Abstract
Translation of small interfering RNA (siRNA)–based approaches into practical therapeutics is limited because of lack of an effective and cell-specific delivery system. Herein, we present a new method of selectively delivering siRNA to dendritic cells (DCs) in vivo using CD40 siRNA-containing immunoliposomes (siILs) that were decorated with DC-specific DEC-205 mAb. Administration of CD40 siILs resulted in DC-specific cell targeting in vitro and in vivo. On treatment with CD40 siILs, the expression of CD40 in DCs, as well allostimulatory activity was inhibited. In vivo administration resulted in selective siRNA uptake into immune organs and functional immune modulation as assessed using a model antigen. In conclusion, this is the first demonstration of DC-specific siRNA delivery and gene silencing in vivo, which highlights the potential of DC-mediated immune modulation and the feasibility of siRNA-based clinical therapy.
Introduction
The discovery of RNA interference (RNAi) has revolutionized molecular biology by allowing selective knock-down of genes in a manner more potent than any other previous technique.1 Induction of RNAi using small interfering RNA (siRNA), composed of double-stranded RNA molecules 19 to 21 base pairs in length, has offered a practical and widely applicable tool for not only research-based investigations but also potential therapeutic interventions. Despite growing interest in the application of siRNA into in vivo systems, the most challenging hurdle has been in vivo delivery in general, and specifically the ability to selectively target siRNA to specific cells.2 Over the past several years, multiple strategies have been developed and assessed in vivo, which can be generally categorized into viral and nonviral methods.1,3 Despite the high transfection efficiency exhibited by viral systems, this strategy was demonstrated to be unsafe for human use because of the associated inflammatory and oncogenic potential.4,5 Thus, to circumvent such side effects, many groups are now using nonviral systems, such as (1) systemic intravenous injection or local administration of naked, unmodified siRNA; (2) complex of siRNA with various cationic molecules, including lipids, liposomes, and peptides; and (3) conjugation of siRNA to natural ligands, such as cholesterol.1
Most strategies of nonviral targeting, however, are considered to be passive and nonspecific in nature because they lack the ability to target any particular cell or tissue type.6 Unfortunately, systemic distribution of nonspecifically targeted siRNA reduces gene-silencing efficiency through allowing siRNA to be taken up indiscriminately by multiple cell and tissue types. Another hurdle is the relatively short half-life of siRNA under physiologic conditions.7 When administered using the aforementioned nonviral methods, siRNA is degraded by endogenous nucleases in the blood and/or body fluids,7,8 and rapidly eliminated from the circulation.6
Dendritic cells (DCs) can act both as potent stimulators of T-cell activation and as regulators of the immune system playing an essential role in preventing autoimmune disease through induction of T regulatory cell generation and other tolerogenic mechanisms. We have previously demonstrated that siRNA manipulation of DC can be used to generate tailor-made tolerogenic DCs.9,10 Although these in vitro gene-silenced DCs have demonstrated therapeutic promise, the clinical use of this strategy is limited because of the associated procedural complexity and technical difficulties associated with ex vivo cell manipulation. An ideal method would be to directly silence DC gene expression in vivo. Here we describe a novel in vivo cell-targeted siRNA delivery system using immunoliposome-mediated targeting of siRNA to DC.
Methods
Animals
Male C57/B6 mice and BALB/c mice (The Jackson Laboratory, Bar Harbor, ME) were kept in filter-top cages at the Animal Facility, University of Western Ontario (London, ON) according to National Canadian Council for Animal Guidelines. This study was approved by the institutional review board of the University of Western Ontario.
Antibody preparation
Hybridomas that produce the NLDC-145 monoclonal antibody (mAb) were a kind gift from Dr Georg Kraal.11 The mAb NLDC-145 (1.5 mg) that is specific for the DC marker DEC-205, was purified from hybridoma supernatant using protein G Sepharose CL-4B (GE Healthcare, Little Chalfont, United Kingdom). For thiolation, 1.5 mg NLDC-145 was dissolved in 0.15 M Na-borate/0.1 mM ethylenediaminetetraacetic acid (pH 8.5) and thiolated for 1 hour at room temperature using 2-iminothiolane (also known as Traut Reagent; Sigma-Aldrich, St Louis, MO) in a 40:1 molar excess ratio. The buffer was then exchanged with 0.05 M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid/0.1 mM ethylenediaminetetraacetic acid (pH 7.0) using an Amnicon ultracentrifugal device with a molecular weight cutoff of 100 kDa (Amnicon, Billerica, MA), and the antibody was immediately used for conjugation to liposomes.
Preparation of siRNA liposomes
Liposomes were generated as previously described by Zhang et al12-16 and Shi et al17,18 using the following lipids: 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (POPC), dimethyldioctadecylammonium bromide (DDAB), distearoylphosphatidylethanolamine-PEG2000 (DSPE-PEG2000), and distearoylphosphatidylethanolamine-PEG2000-maleimide (DSPE-PEG2000-Mal; Avanti Polar Lipids, Alabaster, AL). PEG2000 is a 2000-Da chain of polyethylene glycol. POPC, DDAB, DSPE-PEG2000, and DSPE-PEG2000-Mal were dissolved in chloroform and mixed together in molar ratios of 92:4:3:1 (for neutral liposomes), 91:5:3:1 (for 1 mol% positive liposomes), or 90:6:3:1 (for 2 mol% positive liposomes), respectively. The total amount of lipid used was kept constant at 20.2 μmol. The chloroform-dissolved lipids were mixed together in a conical glass flask, and the chloroform was evaporated using a nitrogen gas stream, leaving a thin lipid film coating the walls of the flask. Lipids were then placed in a vacuum centrifuge for 90 minutes to remove residual chloroform. A total of 250 μg Cy3-labeled siRNA was dissolved in 0.05 M Tris-HCl (pH 8.0) to a final volume of 0.2 mL, which was subsequently added to the lipid film. The dispersion was then vortexed for 5 minutes and sonicated for 2 minutes using a bath sonicator. The dispersion was subjected to freezing by submersion in liquid N2 and thawing at room temperature. The freeze/thaw cycle was repeated 6 times. Liposomes were diluted to a concentration of 40 mM by adding 0.05 M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.0 (0.3 mL) and subsequently extruded, using a LiposoFast Basic extruder (Avestin, Ottawa, ON), through 2 stacked polycarbonate membranes of 400-nm pore size (Avestin). The extrusion was repeated using 200-, 100-, and 50-nm pore size polycarbonate membranes (Avestin). Exteriorized RNA was then degraded using RNase III (New England Biolabs, Ipswich, MA) according to the manufacturer's protocol. Briefly, ShortCut RNase buffer (10% vol/vol), MnCl2 (10% vol/vol), and 20 units of ShortCut RNase III were all added to the liposome/RNA dispersion. The digestion reaction mixture was incubated at 37°C for 2 hours, and the reaction was stopped by adding 20 mM ethylenediaminetetraacetic acid adding 250 mM ethylenediaminetetraacetic acid (10% vol/vol).
Preparation of siILs
Thiolated NLDC-145 was added to the siRNA liposome dispersion, and the mixture was incubated overnight at room temperature. The immunoliposome mixture was passed through a 1.5 × 10 cm Econo column (Bio-Rad, Hercules, CA) filled with Sepharose CL-4B matrix (GE Healthcare) to separate siRNA-bearing immunoliposomes from digested siRNA fragments and nonconjugated antibody. A total of 26 to 30 fractions of 0.5 mL each were collected, and the fluorescence (excitation/emission 550/570 and 650/668) of each eluted fraction was determined using a Cary Eclipse mass spectrofluorometer (Varian, Palo Alto, CA). Fractions corresponding to the first set of overlapping fluorescence peaks, which exhibited comigration of antibody and siRNA, were pooled and concentrated using a Amnicon ultracentrifugal device with a 100-kDa molecular weight cutoff. siRNA fluorescence was measured once again and compared against a standard curve to determine the final concentration of encapsulated siRNA. siRNA-bearing immunoliposomes (siILs) were filter-sterilized using a 0.2-μm filter (Millipore, Billerica, MA). Mean immunoliposome diameter was measured before and after conjugation of liposomes to mAb using a Zetasizer Nano particle sizer (Malvern Instruments, Malvern, United Kingdom) operated in the “volume weighed” mode.
Stability assay of siILs
To assess the stability of siILs in blood plasma in vitro, 40 μL siIL was incubated with 360 μL fresh mouse plasma at 37°C for various time periods. The siRNA inside siILs were extracted by 0.1% Triton X-100 solution. The degradation of siRNA was determined by 12% polyacrylamide gel electrophoresis followed by visualization by a Fluor-S MultiMager (Bio-Rad). To determine the RNase resistance of siIL, 8 μL of siIL was mixed with 40 ng RNase at 37°C for 6 hours. The reaction was terminated with 10% sodium dodecyl sulfate solution. The extraction and detection of siRNA from siIL were the same as described above in this paragraph.
Generation of BMDCs
DCs were prepared from bone marrow progenitors as previously described.19 Briefly, bone marrow cells were flushed from femurs and tibias of C57BL/6 mice, washed, and cultured in 6-well plates (2 × 106 cells/mL) in 4 mL complete medium (RPMI 1640 supplemented with 2 mM l-glutamine, 100 U/mL penicillin, 100 μg streptomycin, 50 μM 2-medroxyestradiol, and 10% fetal calf serum (all from Invitrogen, Burlington, ON) supplemented with recombinant granulocyte-macrophage colony-stimulating factor (10 ng/mL; PeproTech, Rocky Hill, NJ) and recombinant mouse IL-4 (10 ng/mL; PeproTech). All cultures were incubated at 37°C in 5% humidified CO2.
Silencing DCs in vitro
DCs (106 cells) were suspended in 200 μL RPMI 1640 (serum-free) and aliquoted into a 24-well plate (BD Biosciences, Mississauga, ON). Separately, 1 μg CD40 siRNA (sense sequence: UGUUCCACUGGGCUGAGAA) or negative control (nonspecific) siRNA was incubated with 5 μL GenePorter (GP), a transfection reagent (Gene Therapy Systems, San Diego, CA) in a volume of 100 μL RPMI 1640 (serum-free) at room temperature for 5 minutes. The siRNA mixture was then added to the 200 μL DC cell culture. Another negative control group was transfected with 5 μL GP alone. After 4 hours of incubation at 37°C, an equal volume of RPMI 1640 supplemented with 20% fetal calf serum was added to the cells. On day 7, DCs were stimulated with lipopolysaccharide (LPS) (10 ng/mL) and incubated for an additional 24 hours. DC expression of CD40 was determined by flow cytometry on day 8. An identical protocol was used for the transfection of DCs by siRNA-bearing siILs, with the exception that GP-complexed siRNA was substituted with siIL-encapsulated siRNA. Negative control groups were treated with phosphate-buffered saline (PBS) or siIL-encapsulated negative control siRNA.
In vitro DC-specific binding assay
Day 6 bone marrow–derived DCs (BMDCs) were collected (106 cells) and incubated for 30 minutes at 4°C with 3 μg siIL-encapsulated CD40 siRNA. Control groups were treated with an equal amount of CD40 siRNA liposomes or an equal volume of PBS, empty siILs, or CD40 siRNA mixed with GP. Cells were then washed with fresh RPMI medium. The negative control L929 cells were seeded at a density of 2 × 105 in a 24-well plate overnight. The medium was removed, and the cells were incubated with 3 μg siIL-encapsulated CD40 siRNA 30 minutes at 4°C. Control groups for L929 cells were identical to those used for DCs. Cells were washed with RPMI, and both DCs and L929 cells were imaged using a fluorescence microscope (Bio-Rad).
In vivo gene silencing using CD40 siILs
On day 0, 7-week-old C57/B6 mice were subcutaneously injected at the base of the neck with 50 μg keyhole limpet hemocyanin (KLH; Sigma-Aldrich) dissolved in 100 μL PBS and mixed with an equal volume of complete Freund adjuvant (CFA; Sigma-Aldrich). The KLH/CFA emulsion was injected together with 15 μg siIL-encapsulated CD40 siRNA, siIL-encapsulated negative control siRNA, naked CD40 siRNA, or PBS. On day 1, an identical siRNA treatment was administered intravenously. Mice were killed on day 3 to assess the CD40 gene-silencing capacity of CD40 siILs in the spleen and lymph nodes (LNs). Both splenocytes and LN cells were analyzed for CD40 expression by flow cytometry and used for mixed lymphocyte reaction (MLR) and antigen (KLH)–specific recall response assays.
Flow cytometry
For analysis of DC surface molecule expression of CD40 and major histocompatibility complex class II, 106 BMDCs, splenocytes, or LN cells were stained with fluorescein isothiocyanate- or phycoerythrin-conjugated Ab or the appropriate isotype control Ab in a final volume of 100 μL, at 4°C for 30 minutes in the dark (antibodies from Cedarlane, Burlington, ON). This was followed by washing 2 times in 3 mL cold flow cytometry wash buffer (PBS with 1% bovine serum albumin). The cells were then resuspended in 300 μL PBS, and protein expression was analyzed using a FACScan apparatus (BD Biosciences).
Quantitative polymerase chain reaction
Total RNA was isolated from 106 BMDCs or splenocytes using Trizol reagent (Invitrogen) and according to the manufacturer's protocol. The RNA was subsequently reverse-transcribed using an oligo-(dT) primer and reverse transcriptase (Invitrogen). Primers used for the amplification of murine CD40 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an internal loading control, were as follows: CD40, 5′-GATTTGTGCCAGCCAGGAAGC-3′ (forward), and 5′-CCCTTGATTGGGTTCACAGTGTCT-3′ (reverse); and GAPDH, 5′-TGATGACATCAAGAAGGTGGTGAA-3′ (forward) and 5′-TGGGATGGAAATTGTGAGGGAGAT-3′ (reverse).
Quantitative polymerase chain reactions (PCRs) were performed using SYBR Green PCR Master mix (Stratagene, La Jolla, CA) and 100 nM gene-specific forward and reverse primers. The PCR conditions were 95°C for 10 minutes, 95°C for 30 seconds, 58°C for 1 minute, and 72°C for 30 seconds (40 cycles). Amplification was performed according to the manufacturer's cycling protocol and done in triplicate. Gene expression was calculated as 2−ΔΔ(Ct),20 where Ct is cycle threshold, ΔΔ(Ct) = sample 1Δ(Ct) − sample 2Δ(Ct); Δ(Ct) = GAPDH (Ct) − testing gene (Ct). Data were analyzed using MX4000 (Stratagene), Microsoft Excel 2003, and Prism software (GraphPad, San Diego, CA).
Mixed lymphocyte reaction
On day 6 of culture, BMDCs from C57/B6 were transfected by CD40 siILs or other control treatments and activated by LPS/tumor necrosis factor-α, as described in “Generation of BMDCs.” Activated DCs were irradiated (3000 cGy) and seeded in triplicate into a flat-bottom 96-well plate (Corning, Corning, NY) as stimulators. Spleen T cells from BALB/c mice were isolated by gradient centrifugation over Ficoll-Paque (GE Healthcare) and added as responders (2 × 105 cells/well). The mixed lymphocyte culture was incubated at 37°C for 72 hours and was pulsed with 1 μCi/well [3H] thymidine (GE Healthcare) for the last 16 hours of culture. Finally, cells were harvested onto glass fiber filters, and the radioactivity incorporated was measured using a Wallac Betaplate liquid scintillation counter (PerkinElmer Life and Analytical Sciences, Waltham, MA).
Ag-specific T-cell proliferation
Proliferative responses to KLH in subsequent groups of mice were measured with a standard microtiter assay using splenocytes, in the presence of KLH. Splenocytes (105/well) were seeded into a 96-well flat-bottom microtiter plate in triplicate and mixed with serial dilutions of KLH (0-10 μg/well). After 72 hours of incubation, 1 μCi [3H] thymidine was added to each well for 16 hours. The cells were collected onto a glass microfiber filter, and the radioactivity incorporated was measured by a Wallac Betaplate liquid scintillation counter.
Histology
The spleen, kidney, and liver were dissected from mice at various time points (20 minutes, 4 hours, and 48 hours) after intravenous injection with Cy-3–labeled siRNA incorporated within DC-targeting immunoliposomes or within nontargeting liposomes that lack the NLDC-145 mAb. Other controls included naked Cy-3 siRNA and PBS. Organs were frozen in Tissue-Tek OCT compound (Sakura, Torrance, CA), and 5-μm sections were obtained. Sections were analyzed using a fluorescence microscope (Bio-Rad) and Northern Eclipse Software (Empix Imaging, Cheektowaga, NY).
Statistical analysis
Data were expressed as the mean plus or minus SEM. siRNA encapsulation efficiency of different immunoliposome formulations containing 0%, 1%, and 2% DDAB was compared using a one-way analysis of variance followed by the Newman-Keuls test. Data from quantitative PCR, MLR, and KLH recall response assays were also analyzed using analysis of variance. Differences for the value of P less than .05 were considered significant.
Results
Optimization and characterization of siILs
The overall structure of siILs is shown in Figure S1A (available on the Blood website; see the Supplemental Materials link at the top of the online article). In this study, we chose the costimulatory molecule CD40 as a sample target gene, given that it is strictly necessary for bidirectional interaction between T cells and DCs during immune activation.21-24 CD40 siRNA was encapsulated within the aqueous interior of siILs that were conjugated to the NLDC-145, a mAb that targets the DC-specific marker DEC-205. CD40 siILs were prepared by first incorporating Cy3-labeled CD40 siRNA within liposomes composed of the neutral lipid POPC, DDAB, DSPE-PEG2000, and DSPE-PEG2000-maleimide by the freeze/thawing technique.3 Subsequently, liposomes were repeatedly extruded through polycarbonate filter membranes with pore sizes of 400, 200, 100, and 50 nm to produce a liposome population of uniform size. The extrusion step generated siRNA-containing liposomes of approximately 73 nm in diameter, with more than 90% of liposomes ranging from 50 to 92 nm (Figure S1B).
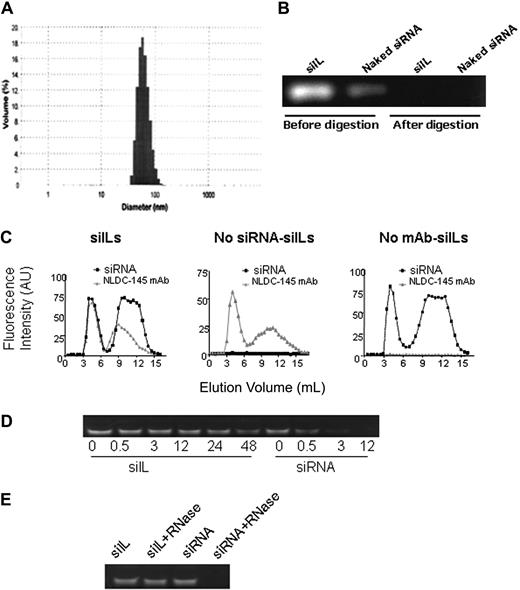
Stealth liposomes can circulate longer in vivo than conventional liposomes but lack a targeting mechanism required for specific delivery. Hence, to achieve DC-specific targeting, NLDC-145 mAbs were coupled to the siRNA-bearing liposomes. The anchoring of NLDC-145 was achieved through the linker lipid DSPE-PEG2000-maleimide, which was incorporated into the liposome formulation. Reacting thiolated NLDC-145 mAb with the maleimide group forms a thioether linkage between the mAb and PEG2000, allowing the mAb to be covalently linked to the liposome surface. The siIL average diameter was once again measured and was found to have increased to 86 nm after conjugation to NLDC-145 mAb (Figure 1A). Unencapsulated siRNA, either in solution or associated with the liposome exterior, was completely removed by RNase III digestion. Digestion was demonstrated by ethidium bromide staining after polyacrylamide gel electrophoresis (Figure 1B).
Characterization of siILs. (A) Measurement of siIL size. siRNA-bearing immunoliposomes (siILs) were prepared as described in “Preparation of siILs.” siIL mean diameter was approximately 86 nm after conjugation of NLDC-145 mAb to liposome surface. (B) Removal of unencapsulated siRNA. Polyacrylamide gel electrophoresis was used to resolve siRNA-containing liposomes before and after RNase III digestion of unencapsulated siRNA. (C) Fraction elution of siILs. After the encapsulation of Cy3-labeled siRNA and conjugation of liposomes to Alexa647-labeled NLDC-145 mAb (anti-DEC205), siILs were separated from digested siRNA fragments of exteriorized siRNA and unconjugated mAb by Sepharose CL-4B gel filtration chromatography. Eluted fractions were analyzed by spectrofluorometry (excitation/emission 550/570 nm for siRNA and 650/668 nm for mAb) for simultaneous detection of encapsulated siRNA and conjugated mAb. (Left panel) siRNA-bearing siILs containing both siRNA and NLDC-145 mAb, which comigrate through the filtration column (first set of overlapping peaks). (Middle panel) Empty siILs containing no siRNA but conjugated to NLDC-145 mAb. (Right panel) Control stealth liposomes containing siRNA but not conjugated to NLDC-145 mAb. (D) Stability assay of siILs in blood plasma. Cy3-labeled CD40-siILs or naked siRNA was incubated with fresh mouse plasma at 37°C for various time periods. After incubation for 0, 0.5, 3, 12, 24, and 48 hours, siRNA from siILs was extracted with Triton X-100 and detected by polyacrylamide gel electrophoresis. (E) RNase resistance assay of siILs. siILs or naked siRNA (30 pmol) was incubated with 40 ng RNase at 37°C for 6 hours. siRNA was extracted from siILs with Triton X-100 and detected by polyacrylamide gel electrophoresis.
Characterization of siILs. (A) Measurement of siIL size. siRNA-bearing immunoliposomes (siILs) were prepared as described in “Preparation of siILs.” siIL mean diameter was approximately 86 nm after conjugation of NLDC-145 mAb to liposome surface. (B) Removal of unencapsulated siRNA. Polyacrylamide gel electrophoresis was used to resolve siRNA-containing liposomes before and after RNase III digestion of unencapsulated siRNA. (C) Fraction elution of siILs. After the encapsulation of Cy3-labeled siRNA and conjugation of liposomes to Alexa647-labeled NLDC-145 mAb (anti-DEC205), siILs were separated from digested siRNA fragments of exteriorized siRNA and unconjugated mAb by Sepharose CL-4B gel filtration chromatography. Eluted fractions were analyzed by spectrofluorometry (excitation/emission 550/570 nm for siRNA and 650/668 nm for mAb) for simultaneous detection of encapsulated siRNA and conjugated mAb. (Left panel) siRNA-bearing siILs containing both siRNA and NLDC-145 mAb, which comigrate through the filtration column (first set of overlapping peaks). (Middle panel) Empty siILs containing no siRNA but conjugated to NLDC-145 mAb. (Right panel) Control stealth liposomes containing siRNA but not conjugated to NLDC-145 mAb. (D) Stability assay of siILs in blood plasma. Cy3-labeled CD40-siILs or naked siRNA was incubated with fresh mouse plasma at 37°C for various time periods. After incubation for 0, 0.5, 3, 12, 24, and 48 hours, siRNA from siILs was extracted with Triton X-100 and detected by polyacrylamide gel electrophoresis. (E) RNase resistance assay of siILs. siILs or naked siRNA (30 pmol) was incubated with 40 ng RNase at 37°C for 6 hours. siRNA was extracted from siILs with Triton X-100 and detected by polyacrylamide gel electrophoresis.
siILs were separated from unconjugated mAb and digested fragments of siRNA by Sepharose CL-4B gel filtration chromatography. Fractions eluted from the gel filtration column were analyzed by mass spectrofluorometry to ensure siRNA encapsulation and antibody conjugation to the liposome. Figure 1C (left panel) shows an overlap of the first siRNA and NLDC-145 mAb fluorescence peaks. Comigration of siRNA and NLDC-145 mAb indicates the incorporation of these 2 components into the same structure. The second set of nonoverlapping peaks in Figure 1C (left panel) corresponds to digested fragments of siRNA and unconjugated antibody. siRNA liposomes (Figure 1C middle panel) or empty immunoliposomes (Figure 1C right panel), which, respectively, lack NLDC-145 mAb or siRNA, were used as controls.
To determine the stability of siILs, siILs and naked siRNA were exposed to mouse plasma that is known to contain numerous RNA-degrading activities, such as various RNases. Naked siRNA degradation was observed after 30 minutes of incubation with plasma. Complete siRNA degradation occurred after 12 hours of exposure. In contrast, siILs displayed significant stability in the presence of plasma with no significant degradation detected at the 48-hour time point (Figure 1D). To confirm the protective effects of siILs, we exposed siILs and naked siRNA to RNase directly. After 6 hours of incubation with RNase, naked siRNA was completely degraded, whereas siILs still retained intact siRNA (Figure 1E). Taken together, these data suggest that the immunoliposome component of the siIL exerts a protective effect on the siRNA leading to extended life span.
Cell-specific targeting by siILs
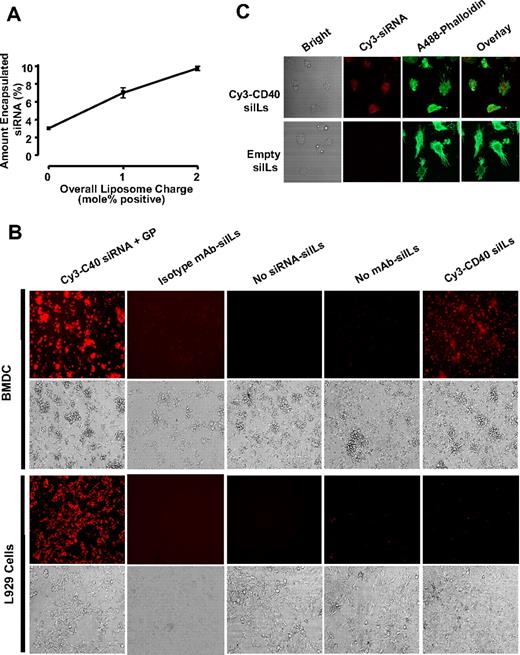
Encapsulation efficiency of siRNA within neutral liposomes was quite low, making it difficult to detect siIL-encapsulated Cy3-siRNA in DC-binding assays, even when large amounts of liposomes were used (data not shown). Previously published observations by Kim et al indicate that altering the liposome formulation by increasing the mol% of cationic lipid DDAB can significantly increase the encapsulation efficiency of anionic nucleic acid within liposomes.25 We therefore generated siILs in which the DDAB content was increased by 1 mol% or 2 mol%. To keep the total lipid amount unaltered at 20.2 μmol, the concentration of neutral lipid (POPC) was decreased relative to DDAB, and the concentrations of the anionic lipids, DSPE-PEG2000 and DSPE-PEG2000-maleimide, were kept constant. Using an initial amount of 250 μg Cy3-labeled siRNA, the encapsulation efficiency was compared between neutral liposomes and liposomes made with 1 mol% or 2 mol% excess DDAB. As shown in Figure 2A, augmenting DDAB content increased the percentage of encapsulated siRNA. Increasing DDAB concentration by 0, 1, and 2 mol% yielded respective siRNA encapsulation efficiencies of approximately 3%, 7%, and 10%. Although the liposome formulation containing 2 mol% excess DDAB exhibited the highest capacity for siRNA encapsulation, it also demonstrated a high degree of nonspecific binding when incubated with BMDCs, most probably a result of electrostatic interactions with the cell membrane because of its increased surface charge (data not shown). Thus, for the duration of this study, we used siILs containing 1 mol% excess DDAB because this formulation demonstrated a greater encapsulation capacity compared with neutral liposomes and did not exhibit significant nonspecific binding to DCs (Figure 2B).
siILs can specifically target DCs in vitro. (A) siRNA encapsulation efficiency within liposomes. The entrapment efficiency of siRNA within neutral and positively charged liposomes was determined using an initial 250 μg amount of Cy3-labeled siRNA. Mass spectrofluorometry (excitation/emission 550/570) was used to measure the encapsulation efficiency of fluorescent siRNA within liposomes formulated using increasing amounts of the positive lipid DDAB. (B) Specific binding of siILs to dendritic cells. BMDCs were incubated for 30 minutes at 4°C with 1 mol/dL positive, DC-specific siILs containing Cy3-labeled CD40 siRNA. Cells were washed and imaged. Cy3-CD40 siRNA was complexed with GP, a commercial transfection reagent, and used as a positive control treatment. Isotype IgG conjugated siILs, or siILs lacking Cy3-CD40 siRNA (Empty siILs), or the no DC-targeting mAb (siRNA liposomes) were used as negative controls. A cell line (L929) that does not express DEC-205 was also used as a negative control. The specificity of siIL binding was determined by epifluorescence microscopy. (C) Confocal imaging of siIL binding to dendritic cells. BMDCs were fixed, permeabilized, stained with Alexa 488–phalloidin, and incubated with empty or Cy3-CD40 siILs for 15 minutes at room temperature. Unbound siILs were washed off using PBS, and cells were imaged by confocal microscopy with a 40× objective.
siILs can specifically target DCs in vitro. (A) siRNA encapsulation efficiency within liposomes. The entrapment efficiency of siRNA within neutral and positively charged liposomes was determined using an initial 250 μg amount of Cy3-labeled siRNA. Mass spectrofluorometry (excitation/emission 550/570) was used to measure the encapsulation efficiency of fluorescent siRNA within liposomes formulated using increasing amounts of the positive lipid DDAB. (B) Specific binding of siILs to dendritic cells. BMDCs were incubated for 30 minutes at 4°C with 1 mol/dL positive, DC-specific siILs containing Cy3-labeled CD40 siRNA. Cells were washed and imaged. Cy3-CD40 siRNA was complexed with GP, a commercial transfection reagent, and used as a positive control treatment. Isotype IgG conjugated siILs, or siILs lacking Cy3-CD40 siRNA (Empty siILs), or the no DC-targeting mAb (siRNA liposomes) were used as negative controls. A cell line (L929) that does not express DEC-205 was also used as a negative control. The specificity of siIL binding was determined by epifluorescence microscopy. (C) Confocal imaging of siIL binding to dendritic cells. BMDCs were fixed, permeabilized, stained with Alexa 488–phalloidin, and incubated with empty or Cy3-CD40 siILs for 15 minutes at room temperature. Unbound siILs were washed off using PBS, and cells were imaged by confocal microscopy with a 40× objective.
To demonstrate that binding of siILs to DCs occurs through liposome-conjugated NLDC-145 mAb, and not through nonspecific electrostatic interactions between siILs and cell membrane, BMDCs and L929 cells were incubated with siILs in culture (Figure 2B). NLDC-145–conjugated siILs were able to bind to BMDCs but did not bind to control L929 cells, which lack expression of DEC-205 (the target of NLDC-145). Furthermore, siRNA-containing stealth liposomes, which are not coupled to NLDC-145 or coupled with isotype IgG, were unable to bind DCs in culture (Figure 2B). The ability of siILs to bind DCs was further confirmed using confocal microscopy (Figure 2C). Taken together, these observations demonstrate a requirement for NLDC-145 in the interaction between siILs and DCs, and indicate that siILs can selectively bind to DCs.
Silencing DCs by CD40 siILs
To confirm the gene-silencing efficacy of the CD40 siRNA sequence we developed, BMDCs were transfected with CD40 siRNA using GP, a cationic lipid transfection reagent. CD40 gene expression was analyzed 48 hours after transfection by flow cytometry (Figure 3A) and quantitative real-time PCR (Figure 3B). Results showed that BMDCs transfected with the CD40 siRNA we generated exhibited a significant decrease in CD40 expression compared with BMDCs transfected with negative control siRNA. Thus, we used this effective CD40 siRNA in preparation of siILs.
Silencing DC expression of CD40 in vitro by CD40 siILs. (A) Gene silencing of CD40 siRNA. Day 6 BMDCs were transfected with CD40 siRNA, or negative control siRNA, complexed with GP transfection reagent. Cells were activated with LPS on day 7 and assayed for protein expression and CD40 mRNA by (A) flow cytometry and (B) quantitative PCR. (C) In vitro silencing of CD40 expression in BMDC using siIL-encapsulated CD40 siRNA. On day 6 of culture, BMDCs were transfected with increasing amounts of siIL-encapsulated CD40 siRNA, siIL-encapsulated negative control (nonspecific) siRNA, or PBS. DCs were activated with LPS on day 7, and CD40 protein expression was measured by flow cytometry on day 8. Data presented are representative of 3 independent experiments (*P < .05).
Silencing DC expression of CD40 in vitro by CD40 siILs. (A) Gene silencing of CD40 siRNA. Day 6 BMDCs were transfected with CD40 siRNA, or negative control siRNA, complexed with GP transfection reagent. Cells were activated with LPS on day 7 and assayed for protein expression and CD40 mRNA by (A) flow cytometry and (B) quantitative PCR. (C) In vitro silencing of CD40 expression in BMDC using siIL-encapsulated CD40 siRNA. On day 6 of culture, BMDCs were transfected with increasing amounts of siIL-encapsulated CD40 siRNA, siIL-encapsulated negative control (nonspecific) siRNA, or PBS. DCs were activated with LPS on day 7, and CD40 protein expression was measured by flow cytometry on day 8. Data presented are representative of 3 independent experiments (*P < .05).
To demonstrate the gene-silencing effectiveness of DC-specific CD40 siILs, BMDCs were transfected with increasing amounts of siIL-encapsulated CD40 siRNA. A dose-dependent gene-silencing response became evident at 24 hours after transfection, was most pronounced at 48 hours (Figure 3C), and persisted until at least 72 hours (data not shown). In contrast, transfection with siIL-encapsulated negative control siRNA did not induce any detectable gene silencing regardless of concentration (Figure 3C). These results show that immunoliposome-encapsulated CD40 siRNA can silence DC expression of CD40 in vitro.
In vivo DC-specific targeting and gene silencing by CD40 siILs
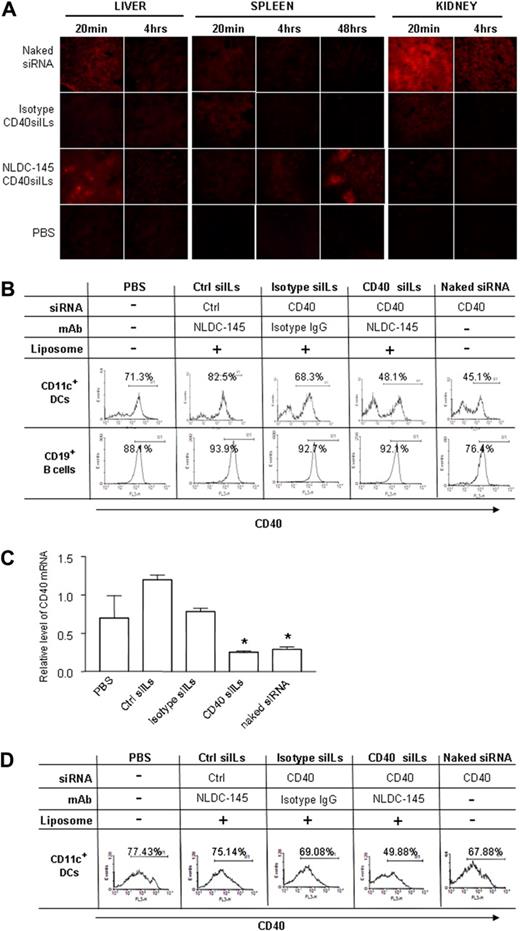
Recent biodistribution studies suggest that the majority of siRNA accumulates in the kidney and is excreted in urine after intravenous administration.6 To compare the biodistribution of siILs with naked siRNA in first pass organs, we injected mice intravenously with 15 μg naked or siIL-encapsulated Cy3-labeled CD40 siRNA that was coupled to NLDC-145 mAb or isotype IgG, respectively. The liver, spleen, and kidney were collected at 20 minutes, 4 hours, and 48 hours after injection. Organs were sectioned and analyzed for the presence of Cy3 siRNA (Figure 4A). Naked siRNA appeared to primarily accumulate in the kidney, to a smaller degree in the liver, and the smallest amount was detected in the spleen. After 48 hours, naked siRNA was still present in detectable amounts within the kidney and liver, but not the spleen. Liposome-encapsulated siRNA was only observed at low levels in the spleen and kidney, 20 minutes after administration. However, when mice were treated with siIL-encapsulated siRNA, an early concentration of siILs was observed at 20 minutes in the liver, which was cleared by 4 hours. Interestingly, siILs began to accumulate within the spleen at 4 hours and were detectable at even higher levels by 48 hours after injection. siIL-encapsulated siRNA was not detected in the kidney at any time point. These observations show that siIL-encapsulated and naked siRNA accumulate in different anatomic locations on intravenous administration. Most naked siRNA seemed to be cleared by the kidney soon after injection, whereas DC-specific siILs accumulated in DC-rich organs, such as the spleen and liver.
Silencing DC expression of CD40 in vivo by CD40 siIL. (A) Organ distribution of CD40 siIL. Mice were intravenously injected with 15 μg naked Cy3-siRNA, isotype IgG-coupled Cy3-siILs, NLDC-145-coupled Cy3-siILs, or with PBS. Liver, spleen, and kidney organs were collected at 20 minutes and 4 hours after siRNA injection. Frozen organ sections were analyzed using fluorescence microscopy. (B) In vivo CD40 gene silencing determined by flow cytometry. Mice were treated with siILs and control reagents as described in panel A. At 48 hours after treatment, splenic CD11c+ DCs and CD19+ B cells were isolated using magnetic-activated cell sorter beads. Cells were stained with Cy5PE-labed anti-CD40 mAb, and the expression of CD40 was detected by flow cytometry. (C) In vivo CD40 gene silencing was determined by quantitative PCR. Splenic CD11c+ DCs from panel B were used for extraction of total RNA. CD40 gene expression at the mRNA level was determined by quantitative PCR as described in “Quantitative polymerase chain reaction.” (D) In vivo CD40 gene silencing on day 12. Mice were treated with siILs as described in panel A. The splenic CD11c+ DCs were isolated on day 12 after siIL treatment. The CD40 expression was determined by flow cytometry as described in panel B. Data presented are representative of 4 independent experiments. Fluorescence-activated cell sorter data and PCR results are representative of 3 or 4 independent experiments with 3 or 4 mice enrolled in each group. *P < .05.
Silencing DC expression of CD40 in vivo by CD40 siIL. (A) Organ distribution of CD40 siIL. Mice were intravenously injected with 15 μg naked Cy3-siRNA, isotype IgG-coupled Cy3-siILs, NLDC-145-coupled Cy3-siILs, or with PBS. Liver, spleen, and kidney organs were collected at 20 minutes and 4 hours after siRNA injection. Frozen organ sections were analyzed using fluorescence microscopy. (B) In vivo CD40 gene silencing determined by flow cytometry. Mice were treated with siILs and control reagents as described in panel A. At 48 hours after treatment, splenic CD11c+ DCs and CD19+ B cells were isolated using magnetic-activated cell sorter beads. Cells were stained with Cy5PE-labed anti-CD40 mAb, and the expression of CD40 was detected by flow cytometry. (C) In vivo CD40 gene silencing was determined by quantitative PCR. Splenic CD11c+ DCs from panel B were used for extraction of total RNA. CD40 gene expression at the mRNA level was determined by quantitative PCR as described in “Quantitative polymerase chain reaction.” (D) In vivo CD40 gene silencing on day 12. Mice were treated with siILs as described in panel A. The splenic CD11c+ DCs were isolated on day 12 after siIL treatment. The CD40 expression was determined by flow cytometry as described in panel B. Data presented are representative of 4 independent experiments. Fluorescence-activated cell sorter data and PCR results are representative of 3 or 4 independent experiments with 3 or 4 mice enrolled in each group. *P < .05.
Next, we sought to determine whether DC expression of CD40 could be specifically silenced in vivo using DC-targeting siILs. Mice were treated with PBS, naked CD40 siRNA, CD40 siILs, or negative control siILs that were coupled with NLDC-145 or isotype IgG. The spleens and draining LNs (DLNs) were collected 48 hours after siIL administration, and CD40 expression was assessed in the CD11c+ (DC-enriched) as well as CD19+ (B cell–enriched) cell populations. Treatment with CD40 siILs or naked CD40 siRNA significantly decreased the surface expression of CD40 in CD11c+ splenic (Figure 4B) and DLN DCs (Figure S2) compared with DCs from mice treated with PBS or negative control siRNA siILs, or isotype-conjugated siILs. Furthermore, gene silencing occurred exclusively in CD11c+ DCs, but not in B cells. CD40 expression on CD11c+ DCs was also determined at the mRNA level by quantitative PCR. A significant decrease in CD40 mRNA levels was observed in mice treated with siIL-encapsulated CD40 siRNA that was coupled with NLDC-145 but not in the mice with siILs that were coupled with isotype IgG (Figure 4C). These data suggest cellular specificity of in vivo silencing by siILs.
To determine the gene-specific silencing, we also monitored the expression of major histocompatibility complex class II in DCs isolated from spleen and DLNs after treatment with CD40-siILs. There was no difference in expression among the different treatment groups in DCs (Figure S3), suggesting that siIL-induced silencing is gene-specific.
Finally, we measured persistence of gene silencing induced by siILs in vivo. According to published literature, the silencing effects of siRNA can last an average of approximately 66 hours26 and up to a maximum of 7 days in mammalian cells.27 Interestingly, CD11c+ DCs from mice treated with CD40 siILs exhibited potent silencing of CD40, even 12 days subsequent to siIL treatment, in comparison with control groups (Figure 4D). In contrast, DCs from mice treated with naked CD40 siRNA, which showed silencing efficacy at 48 hours, did not exhibit any CD40 gene silencing on day 12. These data indicate that gene-silencing effects of siIL-encapsulated siRNA last longer than naked siRNA.
Immune suppression by CD40 siILs
It has been previously reported that CD40-deficient DCs exhibit tolerance-inducing effects.28,29 Having generated CD40-silenced DCs in vitro and in vivo (Figures 3C, 4B) using CD40 siILs, we sought to quantify the tolerogenic potential of these DCs. CD40-silenced BMDCs generated in vitro (as described in Figure 3C) were used as stimulators in an allogeneic MLR. As shown in Figure 5A, DCs transfected with CD40 siRNA siILs were able to inhibit allogeneic T-cell proliferation.
CD40 siIL-induced immune suppression. (A) CD40 siILs inhibit allogenic T-cell proliferation in vitro. CD40 siIL-transfected BMDCs from C57/B6 mice were used as stimulators, and BALB/C splenocytes were used as effectors at 1:5 ratio in MLR. T-cell proliferation was measured by [3H]-thymidine incorporation, as described in “Mixed lymphocyte reaction.” (B) CD40 siILs inhibit KLH-specific recall response. Mice were immunized with KLH emulsified in CFA and simultaneously administered 15 μg CD40 siILs. Control groups included PBS, control siRNA, isotype IgG-coupled siILs. Naked siRNA was used as a positive control. T cells were isolated from the spleens of the siIL-treated mice and control mice, 12 days after KLH immunization and siIL treatment. Ag-specific T-cell response was assessed in the presence of 10 μg KLH antigen. Data presented are representative of 4 independent experiments (n = 3 or 4 mice/group). *P < .05.
CD40 siIL-induced immune suppression. (A) CD40 siILs inhibit allogenic T-cell proliferation in vitro. CD40 siIL-transfected BMDCs from C57/B6 mice were used as stimulators, and BALB/C splenocytes were used as effectors at 1:5 ratio in MLR. T-cell proliferation was measured by [3H]-thymidine incorporation, as described in “Mixed lymphocyte reaction.” (B) CD40 siILs inhibit KLH-specific recall response. Mice were immunized with KLH emulsified in CFA and simultaneously administered 15 μg CD40 siILs. Control groups included PBS, control siRNA, isotype IgG-coupled siILs. Naked siRNA was used as a positive control. T cells were isolated from the spleens of the siIL-treated mice and control mice, 12 days after KLH immunization and siIL treatment. Ag-specific T-cell response was assessed in the presence of 10 μg KLH antigen. Data presented are representative of 4 independent experiments (n = 3 or 4 mice/group). *P < .05.
To test in vivo immune suppression by siILs, mice were immunized with KLH emulsified in CFA and simultaneously injected with CD40 siILs or negative control siILs that were coupled with NLDC-145 or isotype IgG. Coadministration of siILs with KLH Ag allows DCs to take up both siRNA and antigen simultaneously. To enhance the gene silencing, mice were again administered an identical dose of CD40 siILs 24 hours after immunization. Twelve days after immunization with KLH and treatment with CD40 siILs, T cells were isolated and used to evaluate immune suppression of Ag-specific immune responses. As shown in Figure 5B, KLH-specific T-cell proliferation was inhibited in the mice treated with CD40 siILs in an antigen-specific recall response.
Discussion
The major bottleneck in the development of siRNA therapies is the delivery of these macromolecules to the desired cell type, tissue, or organ. Herein, we have shown, for the first time, a novel cell-specific in vivo delivery system for siRNA by modifying liposomal approaches.12-16 Our novel approach, which we termed siRNA immunoliposomes (siILs), demonstrated a potent and long-lasting cell-specific gene-silencing efficiency and can potentially increase the feasibility of RNAi therapy in clinical application.
In this study, some modifications were made to the original formulation to increase gene-silencing potential, encapsulation efficiency, and specifically targeting DCs. The respective changes include: substitution of siRNA-expressing vectors with synthetic duplex siRNA; increased liposomal surface charge; and conjugation of liposomes to a DC-specific antibody. DC-specific siILs were then accordingly characterized to ensure proper size, siRNA encapsulation, and antibody conjugation were as previously reported. Thus, we used siILs containing 1 mol% excess DDAB because this formulation demonstrated a greater encapsulation capacity compared with neutral liposomes and did not exhibit significant nonspecific binding to DCs. Although the encapsulation efficiency of siRNA was relatively low compared with values reported for DNA plasmids,12,17,30 sufficient Cy3-labeled siRNA was incorporated within siILs to be detected during DC binding experiments in the current study. In addition, the encapsulated siRNA was also sufficient for silencing CD40 gene expression.
The liposome surface was decorated with several thousand strands of PEG2000, an inert hydrophilic biopolymer that reduces surface binding of plasma proteins and allows liposomes to persist in vivo by minimizing reticuloendothelial system uptake31 ; 1% to 2% of PEG2000 strands also contain maleimide groups at their distal ends, which are used for attaching the DC-specific mAb NLDC-145. This antibody acts as a targeting mechanism and recognizes the endocytic receptor DEC-205. Pivotal studies performed by Steinman's group have demonstrated that antibodies to this receptor can specifically and efficiently target DCs in vivo.32,33 When mice were subcutaneously injected with αDEC-205 mAb, most CD11c+ DCs, but not macrophages or lymphocytes, internalized the antibody. Furthermore, antigen targeted to DCs in vivo using conjugated αDEC-205 mAb increased the efficiency of antigen capture and presentation by 100- to 1000-fold. In the current study, we demonstrated that NLDC-145–guided, CD40 siILs can bind specifically to DCs in culture and dose-dependently silence CD40 expression (Figure 3C).
It is interesting to note that silencing efficacy of CD40 siILs was lower than that of CD40 siRNA/GP in vitro. This is not surprising because GP, a cationic lipid transfection reagent, binds negatively charged cell membranes through nonspecific electrostatic interactions. The high levels of nonspecific binding exhibited by CD40 siRNA/GP complexes can be seen in Figure 2B, as these bind to both BMDCs and L929 cells equally. Nevertheless, siILs are also capable of delivering siRNA into DCs in a dose-dependent fashion. When the dose increased, higher efficiency of gene silencing can be achieved, as shown in Figure 3C. Despite their efficiency in vitro, cationic lipid complexes have been described as unsafe for in vivo delivery because these particles rapidly aggregate the circulation34 with their unshielded excess positive charges inducing abnormal protein interaction,35,36 possibly being causative of clot formation.35 Moreover, siRNA/GP complexes lack any sort of targeting mechanism and cannot be specifically delivered to certain cells or tissues. Our newly developed siILs are nearly “neutral,” which causes less effective binding to cells and thus may lead to lower efficiency of siRNA transfection in vitro. However, these siILs are conjugated with a DC-specific mAb NLDC-145, which facilitates siRNA delivery specifically to DCs in vivo.
High-volume intravenous (hydrodynamic) injection of naked siRNA is currently the most common methodology for inducing RNAi in laboratory animals, despite the ubiquitous gene-silencing effects and lack of feasibility for human application.37 Consistent with the current literature,38,39 the majority of intravenously injected naked siRNA was concentrated in and cleared by the kidney soon after administration (Figure 4A). In contrast, siILs were primarily found in the liver at early time points and accumulated to high levels in specific anatomic regions of the spleen 48 hours after administration (Figure 4A). These exclusive areas of the spleen, in which siILs seemed to concentrate, may represent T-cell areas that are known to be rich in splenic DCs.40 Furthermore, CD40 gene silencing was observed to persist for at least 12 days after treatment with CD40 siILs. In contrast, previous reports indicate that, in mammalian cells, RNAi persists only for approximately 66 hours.26 Thus, our results suggest that siIL-mediated in vivo delivery may endow siRNA with a time-release property. Because siRNA is protected from endogenous nucleases within the immunoliposome interior and was targeted to DCs through a specific mAb, it is possible that siILs accumulated in the DC-populated areas of the spleen and slowly released their protected siRNA cargo, resulting in a prolonged gene-silencing effect.
An interesting observation was that, although siRNA did not induce long-lasting gene silencing, the transient silencing of DCs at early stage of immune response induction may subsequently induce vigorous immune modulation, as we reported previously.10 In support of this notion, in the current studies we observed that transiently silencing CD40 by siRNA resulted in an inhibition of KLH specific T-cell response (Figure 5). Nonetheless, compared with siRNA, siILs can specifically target DC population, which offers an alternative strategy for in vivo gene silencing when cell-type specificity is required.
One possible drawback of clinical translation of siILs is the reliance on antibodies as targeting agents. Not only chimeric, but also completely humanized, antibodies have been reported to elicit immune responses, sometimes cross-reacting with endogenous molecules and contributing to various adverse effects.41 Indeed, it may be that the concentrated antibodies found on the siILs will cause a more immunogenic reaction. Although in our studies we did not observe hypersensitive reactions to the siILs, this is a concern for clinical translation of the proposed approach. Conceptually, one may overcome this hurdle by use of less immunogenic targeting agents. For example, RNA-based aptamers have been previously used as targeting agents on liposomes as a substitute for antibodies.42 Other possible approaches include decorating the liposome with peptides components with high affinity toward DC-specific receptors.43
In conclusion, this is the first study to demonstrate the induction of DC-specific in vivo gene silencing using an siIL system. These nanovesicles can be used to specifically target CD40 siRNA to DCs in vivo and consequently induce antigen-specific immune suppression. This highlights the potential applications of siILs for in vivo siRNA delivery and DC-based immunotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Gill Strejan and Wayne Flintoff for their critical comments and suggestions and Hongtao Sun, Weihua Liu, Siobhan Ramcherran, and Jacqueline Arp for their technical assistance.
This work was supported by grants from the Heart and Stroke Foundation of Canada (Toronto, ON), Canadian Institutes of Health Science (Ottawa, ON), the Roche Organ Transplantation Research Foundation (Meggen, Switzerland), and the Multi-Organ Transplant Program at the London Health Sciences Centre (London, ON).
Authorship
Contribution: X. Zheng, C.V., X. Zhang, M.S., T.E.I., and M.L. performed research; X. Zheng, C.V., Z.-X.Z., E.C., and W.-P.M. analyzed data and wrote the paper; and W.-P.M., B.G., and A.M.J. designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wei-Ping Min, 339 Windermere Rd, University Hospital C9-136, London, ON, N6A 5A5, Canada; e-mail: weiping.min@uwo.ca.
References
Author notes
*X. Zheng and C.V. contributed equally to this study.


![Figure 5. CD40 siIL-induced immune suppression. (A) CD40 siILs inhibit allogenic T-cell proliferation in vitro. CD40 siIL-transfected BMDCs from C57/B6 mice were used as stimulators, and BALB/C splenocytes were used as effectors at 1:5 ratio in MLR. T-cell proliferation was measured by [3H]-thymidine incorporation, as described in “Mixed lymphocyte reaction.” (B) CD40 siILs inhibit KLH-specific recall response. Mice were immunized with KLH emulsified in CFA and simultaneously administered 15 μg CD40 siILs. Control groups included PBS, control siRNA, isotype IgG-coupled siILs. Naked siRNA was used as a positive control. T cells were isolated from the spleens of the siIL-treated mice and control mice, 12 days after KLH immunization and siIL treatment. Ag-specific T-cell response was assessed in the presence of 10 μg KLH antigen. Data presented are representative of 4 independent experiments (n = 3 or 4 mice/group). *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/12/10.1182_blood-2008-04-151191/4/m_zh80150933280005.jpeg?Expires=1765006577&Signature=xaWZ1m7JedR9NehlSGmRcl7DL3UUCsPSDgDEhPzrNWqfz7XNr3v3HlAsXZUT5NudinuRIvDK-V87aCh5mlcdWuiJGS08~Zdf2QRj~J1Fb6pQ5M4a44~C4HwpmPMMzsm6dO70NqAV9uiAD981TfKQ2VbvcfEnnXqLd5cTSbxoay2D2o5eIlsrZmfa4ZaUZjBBaugbQUTfxoXeRw0iRA-WnyJqTxAGcT3o6h7E2Ga~~nagFZKWJX1rT97-co9n6G~6sdxiPm4MmFQgOvJG4QRFobH84P3qk8X35Qwfs7rEKyYqzFT5x4mL~~nwWNTf~pHpvX2w4lCBJaffFDeIiOKnGw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal