R.MLEGGEDPLY*VAR.R | 534 | Werner helicase interacting protein 1 | 1:1.20 | NP_064520 | 2 | 1 |
| Gene symbol . | Peptide . | PTyr site . | Protein . | *Average H:L . | NCBI access no. . | shALK . | ALK inhibitor . |
|---|---|---|---|---|---|---|---|
| Peptides that decrease when ALK is reduced | |||||||
| Kinases | |||||||
| ALK | R.TSTIMTDYNPNY*CFAGK | 1096 | Anaplastic lymphoma kinase | 1:6.20 | NP_004295 | 1 | 1 |
| ALK | R.NKPTSLWNPTY*GSWFTEKPTK | 1507 | Anaplastic lymphoma kinase | 1:3.72 | NP_004295 | 2 | 1 |
| CDC2 | K.IEKIGEGTY*GVVYK | 15 | Cell division cycle 2 protein | 1:2.7 | NP_203698 | 2 | 1 |
| CDK2 | K.VEKIGEGTY*GVVYK | 15 | Cyclin dependent kinase 2 | >1:6 | NP_001789 | 2 | 1 |
| ERK1/MAPK3 | R.IADPEHDHTGFLTEY*VATR | 187 | Mitogen-activated protein kinase 3 | 1:2.07 | NP_002737 | Not found | 2 |
| p38/MAPK14 | R.HTDDEMTGY*VATR | 182 | Mitogen-activated protein kinase 14 | 1:3.22 | NP_620581 | 2 | 1 |
| Adaptors | |||||||
| HGS | R.VCEPCY*EQLNR | 216 | HGF regulated tyrosine kinase substrate | >1:4 | NP_004703 | Not found | 1 |
| PAG | K.SGQSLTVPESTY*TSIQGDPQR | 341 | Phosphoprotein associated with glycosphingo | 1.47:1 | NP_060910 | Not found | 3 |
| PARD3 | R.DVTIGGSAPIY*VK | 489 | par-3 partitioning defective 3 homolog | >1:4 | NP_062565 | Not found | 2 |
| Cytoskeletal proteins | |||||||
| ACTB | R.KDLY*ANTVLSGGTTMYPGIADR.M | 294 | Actin, β | 1:5.71 | NP_001092 | 1 | Not found |
| ACTB | K.EKLCY*VALDFEQEMATAASSSSLEK.S | 218 | Actin, β | >1:3 | NP_001092 | 1 | 1 |
| CORO1C | R.YFEITDESPY*VHYLNTFSSK.E | 301 | Coronin, actin binding protein, 1C | 1:2.47 | NP_055140 | 3 | 1 |
| VASP | R.VQIY*HNPTANSFR.V | 39 | Vasodilator-stimulated phosphoprotein | >1:9 | NP_003361 | 1 | 1 |
| VIM | R.FANY*IDKVR.F | 117 | Vimentin | >1:6 | NP_003371 | 2 | 1 |
| Chaperones | |||||||
| HSP90AB1 | K.SIY*YITGESK.E | 484 | Heat shock protein 90kDa α | 1:2.74 | NP_031381 | 4 | 1 |
| HSPA2 | R.TTPSY*VAFTDTER.L | 43 | Heat shock 70kDa protein 2 | >1:2 | NP_068814 | Not found | 1 |
| HSPA4 | K.LKKEDIY*AVEIVGGATR.I | 336 | Heat shock 70kDa protein 4 | 1:2.66 | NP_002145 | 2 | 1 |
| TRAP1 | R.NIY*YLCAPNR.H | 498 | TNF receptor-associated protein 1 | 1:2.9 | NP_057376 | 1 | 1 |
| Cytokines | |||||||
| PBEF | K.YLLETSGNLDGLEY*KLHDFGYR.G | 188 | Pre-B-cell colony enhancing factor 1 | 1:2.04 | NP_005737 | 1 | 1 |
| Energy metabolism | |||||||
| RASA1 | K.HFTNPY*CNIYLNSVQVAK.T | 615 | RAS p21 protein activator 1 | 1:3.25 | NP_072179 | ||
| Metabolic enzymes | |||||||
| ACLY | R.TTDGVY*EGVAIGGDRYPGSTFMDHVLR.Y | 682 | ATP cytrate lyase | 1:4.27 | NP_001087 | 1 | 1 |
| ACP1 | K.QLIIEDPYYGNDSDFETVY*QQCVR.C | 143 | Acidic phosphatase 1 | 1:5.2 | NP_004291 | 2 | 1 |
| ATIC | VVACNLyPFVK | 104 | AICAR formyltransferase/IMP Chase | 1:3.70 | NP_004035 | 1 | 1 |
| ELP3 | R.NLHDALSGHTSNNIY*EAVK.Y | 202 | Elongation protein 3 | 1:5.57 | NP_060561 | 2 | 1 |
| ENO1 | K.SFIKDY*PVVSIEDPFDQDDWGAWQK.F | 287 | Enolase 1 | 1:2.91 | NP_001419 | 1 | 1 |
| GBE1 | R.EGDNVNY*DWIHWDPEHSYEFK.H | 173 | Glucan (1,4-α-), branching enzyme 1 | 1:1.86 | NP_000149 | 3 | 1 |
| LDHA | K.QVVESAY*EVIK.L | 239 | Lactate dehydrogenase A | 1:3.03 | NP_005557 | 2 | 1 |
| LDHB | K.MVVESAY*EVIK.L | 240 | Lactate dehydrogenase B | 1:4.00 | NP_002291 | 1 | 1 |
| NIT2 | K.TLSPGDSFSTFDTPY*CR.V | 145 | Nitrilase family, member 2 | 1:4.14 | NP_064587 | 1 | 1 |
| NUDT5 | R.TLHY*ECIVLVK.Q | 74 | Nudix-type motif 5 | 1:10.3 | NP_054861 | 2 | 1 |
| PFKFB3 | R.ISCY*EASYQPLDPDKCDR.D | 194 | 6-Phosphofructo-2-kinase | >1:8 | NP_004557 | Not found | 3 |
| PKM2 | R.TATESFASDPILY*RPVAVALDTKGPEIR.T | 105 | Pyruvate kinase 3 | 1:4.56 | NP_002645 | 1 | 1 |
| SUCLA2 | K.SPDEAY*AIAK.K | 84 | succinate-CoA ligase, ADP-forming, β sub | >1:2.64 | NP_003841 | Not found | 1 |
| Metabolism of nucleic acids | |||||||
| HIST1H4I | R.ISGLIY*EETR.G | 52 | H4 histone family | 1:2.45 | NP_068803 | 4 | 1 |
| HNRPF | K.ATENDIY*NFFSPLNPVR.V | 306 | Heterogeneous nuclear ribonucleoprotein F | >1:3 | NP_004957 | Not found | 2 |
| MKI67IP | R.TGNSKGY*AFVEFESEDVAK.I | 88 | FHA domain interacting nucleolar phosphopro | 1:2.8 | NP_115766 | Not found | 1 |
| PABP 1 | R.SLGY*AYVNFQQPADAER.A | 56 | Poly A binding protein, cytoplasmic 4 | >1:8 | NP_003810 | 2 | 1 |
| POLR2A | R.LTHVY*DLCK.G | 145 | DNA directed RNA pol. II polyp. A | 1:3.58 | NP_000928 | 2 | 1 |
| RNPS1 | K.GY*AYVEFENPDEAEK.A | 205 | RNA binding prot S1, serine-rich domain | >1:4 | NP_542161 | 4 | 1 |
| SFPQ | R.FAQHGTFEY*EYSQR.W | 488 | Splicing factor proline/glutamine rich (polypyri | 1:3.63 | NP_005057 | 2 | 3 |
| SYNCRIP | K.LKDY*AFIHFDERDGAVK.A | 373 | Synaptotagmin binding, cytoplasmic RNA inte | 1:3.88 | NP_006363 | Not found | 2 |
| Regulation of translation | |||||||
| PHF19 | K.LTEGQY*VLCR.W | 45 | PHD finger protein 19 | 1:3.5 | NP_082992 | Not found | 2 |
| PHB2 | R.IPWFQY*PIIYDIR.A | 77 | Prohibitin 2 | 1:3.43 | NP_009204 | Not found | 1 |
| PHB2 | R.LGLDY*EER.V | 128 | Prohibitin 2 | 1:1.93 | NP_009204 | Not found | 3 |
| EEF1A1 | K.STTTGHLIY*K.C | 29 | Eukaryotic elongation factor 1 α 1 | >1:3 | NP_001393 | Not found | 3 |
| EIF3S2 | K.SYSSGGEDGY*VR.I | 308 | Eukaryotic translation initiation factor 3 | >1:13 | NP_003748 | 2 | 1 |
| EIF3S9 | K.NGDY*LCVK.V | 525 | Eukaryotic translation initiation factor 3, subun | 1:3.75 | NP_003742 | 2 | 1 |
| EIF3S7 | R.NLAMEATY*INHNFSQQCLR.M | 318 | Eukaryotic translation initiation factor 3,subun | 1:2.68 | NP_003744 | 4 | 1 |
| KNTC2 | R.AQVY*VPLKELLNETEEEINK.A | 458 | Kinetochore associated 2 | >1:5 | NP_006092 | Not found | 1 |
| Ribosomal proteins | |||||||
| RPL18A | K.SSGEIVY*CGQVFEK.S | 63 | Ribosomal protein L18a | 1:3.16 | NP_000971 | 1 | 1 |
| RPL31 | R.NEDEDSPNKLY*TLVTYVPVTTFK.N | 103 | Ribosomal protein L31 | 1:2.87 | NP_000984 | 1 | 1 |
| RPL7 | R.IVEPY*IAWGYPNLK.S | 139 | Ribosomal protein L7 | 1:2.10 | NP_000962 | 2 | 1 |
| RPL8 | R.ASGNY*ATVISHNPETK.K | 133 | Ribosomal protein L8 | 1:3.15 | NP_000964 | 1 | 1 |
| RPLP0 | K.IIQLLDDY*PK.C | 24 | Ribosomal protein P0 | 1:5.23 | NP_000993 | 4 | 1 |
| RPS10 | R.IAIY*ELLFK.E | 12 | Ribosomal protein S10 | 1:2.52 | NP_001005 | Not found | 1 |
| RPS13 | K.LTSDDVKEQIY*K.L | 38 | Ribosomal protein S13 | 1:1.8 | NP_001008 | Not found | 1 |
| Transcription factors | |||||||
| DCP1A | R.SASPY*HGFTIVNR.L | 64 | DCP1 decapping enzyme homolog A | 1:2.24 | NP_060873 | Not found | 1 |
| STAT3 iso1 | K.YCRPESQEHPEADPGSAAPY*LK.T | 705 | Signal transducer and activator of transcriptio | 1:13.23 | NP_644805 | 1 | 1 |
| STAT3 iso2 | K.YCRPESQEHPEADPGAAPY*LK.T | 704 | Signal transducer and activator of transcriptio | 1:13.57 | NP_003141 | 4 | 1 |
| Ubiquitin-proteasome system | |||||||
| PSMA2 | K.LVQIEY*ALAAVAGGAPSVGIK.A | 98 | Proteasome subunit α type 2 | 1:4.37 | NP_002778 | 2 | 1 |
| PSMA2 | R.KLAQQYY*LVYQEPIPTAQLVQR.V | 24 | Proteasome subunit α type 2 | 1:3.18 | NP_002778 | 2 | 1 |
| Others | |||||||
| ANXA2 | K.SLY*YYIQQDTK.G | 316 | Annexin A2 | 1:1.91 | NP_004030 | 2 | 1 |
| GSDMDC1 | R.SRGDNVY*VVTEVLQTQK.E | 71 | Gasdermin domain containing 1 | 1:3.27 | NP_079012 | Not found | 1 |
| FAM62B | R.NLIAFSEDGSDPY*VR.M | 796 | Family with sequence similarity 62 member B | >1:3 | NP_065779 | 2 | 3 |
| WDR1 | K.AHDGGIY*AISWSPDSTHLLSASGDK.T | 98 | WD repeat-containing protein 1 | 1:3.58 | NP_005103 | 2 | 1 |
| Peptides with no significant change | |||||||
| Kinases | |||||||
| DYRK1A | R.KVYNDGYDDDNY*DYIVK.N | 145 | Dual-specificity tyr. phosphorylation regulated | 1:1.15 | NP_001387 | 2 | 1 |
| DYRK1A | R.IYQY*IQSR.F | 321 | Dual-specificity tyr. phosphorylation regulated | 1:1.36 | NP_001387 | 1 | 1 |
| GSK3 | R.GEPNVSY*ICSR.Y | 216 | Glycogen synthase kinase 3 α | 1:1.15 | NP_063937 | 1 | 1 |
| HIPK1 | K.AVCSTY*LQSR.Y | 352 | Homeodomain interacting protein kinase 1 | 1:1.15 | NP_689909 | 2 | 1 |
| HIPK3 | K.TVCSTY*LQSR.Y | 359 | Homeodomain interacting protein kinase 3 | 1:0.99 | NP_005725 | 3 | 1 |
| PRPF4B | K.LCDFGSASHVADNDITPY*LVSR.F | 849 | Serine/threonine-protein kinase PRP4K | 1:1.02 | NP_003904 | 1 | 1 |
| SRC | R.LIEDNEY*TAR.Q | 419 (416) | v-src sarcoma viral Oncogene homolog | 1:1.10 | NP_005408 | Not found | Not found |
| Adaptors | |||||||
| SHC | R.ELFDDPSY*VNVQNLDK.A | 318 | SHC 1 | 1:1.44 | NP_003020 | 1 | 1 |
| Chaperones | |||||||
| HSPD1 | R.GYISPY*FINTSK.G | 227 | Heat shock 60 kDa protein 1 (chaperonin) | 1:1.35 | NP_955472 | Not found | 1 |
| HSP90AA1 | K.HIY*YITGETK.D | 492 | Heat shock 90 kDa protein | 1:1.18 | NP_005339 | Not found | 1 |
| Cytokines | |||||||
| PBEF | K.VY*SYFECR.E | 34 | Pre-B-cell colony enhancing factor 1 | 1:1.02 | NP_005737 | 2 | 1 |
| Energy metabolism | |||||||
| GDI2 | R.TDDYLDQPCY*ETINR.I | 203 | GDP dissociation inhibitor 2 | 1:1.46 | NP_001485 | 2 | 1 |
| RAN | K.SNY*NFEKPFLWLAR.K | 155 | ras-related nuclear protein | 1:1.12 | NP_006316 | Not found | 1 |
| WRNIP1 | R.MLEGGEDPLY*VAR.R | 534 | Werner helicase interacting protein 1 | 1:1.20 | NP_064520 | 2 | 1 |
Average H:L indicates SILAC quantification performed on samples treated with ALK shRNA.
1Phosphopeptides that are present both in CEP11988 and CEP14083, in shALK− and shALK+.
2Phosphopeptides that are present in CEP11988 and in shALK−, but not in CEP14083 and shALK+.
3Phosphopeptides that are present in CEP14083 and shALK+, but not in CEP11988 and in shALK−.
4Phosphopeptides detected twice with ambiguous results.
NPM-ALK induces ATIC and VASP phosphorylation
To validate new ALK interactors, we selected 2 biologically relevant proteins: ATIC and VASP. ATIC is an enzyme involved in purine biosynthesis and VASP is a protein regulating actin polymerization and cytoskeletal reorganization.3,34 The MS/MS spectra of both phosphopeptides displayed an Xcorr value larger than 2.3 and a well-represented proline peak, suggesting a correct assignment35 (Figure 2A,B). Phosphorylated ATIC and VASP were confirmed by immunoprecipitation and Western blot in all NPM-ALK–positive lines. Notably, ATIC Tyr-phosphorylation was also detectable in control lines (CEM and Jurkat), suggesting that ATIC might be asubstrate of other kinases (Figure 2C). In a subsequent survey of 243 cell lines, we found that p-ATIC (Tyr 104) was detectable in all ALK+ ALCL, and in 1 of 3 non–small cell lung cancer lines carrying p-ALK. Among ALK− nonlymphoid samples, p-ATIC was documented in 5 of 237 lines expressing activated kinases (ABL, EGFR, and PDGFR) and 7 of 148 tumor samples, some of which displayed detectable p-Met.18,36
Tyrosine phosphopeptides are identified in ALK+ ALCL cells. (A) MS/MS spectrum of tyrosine-phosphorylated peptide VVACNLYPFVK, assigned to the protein ATIC (percentage coverage, 1.8% of total sequence). (B) MS/MS spectrum of tyrosine-phosphorylated peptide VQIYHNPTANSFR, assigned to the protein VASP (percentage coverage, 3.4% of total sequence). (C) Total protein expressions (top panel) and phosphorylation status (bottom panel) as assayed by Western blot with the indicated antibodies.
Tyrosine phosphopeptides are identified in ALK+ ALCL cells. (A) MS/MS spectrum of tyrosine-phosphorylated peptide VVACNLYPFVK, assigned to the protein ATIC (percentage coverage, 1.8% of total sequence). (B) MS/MS spectrum of tyrosine-phosphorylated peptide VQIYHNPTANSFR, assigned to the protein VASP (percentage coverage, 3.4% of total sequence). (C) Total protein expressions (top panel) and phosphorylation status (bottom panel) as assayed by Western blot with the indicated antibodies.
To assess whether Tyr-phosphorylation of ATIC and VASP was regulated by the NPM-ALK, both proteins were immunoprecipitated and analyzed by Western blotting. Figure 3A shows that the down-regulation of NPM-ALK in ALCL cell lines by RNAi coincided with a significant decrease of p-ATIC and p-VASP, whereas their expression remained unaffected. Similarly, treatment of cells with the ALK inhibitor, CEP14083, but not the control CEP11988, led to a significant decrease of p-ATIC and p-VASP (Figure 3B).
ATIC and VASP phosphorylation is dependent on NPM-ALK kinase activity. (A) Phospho-tyrosine containing proteins were first immunoprecipitated (IP) with a specific anti (pTyr) antibody and subsequently blotted with the indicated antibodies. (B) Lysates from ALK+ ALCL cell lines (TS and SU-DHL1), treated with small-molecule ALK inhibitor (CEP14083) or a control compound (CEP11988), were immunoprecipitated and blotted with the indicated antibodies. (C) Total lysates from doxycycline-treated TS TTA A5 cells (1 mg/mL for 84 and 96 hours) were IP with a specific anti-ATIC or anti-VASP antibody and blotted with anti-ALK antibody. (D) Total proteins from TS cells (300 nM of CEP14083 or CEP11988) were IP with a specific anti-ATIC or anti-VASP antibody and blotted with anti-ALK antibody.
ATIC and VASP phosphorylation is dependent on NPM-ALK kinase activity. (A) Phospho-tyrosine containing proteins were first immunoprecipitated (IP) with a specific anti (pTyr) antibody and subsequently blotted with the indicated antibodies. (B) Lysates from ALK+ ALCL cell lines (TS and SU-DHL1), treated with small-molecule ALK inhibitor (CEP14083) or a control compound (CEP11988), were immunoprecipitated and blotted with the indicated antibodies. (C) Total lysates from doxycycline-treated TS TTA A5 cells (1 mg/mL for 84 and 96 hours) were IP with a specific anti-ATIC or anti-VASP antibody and blotted with anti-ALK antibody. (D) Total proteins from TS cells (300 nM of CEP14083 or CEP11988) were IP with a specific anti-ATIC or anti-VASP antibody and blotted with anti-ALK antibody.
Having determined that ATIC and VASP phosphorylation is mediated by ALK, we subsequently investigated their physical association with NPM-ALK. ATIC and VASP coprecipitated with NPM-ALK, and this interaction was largely lost when the expression of NPM-ALK was down-regulated via ALK RNAi (Figure 3C) or, alternatively, when its kinase activity was pharmacologically repressed (Figure 3D).
NPM-ALK enhances ATIC enzymatic activity
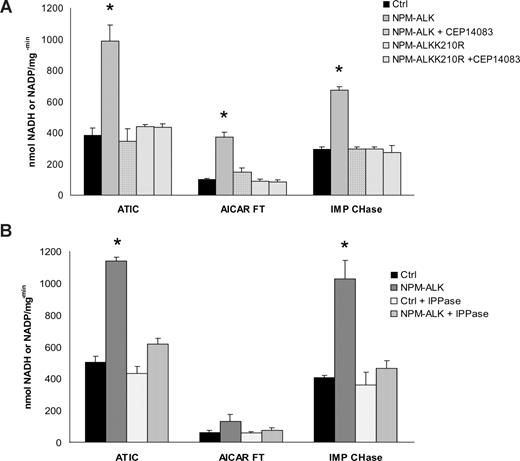
To test whether the Tyr-phosphorylation of ATIC modulates its enzymatic activity, we set 3 different assays, determining the transformylase (AICAR-FT), cyclohydrolase (IMP-CHase), and the total ATIC activity. NPM-ALK was enriched by immunoprecipitation using NPM-ALK HEK-293T-Rex cells and then combined in vitro with wild-type-ATIC from HEK-293T. Figure 4A shows that ATIC activity was enhanced in the presence of NPM-ALK, whereas it remained at the basal level when NPM-ALK was inhibited or when an inactive NPM-ALK (NPM-ALKK210R) was used. The finding that ATIC activity was enhanced by its phosphorylation was further demonstrated using lambda-phosphatase capable to dephosphorylate p-ALK species (Figure 4B).
ATIC activity is enhanced in the presence of NPM-ALK in vitro. (A) Immunoprecipitated ATIC was combined with the active NPM-ALK kinase (isolated from wild-type HEK-293T cells and NPM-ALK+ HEK-293T cells, respectively) or with a mutated NPM-ALKK210R kinase (from NPM-ALKK210R–positive HEK-293T cells), with and without the ALK inhibitor CEP14083 (6 hours). Total or specific ATIC activity was measured as described in “Measurement of AICAR-FT/IMP-CHase activity.” (B) ATIC enzymatic activities were also determined in the presence of lambda phosphatase (1 mg for 1 hour). *P < .001.
ATIC activity is enhanced in the presence of NPM-ALK in vitro. (A) Immunoprecipitated ATIC was combined with the active NPM-ALK kinase (isolated from wild-type HEK-293T cells and NPM-ALK+ HEK-293T cells, respectively) or with a mutated NPM-ALKK210R kinase (from NPM-ALKK210R–positive HEK-293T cells), with and without the ALK inhibitor CEP14083 (6 hours). Total or specific ATIC activity was measured as described in “Measurement of AICAR-FT/IMP-CHase activity.” (B) ATIC enzymatic activities were also determined in the presence of lambda phosphatase (1 mg for 1 hour). *P < .001.
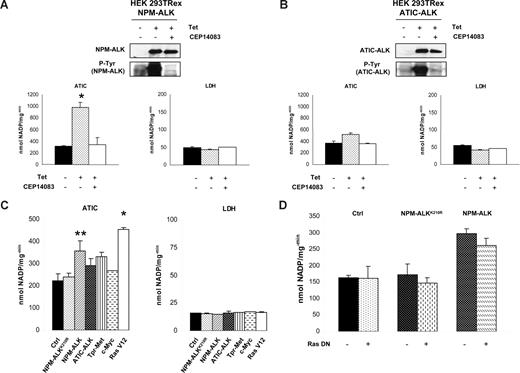
To exclude that the increased activity of ATIC could be the result of the neoplastic phenotype, we measured in parallel the activity of other metabolic enzymes (LDH, G6PD, 6PGD, and ODC). LDH was selected as a control because it was highly phosphorylated in NPM-ALK-positive cells (according to LC-MS/MS data), although it was not modulated by NPM-ALK kinase activity (Figure 5A). Considering that ATIC can be a ALK partner with ATIC-ALK fusion chimera,9 we stably transfected an ATIC-ALK construct into the HEK-293T-Rex cells and showed that the wild-type ATIC activity was also enhanced by this fusion, similar to the NPM-ALK chimera (Figure 5B).
The ATIC activity is enhanced by NPM-ALK in HEK-293T-Rex cells. (A) NPM-ALK inducible HEK-293T-Rex cells were induced with tetracycline with and without CEP14083 ALK inhibitor. p-NPM-ALK was determined by Western blotting using anti–p-ALK antibodies. ATIC and LDH enzymatic activities were evaluated; *P < .001. (B) HEK-293T-Rex cells containing an inducible ATIC-ALK construct were induced with tetracycline and/or with CEP14083. Western blotting analysis confirmed ALK expression and phosphorylation. Enzymatic activity of ATIC and LDH was evaluated. (C) Cell lysates of transiently transfected HEK-293T cells (NPM-ALK, NPM-ALKK210R, ATIC-ALK, TPR-Met, c-myc, and the self-activating form of Ras, Ras V12) were tested for their ATIC and LDH enzymatic activity. *P < .001, **P < .05. (D) Cell lysates from transiently transfected HEK-293T cells with NPM-ALK, NPM-ALKK210R, and/or a Ras DN, respectively, were tested for their ATIC and LDH enzymatic activity.
The ATIC activity is enhanced by NPM-ALK in HEK-293T-Rex cells. (A) NPM-ALK inducible HEK-293T-Rex cells were induced with tetracycline with and without CEP14083 ALK inhibitor. p-NPM-ALK was determined by Western blotting using anti–p-ALK antibodies. ATIC and LDH enzymatic activities were evaluated; *P < .001. (B) HEK-293T-Rex cells containing an inducible ATIC-ALK construct were induced with tetracycline and/or with CEP14083. Western blotting analysis confirmed ALK expression and phosphorylation. Enzymatic activity of ATIC and LDH was evaluated. (C) Cell lysates of transiently transfected HEK-293T cells (NPM-ALK, NPM-ALKK210R, ATIC-ALK, TPR-Met, c-myc, and the self-activating form of Ras, Ras V12) were tested for their ATIC and LDH enzymatic activity. *P < .001, **P < .05. (D) Cell lysates from transiently transfected HEK-293T cells with NPM-ALK, NPM-ALKK210R, and/or a Ras DN, respectively, were tested for their ATIC and LDH enzymatic activity.
Next, to test whether the up-regulation of ATIC activity could be associated with a neoplastic phenotype and/or with other kinases, we transfected the HEK-293T cells with TPR-Met, c-Myc, and Ras expressing constructs. Figure 5C shows that all fusion kinases increased the ATIC activity, but this effect was significantly higher in NPM-ALK and in Ras (RasV12) transfected cells. Because Ras is a downstream target of NPM-ALK,37 we tested whether it was required in the NPM-ALK–mediated ATIC activation. Figure 5D shows that NPM-ALK–driven activation of ATIC was not affected by a dominant negative Ras construct.
To confirm our findings in a model that mimics human ALCL, we treated ALK+ (SU-DHL1, TS, and JB-6) and ALK− (Mac-1) ALCL cell lines with CEP14083, and then we evaluated the activity of ATIC and other metabolic enzymes (G6PD and ODC). All enzymatic measurements were performed shortly after the pharmacologic inhibition and in viable cells, as determined by tetramethylrhodamine methyl ester (TMRM) staining. ATIC activity was gradually reduced over a 6-hour time course after CEP14083 treatment in ALK+ cells, whereas no effect was documented in ALK− cells (Figure 6). Notably, both G6PD and ODC activities remained unaffected under these conditions.
The inhibition of NPM-ALK abrogated the ATIC activity in ALCL cells. The enzymatic activity of ATIC, G6PD, and ODC was measured in ALK+ (TS, SU-DHL1, JB-6) and ALK− (Mac-1) lymphoblastoid cell lines after treatment with a specific ALK inhibitor, as described in “Measurement of AICAR-FT/IMP-CHase activity.”
The inhibition of NPM-ALK abrogated the ATIC activity in ALCL cells. The enzymatic activity of ATIC, G6PD, and ODC was measured in ALK+ (TS, SU-DHL1, JB-6) and ALK− (Mac-1) lymphoblastoid cell lines after treatment with a specific ALK inhibitor, as described in “Measurement of AICAR-FT/IMP-CHase activity.”
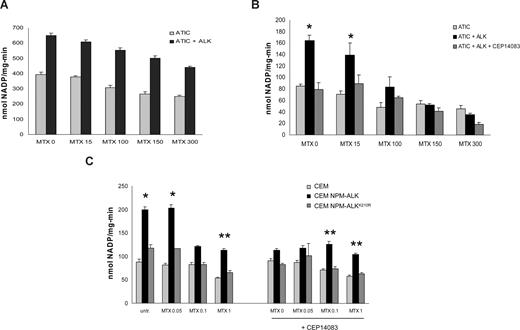
NPM-ALK protects cancer cells from the effects of MTX
To test whether the enzymatic modulation of ALK-mediated ATIC activity could be biologically relevant, we studied ATIC inhibition via MTX, a folic acid analog.38,39 MTX decreased the ATIC transformylase and the total enzyme activity, whereas its cyclohydrolase function remained unaffected (Figure 7A). In the presence of NPM-ALK, the effect of the MTX was partially abrogated, and higher doses of MTX were required to produce similar inhibitory rates compared with samples lacking NPM-ALK. To demonstrate that the NPM-ALK activity was necessary, we inhibited ALK kinase activity by CEP14083, and this led to a reduced ATIC transformylase activity in the presence of MTX (Figure 7B). Finally, to validate these findings in a cell-based assay, NPM-ALK or NPM-ALKK210R retroviral particles were transduced into T-cell lymphoma cells (CEM). Cells were then treated with the MTX and ATIC activity was determined. ATIC was impaired by MTX, whereas other cellular enzymes (LDH, G6PD, and 6PGD) were not affected (data not shown), either in control or NPM-ALKK210R cells. On the contrary, the inhibition of ATIC by MTX was less pronounced in ALK+ CEM cells (Figure 7C).
NPM-ALK attenuates the effect of methotrexate in lymphoma cells. (A) ATIC enzyme and NPM-ALK kinase were immunoprecipitated from 2 mg of HEK-293T cell lysates and then combined in vitro with increasing concentrations of MTX. Enzymatic activity of total ATIC was measured in vitro. P < .005. (B) ATIC enzyme and NPM-ALK kinase were combined in vitro in the presence of increasing concentrations of MTX and 300 nM of CEP14083 and AICAR-FT activity was measured. *P < .05. (C) CCRF-CEM cells were infected with NPM-ALK or the kinase dead mutant NPM-ALKK210R and treated in culture with increasing concentrations of MTX for 24 hours. Where indicated, the cells were pretreated with 300 nM of CEP14083 for 30 minutes before MTX treatment. AICAR-FT activity was measured. *P < .001, **P < .005.
NPM-ALK attenuates the effect of methotrexate in lymphoma cells. (A) ATIC enzyme and NPM-ALK kinase were immunoprecipitated from 2 mg of HEK-293T cell lysates and then combined in vitro with increasing concentrations of MTX. Enzymatic activity of total ATIC was measured in vitro. P < .005. (B) ATIC enzyme and NPM-ALK kinase were combined in vitro in the presence of increasing concentrations of MTX and 300 nM of CEP14083 and AICAR-FT activity was measured. *P < .05. (C) CCRF-CEM cells were infected with NPM-ALK or the kinase dead mutant NPM-ALKK210R and treated in culture with increasing concentrations of MTX for 24 hours. Where indicated, the cells were pretreated with 300 nM of CEP14083 for 30 minutes before MTX treatment. AICAR-FT activity was measured. *P < .001, **P < .005.
Discussion
We used 2 proteomic methods, together with a functional validation approach, to identify novel molecules downstream to NPM-ALK. Performing global HTP posttranscriptional analyses offers a new and powerful approach to dissect pathogenic mechanisms and to apply novel technologies for the diagnosis of malignancy, identification of biomarkers, discovery of targets, and design of tailored therapies.2-4,40
We confirmed that ALK signaling is associated with a well-defined transcriptional signature13,14 and that ALK+ cells preferentially express phosphorylated proteins with specific functions.10 Our data revealed some proteins in common with those reported in previous HTP proteomic studies and identified new targets.16,41-43 Relevant proteins were either known mediators of the ALK-signaling or novel proteins, including transcription factors, cytoskeletal/structural, motor/adhesion, and ribosomal proteins or proteins involved in the nucleic acid and/or protein metabolism.
Proteomic studies are successful tools for the diagnosis of ALCL42 and for the identification of ALK-related proteins and/or adaptor molecules.26,44 Our findings are in line with those of Rush et al, who have first demonstrated that HTP proteomics of Tyr-phosphorylated peptides provide selective signatures, identifying key regulators and predicting signaling pathways.15 Although proteomic studies have unveiled new players within kinase-driven signaling, it is well established that any molecular signature is not fully representative of the tumorigenic events leading to cellular transformation. Indeed, each genetic alteration may be simply associated with a given neoplastic phenotype but may lack any pathogenetic role; these lesions are now referred to as “passengers.” Although these defects do not maintain the neoplastic phenotype, they often represent “tumor-associated biomarkers” capable of better stratifying tumors or patients allowing better patient follow-up during treatment. Pathogenetically relevant lesions, referred to as “drivers,” have a more pertinent impact and represent ideal targets for novel therapies. Thus, it has become imperative to design strategies dissecting “passengers” from “drivers.”
Although proteomic approaches may be more informative than genomic analyses, functional validation studies are required to dissect the relationships between causal events and phenotypes and to define the tumorigenic contribution of each change.
Because HTP analyses are often descriptive, we adopted 2 alternative strategies to gain more insights into ALK-mediated transformation. First, we have compared the phospho-profile obtained from ALK+ ALCL cell lines with those derived from cells ectopically expressing ALK fusions, in the presence or in the absence of ALK signaling. These models have revealed a restricted set of phosphoproteins shared by ALK cells, in different cellular contexts. Although this may be restrictive and could result in the loss of relevant ALK-associated targets, it defines a limited number of proteins, whose phosphorylation status is highly reproducible and strictly dependent on ALK signaling. Second, to identify a biologically relevant set of ALK-associated proteins, we used an ALK inhibitor and performed a quantitative determination of phosphorylation changes. These studies have shown that determining the level of protein phosphorylation is necessary to identify appropriate targets, many of them undetectable using semiquantitative approaches.
To validate newly defined ALK-associated proteins, we performed biochemical studies, which confirmed that VASP and ATIC were associated with NPM-ALK and phosphorylated. More importantly, we demonstrated that ATIC phosphorylation enhanced its enzymatic activity. This is the first demonstration that ALK-mediated phosphorylation leads to an enhanced metabolic activity of a substrate, which may contribute to cell transformation. Enhanced purine synthesis sustains cell proliferation, and its therapeutic inhibition has been adopted to hamper tumor growth in many human malignancies. The discovery that ALK posttranscriptional modifications may change the activity of a key enzyme is important because this observation may be applied to other kinase-driven tumors. This hypothesis is supported by the detection of p-ATIC in many tumor cell lines and fresh samples.18,36 Thus, it is tempting to speculate that the detection of p-ATIC may be useful to predict the response to antifolate agents and facilitate the design of more efficacious therapeutic protocols. The identification of tumorigenic molecules has served as a platform to improve chemotherapeutic protocols using conventional cancer agents and to open a new era in anticancer research in which new targeted compounds are being developed to supplant the more toxic traditional anticancer drugs.
Here we provided new evidence on a posttranscriptional modification of a key purine synthesis enzyme, which may predict response to antifolate analogs. Detailed knowledge of ATIC enzyme activity in human tumors may allow the identification of low responders and thus the application of ad hoc therapeutic strategies or patient-related dose regimens.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Paola Bernabei and Andrea Manazza for their technical help and management the flow cytometry and image cores.
This work was supported in part by the National Institutes of Health (Bethesda, MD; R01-CA90773), the Sixth Research Framework Programme of the European Union, Project RIGHT (LSHB-CT-2004-005276), Ministero dell'Università e Ricerca Scientifica, Regione Piemonte, Compagnia di San Paolo, Torino (Progetto Oncologia), Associazione Italiana per la Ricerca sul Cancro and Fondazione Guido Berlucchi. C.V. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro (FIRC, Milano, Italy). F.E.B. is the recipient of a Research Fellowship from the Fondazione Internazionale di Ricerca in Medicina Sperimentale (FIRMS, Torino, Italy).
National Institutes of Health
Authorship
Contribution: C.V., R.D.P., R.C., A.B., M.J.C., and G.I. designed research; F.E.B., C.R., L.D., L.R., and V.L.G. performed research; M.C., B.R., J.R., and O.N.J. contributed reagents/analytic tools; K.L. and J.N. analyzed and interpreted data; and F.E.B., C.V., and G.I. wrote the paper.
Conflict-of-interest disclosure: G.I. receives research support from Cephalon. The remaining authors declare no competing financial interests.
Correspondence: Giorgio Inghirami, Center for Experimental Research and Medical Studies, Department of Biomedical Sciences and Human Oncology, University of Torino, Via Santena 5, 10126 Torino, Italy; e-mail: giorgio.inghirami@unito.it.
References
Author notes
*F.E.B. and C.V. contributed equally to this manuscript.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal