Abstract
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder, characterized by the presence of BCR/ABL fusion gene. It is unclear which cellular events drive BCR/ABL gene translocation or initiate leukemogenesis in CML. Bcl-2 promotes survival of hematopoietic stem cells. Accordingly, apoptosis-related pathway may involve in the leukemogenesis of CML. In the current study, we evaluated 80 single nucleotide polymorphism (SNP) markers involved in the pathways of apoptosis (n = 30), angiogenesis (n = 7), myeloid cell growth (n = 14), xenobiotic metabolism (n = 13), WT1 signaling (n = 7), interferon signaling (n = 4), and others (n = 5) in 170 CML patients and 182 healthy controls. In a single-marker analysis, the following SNPs were identified including VEGFA, BCL2, CASP7, JAK3, CSF3, and HOCT1. In the multivariate logistic model with these SNPs and covariates, only BCL2 (rs1801018) was significantly associated with the susceptibility to CML (P = .05; odds ratio [OR] 2.16 [1.00-4.68]). In haplotype analyses, haplotype block of BCL2 consistently showed significant association with the susceptibility to CML. Risk allele analysis showed that a greater number of risk alleles from BCL2 SNP correlated to increasing risk of CML (overall P = .1, OR 1.84 [1.06-3.22] for 3-4 risk alleles vs 0-1 risk alleles). The current study indicated that BCL2 SNP seemed to be associated with increasing susceptibility to CML.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder, characterized by enhanced proliferative capacity and prolonged survival of hematopoietic stem cells (HSCs), reduced apoptosis, and altered cell adhesion properties. The causative event in the leukemogenesis of CML is formation of the BCR/ABL oncogene, which codes for a constitutively active bcr/abl fusion tyrosine kinase (FTK) on the Philadelphia chromosome (Ph). Although bcr/abl FTK is a key molecular marker of CML, it still remains to be understood which molecular or cellular events drive translocation of BCR/ABL gene or initiate leukemogenesis of CML. Previous observations showing the intriguing detection of BCR/ABL fusion gene at a very low level in the blood of healthy people1,2 suggest that only a minor fraction of spontaneous Ph translocations progress to CML, or that BCR/ABL fusion gene is essential but not sufficient to lead to the leukemogenesis without additional second cellular or molecular events
Several mechanisms have been proposed inducing leukemogenesis in CML. In the present study, we focused on 6 major candidate pathways that have been proposed to provoke leukemogenesis in CML: (1) apoptosis-related pathway, (2) myeloid cell growth pathway, (3) angiogenesis pathway, (4) multidrug resistance pathway, (5) WT1 (Wilms tumor gene) pathway, and (6) interferon signaling pathway.
For apoptosis-related pathway, several clinical and molecular studies have suggested that the expression levels of bcr/abl FTK may play a major role in resistance to apoptosis.3,4 Intrinsic-, extrinsic-, and Granzyme B-inducing signaling pathways are involved in the pathway of programmed cell death. The present study included several single nucleotide polymorphisms (SNPs) in the genes, including the bcl-2 family members (ie, BCL2, BCL6, BAX, BCL2L2, and BCL2L11), caspases (ie, CASP1, -3, -7, -8, -9, and -10), FAS, APAF1, tumor necrosis factor (TNF) receptor, Granzyme B, survivin (BIRC5), and PDCD1.
With features of CML characterized by a clonal proliferation and growth factor–independent myeloid cell growth, several candidate genes in the myeloid cell growth pathway were included into the study, such as CSF2 (G-CSF),5 CSF3 (GM-CSF),6,7 FLT3,8 JAK3,9,10 and IL1.11 In addition, with evidence that (1) increasing angiogenesis and higher vascular endothelial growth factor (VEGF) production was noted in CML patients,12–14 (2) ABCB1 (multidrug resistance-1; MDR1),15–17 and ABCG2 (breast cancer resistance protein [BCRP]) is highly expressed in CD34+ progenitor cells in CML patients,18 and (3) the WT1 gene may be involved in the leukemogenesis of CML,19 the SNPs in the pathways of angiogenesis, multidrug resistance, and WT1 signaling have been included into the study.
Recent advances in molecular epidemiologic study demonstrated the usefulness of SNP approaches to identify key molecular events of certain diseases. A few association studies had been pursued to identify the susceptible genetic variants for CML, but in a nonsystemic approach.20–22 In the present study, we included and analyzed multiple candidate gene SNPs for 80 polymorphisms in 6 major pathways associated with leukemogenesis of CML including apoptosis, angiogenesis, myeloid cell growth, multidrug resistance, cytokine, and WT1 signaling pathways in 170 CML patients and 182 healthy controls.
Methods
Patients and control population
The original case group was 170 white patients with Ph+ CML confirmed by BCR/ABL fusion gene mRNA reverse transcription polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) technique at the Princess Margaret Hospital (Toronto, ON) between August 2000 and December 2006. Sample collection was performed between March 2006 and April 2007 under the approval of the Research Ethics Board of the University Health Network, University of Toronto. All consecutive patients confirmed as CML at the Princess Margaret Hospital since January 2005 were enrolled into the sample collection that was done (n = 66); we denominated them the “de novo group.” Remaining patients were diagnosed before 2005 and were on therapy (n = 104) and were denominated the “on therapy group.” Patients' and disease characteristics at the time of blood sampling are shown in Table 1. There was no difference of achieving rates of complete cytogenetic or major molecular response between the de novo group and on therapy group after imatinib therapy, as shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The control group of 182 healthy white donors served as a control group. All blood samples were collected after informed consent had been obtained from the patient or donor in accordance with the Declaration of Helsinki.
Patient and disease characteristics
| Characteristic at time of sampling, n (%) . | All patients,170 (100) . | De novo,66 (39) . | On therapy,104 (61) . |
|---|---|---|---|
| Female/male | 72/98 (42/58) | 26/40 (39/61) | 46/58 (44/56) |
| Median age at diagnosis, y | 51.5 | 53.5 | 50.0 |
| Disease stage | |||
| Chronic phase | 151 (89) | 61 (92) | 90 (87) |
| Accelerated phase | 16 (9) | 5 (8) | 11 (10) |
| Blastic crisis | 3 (2) | 0 (0) | 3 (3) |
| Cytogenetics | |||
| t(9;22) only | 146 (86) | 55 (83) | 91 (87) |
| Additional abnormalities | 24 (14) | 11 (17) | 13 (13) |
| Median disease duration, mo | 31 | 8 | 52 |
| Median duration on imatinib, mo | 43 | 7 | 47 |
| Treatment | |||
| Imatinib mesylate | 170 (100) | 66 (100) | 104 (100) |
| Interferon | 34 (20) | 0 (0) | 34 (33) |
| Bone marrow transplantation | 11 (6) | 1 (2) | 10 (10) |
| Characteristic at time of sampling, n (%) . | All patients,170 (100) . | De novo,66 (39) . | On therapy,104 (61) . |
|---|---|---|---|
| Female/male | 72/98 (42/58) | 26/40 (39/61) | 46/58 (44/56) |
| Median age at diagnosis, y | 51.5 | 53.5 | 50.0 |
| Disease stage | |||
| Chronic phase | 151 (89) | 61 (92) | 90 (87) |
| Accelerated phase | 16 (9) | 5 (8) | 11 (10) |
| Blastic crisis | 3 (2) | 0 (0) | 3 (3) |
| Cytogenetics | |||
| t(9;22) only | 146 (86) | 55 (83) | 91 (87) |
| Additional abnormalities | 24 (14) | 11 (17) | 13 (13) |
| Median disease duration, mo | 31 | 8 | 52 |
| Median duration on imatinib, mo | 43 | 7 | 47 |
| Treatment | |||
| Imatinib mesylate | 170 (100) | 66 (100) | 104 (100) |
| Interferon | 34 (20) | 0 (0) | 34 (33) |
| Bone marrow transplantation | 11 (6) | 1 (2) | 10 (10) |
Sequenom MassARRAY genotyping system
The candidate genotypes have been selected by literature review and by choosing the SNPs in nonsynonymous SNPs in exon region with minor allele frequency greater than 0.05 to 0.1. If frequency was not available, the reported frequency in the Entrez SNP site (http://www.ncbi.nlm.nih.gov/sites/entrez) was used.
Genotyping was undertaken using the Sequenom iPLEX platform, according to the manufacturer's instructions (Sequenom, San Diego, CA). Whole blood samples were collected and informed consent was obtained in accordance with the Declaration of Helsinki. DNA was extracted using the Puregene DNA purification kit (Gentra Systems, Minneapolis, MN). The detection of SNPs was performed by the analysis of primer extension products generated from previously amplified genomic DNA using a Sequenom chip-based matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry platform. Multiplex SNP assays were designed using SpectroDesigner software (Sequenom). Ninety-six–well plates containing 2.5 ng DNA in each well were amplified by PCR following the specifications of Sequenom. Unincorporated nucleotides in the PCR product were deactivated using shrimp alkaline phosphatase. Allele discrimination reactions were conducted by adding the extension primer(s), DNA polymerase, and a cocktail mixture of deoxynucleotide triphosphates and dideoxynucleotide triphosphates to each well. MassExtend clean resin (Sequenom) was added to the mixture to remove extraneous salts that could interfere with MALDI-TOF analysis. The primer extension products were then cleaned and spotted onto a SpectroChip (Sequenom). Genotypes were determined by spotting an aliquot of each sample onto a 384 SpectroChip (Sequenom), which was subsequently read by the MALDI-TOF mass spectrometer. Duplicate samples and negative controls were included to check genotyping quality. The sequences of primers are listed in Table S1.
Preliminary analysis
Genotype and clinical information was maintained in our database. Genotype errors and genotype frequencies were summarized using Haploview version 3.32 (Broad Institute, Cambridge, MA; available at http://www.broad.mit.edu/mpg/haploview). The SNP ABCB1 (rs2032582) had more than 2 alleles and was coded in 2 ways: (1) GG, G/−, −/− and (2) T/T, T/−, −/−. The Hardy-Weinberg equilibrium was tested using the Pearson χ2 test. Descriptive statistics of demographic variables were generated using SAS version 9.1 (SAS Institute, Cary, NC). The Student t test and χ2 test were used to identify differences in demographic variables between the case and control cohorts.
Summary of 80 candidate gene single-nucleotide polymorphisms
| Gene . | SNP ID . | Gene description . | Chromosome . | Minor allele frequency . | Minor allele . |
|---|---|---|---|---|---|
| Apoptosis pathway (n = 30) | |||||
| BCL2 | rs1801018 | B-cell CLL/lymphoma 2 | 18 | 0.414 | G |
| BCL2 | rs2279115 | B-cell CLL/lymphoma 2 | 18 | 0.486 | C |
| BAX | rs11667351 | BCL2-associated X protein | 19 | 0.129 | G |
| BCL2L2 | rs7042474 | BCL2-like 2 | 9 | 0.127 | T |
| BCL6 | rs1056932 | B-cell CLL/lymphoma 6 | 3 | 0.341 | C |
| BCL6 | rs11545363 | B-cell CLL/lymphoma 6 | 3 | 0 | C |
| BCL2L11 | rs6746608 | BCL2-like 11 | 2 | 0.450 | A |
| BCL2L11 | rs12613243 | BCL2-like 11 | 2 | 0.072 | C |
| BIRC5 | rs9904341 | Survivin | 17 | 0.328 | C |
| BIRC5 | rs2071214 | Survivin | 17 | 0.057 | G |
| CASP1 | rs580253 | Caspase 1 | 11 | 0.184 | T |
| CASP3 | rs1049253 | Caspase 3 | 4 | 0.167 | C |
| CASP7 | rs7922608 | Caspase 7 | 10 | 0.313 | G |
| CASP8 | rs1045485 | Caspase 8 | 2 | 0.126 | C |
| CASP8 | rs3769818 | Caspase 8 | 2 | 0.276 | T |
| CASP8 | rs3834129 | Caspase 8 | 2 | 0.461 | C |
| CASP9 | rs4645981 | Caspase 9 | 1 | 0 | C |
| CASP10 | rs13010627 | Caspase 10 | 2 | 0.074 | A |
| CASP10 | rs13006529 | Caspase 10 | 2 | 0.458 | A |
| FASL | rs763110 | Fas ligand (TNF superfamily 6) | 1 | 0.383 | T |
| FAS | rs2234767 | Fas (TNF receptor superfamily 6) | 10 | 0.114 | A |
| FAS | rs1800682 | Fas (TNF receptor superfamily 6) | 10 | 0.473 | C |
| FAS | rs3218619 | Fas (TNF receptor superfamily 6) | 10 | 0.003 | A |
| FAS | rs3218612 | Fas (TNF receptor superfamily 6) | 10 | 0.042 | G |
| FAS | rs2234978 | Fas (TNF receptor superfamily 6) | 10 | 0.331 | T |
| APAF1 | rs1439123 | Apoptotic peptidase activating factor 1 | 12 | 0.224 | C |
| APAF1 | rs2288713 | Apoptotic peptidase activating factor 1 | 12 | 0.103 | G |
| TNFR2 | rs1061622 | TNF receptor superfamily, member 1B | 1 | 0.22 | G |
| PDCD1 | rs2227981 | Programmed cell death 1 | 2 | 0.418 | T |
| GZMB | rs7144366 | Granzyme B | 14 | 0.391 | T |
| Angiogenesis (n = 7) | |||||
| VEGFA | rs699947 | VEGFA | 6 | 0.476 | A |
| VEGFA | rs833061 | VEGFA | 6 | 0.480 | C |
| VEGFA | rs2010963 | VEGFA | 6 | 0.335 | C |
| VEGFA | rs3025039 | VEGFA | 6 | 0.142 | T |
| VEGFR2 | rs1531289 | VEGF receptor 2 | 4 | 0.302 | A |
| VEGFR2 | rs1870377 | VEGF receptor 2 | 4 | 0.228 | A |
| VEGFR2 | rs2305948 | VEGF receptor 2 | 4 | 0.092 | T |
| Myeloid growth (n = 14) | |||||
| FLT3 | rs35602083 | fms-related tyrosine kinase 3 | 13 | 0.035 | T |
| CSF3 | rs25645 | G-CSF | 17 | 0.389 | A |
| CSF3 | rs1042658 | G-CSF | 17 | 0.337 | T |
| CSF2 | rs25882 | GM-CSF | 5 | 0.225 | C |
| JAK3 | rs3008 | Janus kinase 3 | 19 | 0.447 | C |
| JAK3 | rs3212713 | Janus kinase 3 | 19 | 0.341 | A |
| IL1A | rs17561 | Interleukin 1 α | 2 | 0.274 | T |
| IL1A | rs1800587 | Interleukin 1 α | 2 | 0.277 | T |
| IL1B | rs1143634 | Interleukin 1 β | 2 | 0.222 | T |
| IL1B | rs1143633 | Interleukin 1 β | 2 | 0.358 | A |
| IL1B | rs1143627 | Interleukin 1 β | 2 | 0.371 | C |
| IL1B | rs16944 | Interleukin 1 β | 2 | 0.371 | A |
| IL1R | rs2228139 | Interleukin 1 receptor, type I | 2 | 0.060 | G |
| IL1R | rs315952 | Interleukin 1 receptor antagonist | 2 | 0.282 | C |
| WT1 signaling (n = 7) | |||||
| WT1 | rs1799937 | Wilms tumor 1 | 11 | 0.244 | C |
| WT1 | rs2234591 | Wilms tumor 1 | 11 | 0.006 | G |
| WT1 | rs2234590 | Wilms tumor 1 | 11 | 0.003 | G |
| WT1 | rs2301254 | Wilms tumor 1 | 11 | 0.380 | C |
| WT1 | rs2301252 | Wilms tumor 1 | 11 | 0.380 | T |
| WT1 | rs2301250 | Wilms tumor 1 | 11 | 0.380 | T |
| WT1 | rs6508 | Wilms tumor 1 | 11 | 0.067 | A |
| Xenobiotic metabolism (n = 13) | |||||
| ABCB1 | rs1045642 | Multidrug resistance 1 | 7 | 0.474 | C |
| ABCB1 | rs2032582 | Multidrug resistance 1 | 7 | 0.466 | T/G/A |
| ABCB1 | rs1128503 | Multidrug resistance 1 | 7 | 0.440 | T |
| ABCG2 | rs2231142 | Breast cancer resistance protein | 4 | 0.102 | A |
| ABCG2 | rs2231137 | Breast cancer resistance protein | 4 | 0.057 | A |
| CYP3A5 | rs776746 | Cytochrome P450, family 3, A5 | 7 | 0.060 | A |
| CYP3A5 | rs28383469 | Cytochrome P450, family 3, A5 | 7 | 0 | G |
| CYP3A5 | rs28383468 | Cytochrome P450, family 3, A5 | 7 | 0.006 | T |
| HOCT1 | rs1867351 | Solute carrier family 22, member 1 | 6 | 0.227 | G |
| HOCT1 | rs12208357 | Solute carrier family 22, member 1 | 6 | 0.070 | T |
| HOCT1 | rs683369 | Solute carrier family 22, member 1 | 6 | 0.201 | G |
| HOCT1 | rs2282143 | Solute carrier family 22, member 1 | 6 | 0.019 | T |
| HOCT1 | rs628031 | Solute carrier family 22, member 1 | 6 | 0.374 | A |
| Interferon signaling (n = 4) | |||||
| IFNG | rs1861494 | Interferon γ | 12 | 0.274 | C |
| IFNG | rs2069705 | Interferon γ | 12 | 0.326 | C |
| IFNGR1 | rs3799488 | Interferon γ receptor 1 | 6 | 0.105 | C |
| IFNGR2 | rs9808753 | Interferon γ receptor 2 | 21 | 0.164 | G |
| Other (n = 5) | |||||
| GNB3 | rs5443 | G protein β polypeptide 3 | 12 | 0.338 | T |
| ULK3 | rs2290573 | Unc-51–like kinase 3 | 15 | 0.449 | T |
| ORM | rs1126724 | Orosomucoid 1 | 9 | 0 | G |
| ORM | rs3182041 | Orosomucoid 1 | 9 | 0.003 | G |
| PTK2 | rs4554515 | PTK2 protein tyrosine kinase 2 | 8 | 0.452 | G |
| Gene . | SNP ID . | Gene description . | Chromosome . | Minor allele frequency . | Minor allele . |
|---|---|---|---|---|---|
| Apoptosis pathway (n = 30) | |||||
| BCL2 | rs1801018 | B-cell CLL/lymphoma 2 | 18 | 0.414 | G |
| BCL2 | rs2279115 | B-cell CLL/lymphoma 2 | 18 | 0.486 | C |
| BAX | rs11667351 | BCL2-associated X protein | 19 | 0.129 | G |
| BCL2L2 | rs7042474 | BCL2-like 2 | 9 | 0.127 | T |
| BCL6 | rs1056932 | B-cell CLL/lymphoma 6 | 3 | 0.341 | C |
| BCL6 | rs11545363 | B-cell CLL/lymphoma 6 | 3 | 0 | C |
| BCL2L11 | rs6746608 | BCL2-like 11 | 2 | 0.450 | A |
| BCL2L11 | rs12613243 | BCL2-like 11 | 2 | 0.072 | C |
| BIRC5 | rs9904341 | Survivin | 17 | 0.328 | C |
| BIRC5 | rs2071214 | Survivin | 17 | 0.057 | G |
| CASP1 | rs580253 | Caspase 1 | 11 | 0.184 | T |
| CASP3 | rs1049253 | Caspase 3 | 4 | 0.167 | C |
| CASP7 | rs7922608 | Caspase 7 | 10 | 0.313 | G |
| CASP8 | rs1045485 | Caspase 8 | 2 | 0.126 | C |
| CASP8 | rs3769818 | Caspase 8 | 2 | 0.276 | T |
| CASP8 | rs3834129 | Caspase 8 | 2 | 0.461 | C |
| CASP9 | rs4645981 | Caspase 9 | 1 | 0 | C |
| CASP10 | rs13010627 | Caspase 10 | 2 | 0.074 | A |
| CASP10 | rs13006529 | Caspase 10 | 2 | 0.458 | A |
| FASL | rs763110 | Fas ligand (TNF superfamily 6) | 1 | 0.383 | T |
| FAS | rs2234767 | Fas (TNF receptor superfamily 6) | 10 | 0.114 | A |
| FAS | rs1800682 | Fas (TNF receptor superfamily 6) | 10 | 0.473 | C |
| FAS | rs3218619 | Fas (TNF receptor superfamily 6) | 10 | 0.003 | A |
| FAS | rs3218612 | Fas (TNF receptor superfamily 6) | 10 | 0.042 | G |
| FAS | rs2234978 | Fas (TNF receptor superfamily 6) | 10 | 0.331 | T |
| APAF1 | rs1439123 | Apoptotic peptidase activating factor 1 | 12 | 0.224 | C |
| APAF1 | rs2288713 | Apoptotic peptidase activating factor 1 | 12 | 0.103 | G |
| TNFR2 | rs1061622 | TNF receptor superfamily, member 1B | 1 | 0.22 | G |
| PDCD1 | rs2227981 | Programmed cell death 1 | 2 | 0.418 | T |
| GZMB | rs7144366 | Granzyme B | 14 | 0.391 | T |
| Angiogenesis (n = 7) | |||||
| VEGFA | rs699947 | VEGFA | 6 | 0.476 | A |
| VEGFA | rs833061 | VEGFA | 6 | 0.480 | C |
| VEGFA | rs2010963 | VEGFA | 6 | 0.335 | C |
| VEGFA | rs3025039 | VEGFA | 6 | 0.142 | T |
| VEGFR2 | rs1531289 | VEGF receptor 2 | 4 | 0.302 | A |
| VEGFR2 | rs1870377 | VEGF receptor 2 | 4 | 0.228 | A |
| VEGFR2 | rs2305948 | VEGF receptor 2 | 4 | 0.092 | T |
| Myeloid growth (n = 14) | |||||
| FLT3 | rs35602083 | fms-related tyrosine kinase 3 | 13 | 0.035 | T |
| CSF3 | rs25645 | G-CSF | 17 | 0.389 | A |
| CSF3 | rs1042658 | G-CSF | 17 | 0.337 | T |
| CSF2 | rs25882 | GM-CSF | 5 | 0.225 | C |
| JAK3 | rs3008 | Janus kinase 3 | 19 | 0.447 | C |
| JAK3 | rs3212713 | Janus kinase 3 | 19 | 0.341 | A |
| IL1A | rs17561 | Interleukin 1 α | 2 | 0.274 | T |
| IL1A | rs1800587 | Interleukin 1 α | 2 | 0.277 | T |
| IL1B | rs1143634 | Interleukin 1 β | 2 | 0.222 | T |
| IL1B | rs1143633 | Interleukin 1 β | 2 | 0.358 | A |
| IL1B | rs1143627 | Interleukin 1 β | 2 | 0.371 | C |
| IL1B | rs16944 | Interleukin 1 β | 2 | 0.371 | A |
| IL1R | rs2228139 | Interleukin 1 receptor, type I | 2 | 0.060 | G |
| IL1R | rs315952 | Interleukin 1 receptor antagonist | 2 | 0.282 | C |
| WT1 signaling (n = 7) | |||||
| WT1 | rs1799937 | Wilms tumor 1 | 11 | 0.244 | C |
| WT1 | rs2234591 | Wilms tumor 1 | 11 | 0.006 | G |
| WT1 | rs2234590 | Wilms tumor 1 | 11 | 0.003 | G |
| WT1 | rs2301254 | Wilms tumor 1 | 11 | 0.380 | C |
| WT1 | rs2301252 | Wilms tumor 1 | 11 | 0.380 | T |
| WT1 | rs2301250 | Wilms tumor 1 | 11 | 0.380 | T |
| WT1 | rs6508 | Wilms tumor 1 | 11 | 0.067 | A |
| Xenobiotic metabolism (n = 13) | |||||
| ABCB1 | rs1045642 | Multidrug resistance 1 | 7 | 0.474 | C |
| ABCB1 | rs2032582 | Multidrug resistance 1 | 7 | 0.466 | T/G/A |
| ABCB1 | rs1128503 | Multidrug resistance 1 | 7 | 0.440 | T |
| ABCG2 | rs2231142 | Breast cancer resistance protein | 4 | 0.102 | A |
| ABCG2 | rs2231137 | Breast cancer resistance protein | 4 | 0.057 | A |
| CYP3A5 | rs776746 | Cytochrome P450, family 3, A5 | 7 | 0.060 | A |
| CYP3A5 | rs28383469 | Cytochrome P450, family 3, A5 | 7 | 0 | G |
| CYP3A5 | rs28383468 | Cytochrome P450, family 3, A5 | 7 | 0.006 | T |
| HOCT1 | rs1867351 | Solute carrier family 22, member 1 | 6 | 0.227 | G |
| HOCT1 | rs12208357 | Solute carrier family 22, member 1 | 6 | 0.070 | T |
| HOCT1 | rs683369 | Solute carrier family 22, member 1 | 6 | 0.201 | G |
| HOCT1 | rs2282143 | Solute carrier family 22, member 1 | 6 | 0.019 | T |
| HOCT1 | rs628031 | Solute carrier family 22, member 1 | 6 | 0.374 | A |
| Interferon signaling (n = 4) | |||||
| IFNG | rs1861494 | Interferon γ | 12 | 0.274 | C |
| IFNG | rs2069705 | Interferon γ | 12 | 0.326 | C |
| IFNGR1 | rs3799488 | Interferon γ receptor 1 | 6 | 0.105 | C |
| IFNGR2 | rs9808753 | Interferon γ receptor 2 | 21 | 0.164 | G |
| Other (n = 5) | |||||
| GNB3 | rs5443 | G protein β polypeptide 3 | 12 | 0.338 | T |
| ULK3 | rs2290573 | Unc-51–like kinase 3 | 15 | 0.449 | T |
| ORM | rs1126724 | Orosomucoid 1 | 9 | 0 | G |
| ORM | rs3182041 | Orosomucoid 1 | 9 | 0.003 | G |
| PTK2 | rs4554515 | PTK2 protein tyrosine kinase 2 | 8 | 0.452 | G |
Single-marker association
Several methods and models were used to test for single-marker associations between CML cases and controls. We evaluated individual SNPs using the χ2 test and logistic regression for the additive, dominant, and recessive genetic models, with and without adjusting for covariates. In addition, a permutation test for the additive model only (with 500 permutations per SNP) was used to confirm the single-marker SNP results using software WHAP version 2.09 (http://pngu.mgh.harvard.edu/∼purcell/whap/). Univariate logistic regression on each SNP using the additive model with case/control as the outcome was performed using SAS. Statistically significant and near-significant SNPs (P < .1) from the univariate logistic regression were entered into a multivariate logistic model with and without covariates. Odds ratios (OR) and 95% confidence intervals (CI) were estimated on the risk group compared with the referent group.
Haplotype analysis
The linkage disequilibrium (LD) plot was generated using Haploview. Haplotype blocks containing significantly associated SNPs from the single-marker association were selected for further analysis. The reconstructions of haplotype and the estimation of haplotype frequencies were using WHAP. Two methods were used to test the association of each haplotype block: (1) the Omnibus test and (2) the haplotype-specific test. The Omnibus test compares the effect of all haplotype within a block between cases and controls. The haplotype-specific test compares each haplotype versus all other haplotypes within a block, using the additive, dominant, and recessive models. The results were adjusted by covariates.
Risk allele analysis for each risk block was performed using SAS. For each block, we counted the total number of risk alleles in each block per patient. The number of risk alleles can range from 0 to 2, multiplied by the number of SNPs in the block. For example, Block 2 has 2 SNPs; thus, the number of risk alleles can range from 0 to 4. Then we recategorized them into 3 groups for each block. Logistic regression was used to test for the association of the number of risk alleles with CML susceptibility for each block as separate predictors. We assigned the group with the lowest number of risk alleles as the referent group. The results were also adjusted by covariates of age and sex.
Results
Study cohort
The study cohort consisted of 352 white persons, of which 170 (48.3%) were CML cases and 182 (51.7%) were controls. Overall, there were 199 (56.5%) females, and the mean age at diagnosis was 61.2 years (range, 13.7-102.0). However, a significantly greater proportion of controls (127/170, 69.8%; P < .001) were female compared with cases (72/182, 42.4%). In addition, the controls (median age, 72.4 years; range, 33.0-102.0 years) were significantly older than patients (median age, 49.1 years, range 13.7-77.1 years; P < .001). Accordingly, both sex and age were considered as important covariates in the analysis.
Genotype frequencies
A total of 80 SNP markers were included for analysis in the genes involved in the pathways of apoptosis (n = 30), angiogenesis (n = 7), myeloid cell growth (n = 14), xenobiotic metabolism (n = 13), WT1 signaling (n = 7), and interferon signaling (n = 4) and others (n = 5). The genotype frequencies and allele information for 80 SNPs are summarized in Table 2. From the 80 SNP markers, 4 SNPs were monomorphic in the current population: CASP9 (rs4645981), CYP3A5 (rs28383469), ORM (rs1126724), and BCL6 (rs11545363). These 4 SNPs were excluded from the analysis. Thus, the remaining 76 SNP markers were included in the study. All SNP genotypes satisfied the Hardy-Weinberg equilibrium.
Single-marker association
The adjusted and unadjusted univariate logistic regression results are presented in Table 3. Four SNPs were significantly associated to the cases: VEGFA (rs699947; P = .01; OR 1.47, [1.08-1.99]), VEGFA (rs833061; P = .02; OR 1.42, [1.05-1.92]), BCL2 (rs2279115; P = .03; OR 1.40, [1.03-1.9]), CASP7 (rs7922608; P = .05; OR 1.39, [1.00-1.93]). In addition, 5 other SNPs showed marginal significance of association (P < .1): JAK3 (rs3008), CSF3 (rs1042658), VEGFA (rs2010963), BCL2 (rs1801018), and HOCT1 (rs2282143). After adjusting the single-SNP analysis for covariates, only BCL2, rs1801018 (P = .03; adjusted OR [adjOR] 1.69, [1.06-2.72]) remained significant, whereas BCL2, rs2279115 was near significant (P = .09; adjOR 1.51, [0.94-2.43]). In the multivariate logistic model including 9 SNPs and the covariates, only BCL2, rs1801018 was significantly associated with the susceptibility to CML (P = .05; adjOR 2.16, [1.00-4.68]). Detailed results of single-marker analyses are presented in the Table S2.
Association of single SNPs with the susceptibility to chronic myeloid leukemia in univariate logistic regression analysis
| Gene . | SNP ID . | Assoc. allele . | Common allele . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|---|---|---|
| P . | OR (95% CI) . | P . | OR (95% CI) . | ||||
| JAK3 | rs3008 | T | C | .09* | 1.30 (0.96-1.75) | .90 | 0.97 (0.60-1.56) |
| CSF3 | rs1042658 | C | T | .07* | 1.34 (0.98-1.83) | .46 | 1.20 (0.74-1.92) |
| VEGFA | rs699947 | A | C | .01† | 1.47 (1.08-1.99) | .13 | 1.45 (0.90-2.35) |
| VEGFA | rs833061 | C | T | .02† | 1.42 (1.05-1.92) | .18 | 1.39 (0.86-2.23) |
| VEGFA | rs2010963 | G | C | .09* | 1.32 (0.96-1.81) | .49 | 1.20 (0.72-1.99) |
| BCL2 | rs1801018 | A | G | .10* | 1.29 (0.96-1.75) | .03† | 1.69 (1.06-2.72) |
| BCL2 | rs2279115 | C | A | .03† | 1.40 (1.03-1.90) | .09* | 1.51 (0.94-2.43) |
| CASP7 | rs7922608 | T | G | .05† | 1.39 (1.00-1.93) | .13 | 1.52 (0.89-2.61) |
| HOCT1 | rs2282143 | C | T | .08* | 3.27 (0.89-12.11) | .48 | 1.83 (0.35-9.70) |
| Gene . | SNP ID . | Assoc. allele . | Common allele . | Unadjusted . | Adjusted . | ||
|---|---|---|---|---|---|---|---|
| P . | OR (95% CI) . | P . | OR (95% CI) . | ||||
| JAK3 | rs3008 | T | C | .09* | 1.30 (0.96-1.75) | .90 | 0.97 (0.60-1.56) |
| CSF3 | rs1042658 | C | T | .07* | 1.34 (0.98-1.83) | .46 | 1.20 (0.74-1.92) |
| VEGFA | rs699947 | A | C | .01† | 1.47 (1.08-1.99) | .13 | 1.45 (0.90-2.35) |
| VEGFA | rs833061 | C | T | .02† | 1.42 (1.05-1.92) | .18 | 1.39 (0.86-2.23) |
| VEGFA | rs2010963 | G | C | .09* | 1.32 (0.96-1.81) | .49 | 1.20 (0.72-1.99) |
| BCL2 | rs1801018 | A | G | .10* | 1.29 (0.96-1.75) | .03† | 1.69 (1.06-2.72) |
| BCL2 | rs2279115 | C | A | .03† | 1.40 (1.03-1.90) | .09* | 1.51 (0.94-2.43) |
| CASP7 | rs7922608 | T | G | .05† | 1.39 (1.00-1.93) | .13 | 1.52 (0.89-2.61) |
| HOCT1 | rs2282143 | C | T | .08* | 3.27 (0.89-12.11) | .48 | 1.83 (0.35-9.70) |
P < .1.
P < .05.
The single-marker analysis results incorporating the additive, dominant, and recessive genetic models are presented in Table 4. SNPs having at least one significant association or SNPs belonging to a haplotype block are displayed. The 2 BCL2 SNPs (rs1801018 and rs2279115) were near significantly associated to increasing susceptibility to CML (P = .09 and P = .03, respectively). They remained near significant even after adjusting for sex and age (P = .06 and P = .07). The VEGFA SNPs rs699947 (P = .01), rs833061 (P = .02), and rs2010963 (P = .08) belong to haplotype block 3, and they had strong associations with CML cases without adjusting for covariates. However, after adjustment, only VEGFA, rs699947 was marginally significant (P = .10). As an internal validation of the single-marker results, the single SNP permutation test results, only for the additive model, are presented in the Table S3. Overall, the permutation test results are in agreement with the model-based results.
Association of single SNPs using the WHAP method
| Gene . | SNP ID . | P by WHAP . | Adjusted P by WHAP . | ||||
|---|---|---|---|---|---|---|---|
| Additive model . | Dominant model . | Recessive model . | Additive model . | Dominant model . | Recessive model . | ||
| BCL2 | rs1801018 | .09* | .15 | .20 | .06* | .12 | .10 |
| BCL2 | rs2279115 | .03† | .12 | .05* | .07* | .28 | .07* |
| BAX | rs11667351 | .14 | .24 | .16 | .05* | .06* | .24 |
| BCL6 | rs1056932 | .38 | .63 | .28 | .56 | .65 | .07* |
| CASP3 | rs1049253 | .16 | .09* | .92 | .09* | .16 | .11 |
| CASP7 | rs7922608 | .05† | .12 | .09* | .04* | .14 | .05* |
| CASP8 | rs1045485 | .57 | .27 | .26 | .56 | .30 | .44 |
| CASP8 | rs3769818 | .70 | .29 | .26 | .37 | .89 | .04† |
| CASP8 | RS3834129 | .36 | .66 | .28 | .12 | .18 | .24 |
| CASP10 | rs13010627 | .35 | .32 | .96 | .05* | .04† | .79 |
| CASP10 | rs13006529 | .29 | .58 | .22 | .09* | .16 | .18 |
| FAS | rs2234767 | .91 | .68 | .37 | .87 | .76 | .20 |
| FAS | rs1800682 | .22 | .09* | .80 | .64 | .17 | .50 |
| VEGFA | rs699947 | .01† | .05† | .04† | .10* | .47 | .04† |
| VEGFA | rs833061 | .02† | .07* | .06* | .16 | .60 | .07* |
| VEGFA | rs2010963 | .08* | .12 | .21 | .30 | .48 | .28 |
| VEGFR2 | rs1531289 | .22 | .13 | .86 | <.001† | .55 | <.001† |
| VEGFR2 | rs1870377 | .96 | .74 | .40 | .92 | .90 | .62 |
| VEGFR2 | rs2305948 | .17 | .25 | .10 | .43 | .47 | .52 |
| CSF3 | rs25645 | .60 | .38 | .80 | .30 | .26 | .66 |
| CSF3 | rs1042658 | .06* | .09* | .21 | .65 | .82 | .54 |
| JAK3 | rs3008 | .09* | .15 | .18 | .77 | .80 | .41 |
| JAK3 | rs3212713 | .15 | .54 | .04† | .52 | .44 | .93 |
| IL1R | rs2228139 | .45 | .63 | .10 | .26 | .42 | .09* |
| WT1 | rs1799937 | .16 | .24 | .26 | .09* | .18 | .13 |
| CYP3A5 | rs776746 | .11 | .13 | .34 | .03† | .03† | .32 |
| HOCT1 | rs1867351 | .76 | .83 | .21 | .78 | .82 | .81 |
| HOCT1 | rs12208357 | .26 | .31 | .25 | .36 | .37 | .77 |
| HOCT1 | rs683369 | .85 | .94 | .40 | .76 | .89 | .56 |
| HOCT1 | rs2282143 | .05* | .05* | >.99 | .54 | .54 | >.99 |
| HOCT1 | rs628031 | .72 | .64 | .96 | .47 | .89 | .13 |
| Gene . | SNP ID . | P by WHAP . | Adjusted P by WHAP . | ||||
|---|---|---|---|---|---|---|---|
| Additive model . | Dominant model . | Recessive model . | Additive model . | Dominant model . | Recessive model . | ||
| BCL2 | rs1801018 | .09* | .15 | .20 | .06* | .12 | .10 |
| BCL2 | rs2279115 | .03† | .12 | .05* | .07* | .28 | .07* |
| BAX | rs11667351 | .14 | .24 | .16 | .05* | .06* | .24 |
| BCL6 | rs1056932 | .38 | .63 | .28 | .56 | .65 | .07* |
| CASP3 | rs1049253 | .16 | .09* | .92 | .09* | .16 | .11 |
| CASP7 | rs7922608 | .05† | .12 | .09* | .04* | .14 | .05* |
| CASP8 | rs1045485 | .57 | .27 | .26 | .56 | .30 | .44 |
| CASP8 | rs3769818 | .70 | .29 | .26 | .37 | .89 | .04† |
| CASP8 | RS3834129 | .36 | .66 | .28 | .12 | .18 | .24 |
| CASP10 | rs13010627 | .35 | .32 | .96 | .05* | .04† | .79 |
| CASP10 | rs13006529 | .29 | .58 | .22 | .09* | .16 | .18 |
| FAS | rs2234767 | .91 | .68 | .37 | .87 | .76 | .20 |
| FAS | rs1800682 | .22 | .09* | .80 | .64 | .17 | .50 |
| VEGFA | rs699947 | .01† | .05† | .04† | .10* | .47 | .04† |
| VEGFA | rs833061 | .02† | .07* | .06* | .16 | .60 | .07* |
| VEGFA | rs2010963 | .08* | .12 | .21 | .30 | .48 | .28 |
| VEGFR2 | rs1531289 | .22 | .13 | .86 | <.001† | .55 | <.001† |
| VEGFR2 | rs1870377 | .96 | .74 | .40 | .92 | .90 | .62 |
| VEGFR2 | rs2305948 | .17 | .25 | .10 | .43 | .47 | .52 |
| CSF3 | rs25645 | .60 | .38 | .80 | .30 | .26 | .66 |
| CSF3 | rs1042658 | .06* | .09* | .21 | .65 | .82 | .54 |
| JAK3 | rs3008 | .09* | .15 | .18 | .77 | .80 | .41 |
| JAK3 | rs3212713 | .15 | .54 | .04† | .52 | .44 | .93 |
| IL1R | rs2228139 | .45 | .63 | .10 | .26 | .42 | .09* |
| WT1 | rs1799937 | .16 | .24 | .26 | .09* | .18 | .13 |
| CYP3A5 | rs776746 | .11 | .13 | .34 | .03† | .03† | .32 |
| HOCT1 | rs1867351 | .76 | .83 | .21 | .78 | .82 | .81 |
| HOCT1 | rs12208357 | .26 | .31 | .25 | .36 | .37 | .77 |
| HOCT1 | rs683369 | .85 | .94 | .40 | .76 | .89 | .56 |
| HOCT1 | rs2282143 | .05* | .05* | >.99 | .54 | .54 | >.99 |
| HOCT1 | rs628031 | .72 | .64 | .96 | .47 | .89 | .13 |
P < .1.
P < .05.
To consider potential effect of certain genotype influencing on the disease progression, we compared the genotype and allele frequencies of all the SNPs between the de novo versus the on therapy groups (Table S4A), but without significant statistical differences on the critical (Table S4B) and between on therapy group versus control group (Table S4C). The effect size (OR) of both subgroup analysis was very close to the original analysis with all the subjects (Table S5).
Haplotype analysis
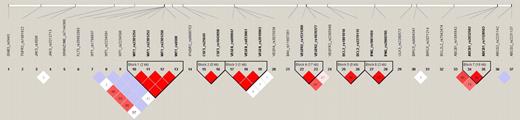
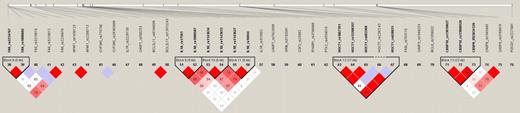
Haplotype blocks were identified from the LD plot (Figure 1). Five haplotype blocks containing significantly associated single SNPs were selected for further haplotype analysis as shown in Figure 1 (blocks 2, 3, 5, 8, and 15). The reconstructed haplotypes and their estimated haplotype frequencies are displayed in Table 5. The results of the omnibus test (Omni) and the haplotype-specific tests for the 3 genetic models are presented. Haplotype blocks 2 (CSF3), 3 (VEGFA), and 5 (BCL2) had significant associations before adjusting for covariates. After adjustment, blocks 5 (BCL2) and 13 (CASP10) had marginally significant associations.
Linkage disequilibrium plot of 76 single-nucleotide polymorphisms.
The outcomes of haplotype analysis and its potential association with the susceptibility to chronic myeloid leukemia
| Block . | Gene . | SNP ID . | Haplo-type . | Frequency . | Unadjusted . | Adjusted . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 . | SNP2 . | SNP3 . | Overall . | Case . | Control . | Omnibus P . | Haplotype-specific P . | Omnibus P . | Haplotype-specific P . | |||
| 2 | CSF3 | rs25645 | rs1042658 | |||||||||
| A | C | AC | 0.389 | 0.399 | 0.380 | .15 | .58 | .55 | .28 | |||
| G | T | GT | 0.338 | 0.303 | 0.370 | .06* | .64 | |||||
| G | C | GC | 0.273 | 0.297 | 0.250 | .14 | .53 | |||||
| 3 | VEGFA | rs699947 | rs833061 | rs2010963 | ||||||||
| A | C | G | ACG | 0.478 | 0.525 | 0.433 | .04† | .01† | .30 | .12 | ||
| C | T | C | CTC | 0.335 | 0.304 | 0.364 | .09* | .28 | ||||
| C | T | G | CTG | 0.187 | 0.171 | 0.203 | .27 | .51 | ||||
| 5 | BCL2 | rs1801018 | rs2279115 | |||||||||
| A | C | AC | 0.483 | 0.523 | 0.447 | .12 | .04† | .16 | .09* | |||
| G | A | GA | 0.409 | 0.379 | 0.436 | .12 | .06* | |||||
| A | A | AA | 0.108 | 0.098 | 0.117 | .43 | .74 | |||||
| 8 | FAS | rs2234767 | rs1800682 | |||||||||
| G | T | GT | 0.528 | 0.505 | 0.551 | .39 | .22 | 1.00 | .65 | |||
| G | C | GC | 0.357 | 0.382 | 0.334 | .18 | .70 | |||||
| A | C | AC | 0.114 | 0.113 | 0.115 | .93 | .87 | |||||
| 13 | CASP10 | rs13010627 | rs13006529 | rs3834129 | ||||||||
| G | T | A | GTA | 0.54 | 0.518 | 0.558 | .49 | .30 | .08* | .09* | ||
| G | A | C | GAC | 0.386 | 0.398 | 0.376 | .56 | .51 | ||||
| A | A | C | AAC | 0.074 | 0.084 | 0.066 | .37 | .05* | ||||
| Block . | Gene . | SNP ID . | Haplo-type . | Frequency . | Unadjusted . | Adjusted . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 . | SNP2 . | SNP3 . | Overall . | Case . | Control . | Omnibus P . | Haplotype-specific P . | Omnibus P . | Haplotype-specific P . | |||
| 2 | CSF3 | rs25645 | rs1042658 | |||||||||
| A | C | AC | 0.389 | 0.399 | 0.380 | .15 | .58 | .55 | .28 | |||
| G | T | GT | 0.338 | 0.303 | 0.370 | .06* | .64 | |||||
| G | C | GC | 0.273 | 0.297 | 0.250 | .14 | .53 | |||||
| 3 | VEGFA | rs699947 | rs833061 | rs2010963 | ||||||||
| A | C | G | ACG | 0.478 | 0.525 | 0.433 | .04† | .01† | .30 | .12 | ||
| C | T | C | CTC | 0.335 | 0.304 | 0.364 | .09* | .28 | ||||
| C | T | G | CTG | 0.187 | 0.171 | 0.203 | .27 | .51 | ||||
| 5 | BCL2 | rs1801018 | rs2279115 | |||||||||
| A | C | AC | 0.483 | 0.523 | 0.447 | .12 | .04† | .16 | .09* | |||
| G | A | GA | 0.409 | 0.379 | 0.436 | .12 | .06* | |||||
| A | A | AA | 0.108 | 0.098 | 0.117 | .43 | .74 | |||||
| 8 | FAS | rs2234767 | rs1800682 | |||||||||
| G | T | GT | 0.528 | 0.505 | 0.551 | .39 | .22 | 1.00 | .65 | |||
| G | C | GC | 0.357 | 0.382 | 0.334 | .18 | .70 | |||||
| A | C | AC | 0.114 | 0.113 | 0.115 | .93 | .87 | |||||
| 13 | CASP10 | rs13010627 | rs13006529 | rs3834129 | ||||||||
| G | T | A | GTA | 0.54 | 0.518 | 0.558 | .49 | .30 | .08* | .09* | ||
| G | A | C | GAC | 0.386 | 0.398 | 0.376 | .56 | .51 | ||||
| A | A | C | AAC | 0.074 | 0.084 | 0.066 | .37 | .05* | ||||
P < .1.
P < .05.
Risk allele frequencies for the 5 risk blocks are displayed in Table 6. The frequencies were categorized into 3 groups, indicated by white, light gray, and dark gray shading. The logistic regression results for the impact of number of alleles on the susceptibility of CML are presented in Table 7. Having a greater number of risk alleles from block 5 for BCL2 was near significantly associated to cases (overall P = .10; 3-4 versus 0-1 risk alleles, OR 1.84, 95% CI [1.06-3.22]). However, after adjusting for age and sex, these effects were no longer statistically significant.
Risk allele frequencies in each haplotype blocks
| SNPs and risk allele . | Frequency of risk alleles (%) . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Block . | Gene . | Chromo-some . | SNP ID . | Position . | Risk allele . | 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
| 2 | CSF3 | 17 | rs25645 | 35426669 | A | 46* | 54* | 119† | 90‡ | 43‡ | ||
| rs1042658 | 35427428 | C | (13.1) | (15.3) | (33.8) | (25.6) | (12.2) | |||||
| 3 | VEGFA | 6 | rs699947 | 43844367 | A | 43* | 41* | 29* | 100† | 61‡ | 1‡ | 77‡ |
| rs833061 | 43845464 | C | (12.2) | (11.6) | (8.2) | (28.4) | (17.3) | (0.3) | (21.9) | |||
| rs2010963 | 43846328 | G | ||||||||||
| 5 | BCL2 | 18 | rs1801018 | 59136859 | A | 59* | 32* | 144† | 41‡ | 76‡ | ||
| rs2279115 | 59137817 | C | (16.8) | (9.1) | (40.9) | (11.6) | (21.6) | |||||
| 8 | FAS | 10 | rs2234767 | 90739236 | G | 2* | 4* | 145† | 159‡ | 42‡ | ||
| rs1800682 | 90739943 | C | (0.6) | (1.1) | (41.2) | (45.2) | (11.9) | |||||
| 13 | CASP10 | 2 | rs13010627 | 201782343 | A | 104* | 4* | 141† | 28† | 54‡ | 19‡ | 2‡ |
| rs13006529 | 201790704 | A | (29.5) | (1.1) | (40.1) | (8.0) | (15.3) | (5.4) | (0.6) | |||
| rs3834129 | 201805777 | C | ||||||||||
| SNPs and risk allele . | Frequency of risk alleles (%) . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Block . | Gene . | Chromo-some . | SNP ID . | Position . | Risk allele . | 0 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . |
| 2 | CSF3 | 17 | rs25645 | 35426669 | A | 46* | 54* | 119† | 90‡ | 43‡ | ||
| rs1042658 | 35427428 | C | (13.1) | (15.3) | (33.8) | (25.6) | (12.2) | |||||
| 3 | VEGFA | 6 | rs699947 | 43844367 | A | 43* | 41* | 29* | 100† | 61‡ | 1‡ | 77‡ |
| rs833061 | 43845464 | C | (12.2) | (11.6) | (8.2) | (28.4) | (17.3) | (0.3) | (21.9) | |||
| rs2010963 | 43846328 | G | ||||||||||
| 5 | BCL2 | 18 | rs1801018 | 59136859 | A | 59* | 32* | 144† | 41‡ | 76‡ | ||
| rs2279115 | 59137817 | C | (16.8) | (9.1) | (40.9) | (11.6) | (21.6) | |||||
| 8 | FAS | 10 | rs2234767 | 90739236 | G | 2* | 4* | 145† | 159‡ | 42‡ | ||
| rs1800682 | 90739943 | C | (0.6) | (1.1) | (41.2) | (45.2) | (11.9) | |||||
| 13 | CASP10 | 2 | rs13010627 | 201782343 | A | 104* | 4* | 141† | 28† | 54‡ | 19‡ | 2‡ |
| rs13006529 | 201790704 | A | (29.5) | (1.1) | (40.1) | (8.0) | (15.3) | (5.4) | (0.6) | |||
| rs3834129 | 201805777 | C | ||||||||||
Frequencies are categorized into 3 groups:
indicates low-risk or reference group;
, intermediate-risk group; and
, high-risk group.
Impact of the number of risk alleles on susceptibility to chronic myeloid leukemia
| Block, gene . | Number of risk alleles . | Unadjusted . | Adjusted . | |||
|---|---|---|---|---|---|---|
| Risk group . | Referent group . | P . | OR (95% CI) . | P . | OR (95% CI) . | |
| 2, CSF3 | .28 | .76 | ||||
| 2 | 0-1 | .84 | 1.18 (0.69-2.01) | .88 | 1.22 (0.54-2.73) | |
| 3-4 | 0-1 | .13 | 1.52 (0.90-2.56) | .55 | 1.34 (0.62-2.93) | |
| 3, VEGFA | .49 | .73 | ||||
| 3 | 0-2 | 1.00 | 1.16 (0.68-2.00) | .99 | 1.15 (0.52-2.54) | |
| 4-6 | 0-2 | .30 | 1.35 (0.82-2.23) | .49 | 1.34 (0.64-2.81) | |
| 5, BCL2 | .10 | .23 | ||||
| 2 | 0-1 | .78 | 1.45 (0.85-2.46) | .95 | 1.4 (0.62-3.15) | |
| 3-4 | 0-1 | .06 | 1.84 (1.06-3.22) | .10 | 2.06 (0.89-4.75) | |
| 8, FAS | .29 | .58 | ||||
| 2 | 0-1 | .87 | 1.58 (0.28-8.90) | .94 | 1.78 (0-1183.53) | |
| 3-4 | 0-1 | .25 | 2.14 (0.38-11.97) | .71 | 2.47 (0-1632.03) | |
| 13, CASP10 | .46 | .25 | ||||
| 2-3 | 0-1 | .49 | 1.02 (0.63-1.65) | .99 | 1.45 (0.70-2.99) | |
| 4-6 | 0-1 | .21 | 1.40 (0.77-2.53) | .15 | 2.12 (0.87-5.17) | |
| Block, gene . | Number of risk alleles . | Unadjusted . | Adjusted . | |||
|---|---|---|---|---|---|---|
| Risk group . | Referent group . | P . | OR (95% CI) . | P . | OR (95% CI) . | |
| 2, CSF3 | .28 | .76 | ||||
| 2 | 0-1 | .84 | 1.18 (0.69-2.01) | .88 | 1.22 (0.54-2.73) | |
| 3-4 | 0-1 | .13 | 1.52 (0.90-2.56) | .55 | 1.34 (0.62-2.93) | |
| 3, VEGFA | .49 | .73 | ||||
| 3 | 0-2 | 1.00 | 1.16 (0.68-2.00) | .99 | 1.15 (0.52-2.54) | |
| 4-6 | 0-2 | .30 | 1.35 (0.82-2.23) | .49 | 1.34 (0.64-2.81) | |
| 5, BCL2 | .10 | .23 | ||||
| 2 | 0-1 | .78 | 1.45 (0.85-2.46) | .95 | 1.4 (0.62-3.15) | |
| 3-4 | 0-1 | .06 | 1.84 (1.06-3.22) | .10 | 2.06 (0.89-4.75) | |
| 8, FAS | .29 | .58 | ||||
| 2 | 0-1 | .87 | 1.58 (0.28-8.90) | .94 | 1.78 (0-1183.53) | |
| 3-4 | 0-1 | .25 | 2.14 (0.38-11.97) | .71 | 2.47 (0-1632.03) | |
| 13, CASP10 | .46 | .25 | ||||
| 2-3 | 0-1 | .49 | 1.02 (0.63-1.65) | .99 | 1.45 (0.70-2.99) | |
| 4-6 | 0-1 | .21 | 1.40 (0.77-2.53) | .15 | 2.12 (0.87-5.17) | |
Discussion
The key finding of the current study is that the genetic variant of BCL2 gene is significantly associated with increasing susceptibility to CML. The bcr/abl fusion tyrosine kinase contributes to increasing proliferative capacity, diminished programmed cell death potential, mitogenic activation, and growth factor independence.23,24
Several studies attempted to generate CML model in mice by introducing p210BCR/ABL fusion gene to hematopoietic cells using retroviral transduction or germ line transgenesis. Retroviral transduction of BCR/ABL into irradiated mouse bone marrow (BM) generated a CML-like disease. But, it often showed a lack of chronic phase disease, predominance of lymphoid malignancies.25–30 In animal models, bcr/abl was shown to be sufficient for inducing leukemia31 and necessary for its maintenance.32 However, the intriguing detection of BCR/ABL fusion gene at a very low level in the blood of healthy people1,2 suggest that only a minor fraction of spontaneous Ph translocation terminate in a myeloproliferative disorder. It is plausible to explain that a successful initiation of tumorigenesis may require other predisposing cellular or molecular events. Accordingly, BCR/ABL chimeric gene alone is insufficient for the development of CML. Jaiswal et al30 reported that BCR/ABL transgenic mice developed a chronic myeloproliferative disorder. But when they crossed bcr/abl transgenic mice to apoptosis-resistant mice with bcl-2, transition of myeloproliferative disorder to myeloid blastic crisis was more frequently observed.30 Bcl-2 may promote the survival of bcr/abl-expressing cells, enabling them to accumulate additional genetic lesions and predominate among HSCs.
HSC survival is regulated by apoptosis. One of the important functions of bcr/abl fusion tyrosine kinase is to inhibit apoptosis and prolong survival of myeloid cells.3 In addition, deregulated BCL2 gene expression selectively prolongs survival of growth factor-independent HSCs.33 In addition, bcr/abl tyrosine kinase has been reported to alter the ratio of pro- versus antiapoptotic bcl-2 family members.34 Sanchez-Garcia et al35 showed that BCR/ABL chimeric gene prevented apoptotic death by inducting bcl-2 expressing pathway and that bcr/abl-expressing cells revert to growth factor dependence and nontumorigenicity after bcl-2 expression was suppressed. A report also showed an excellent correlation between the amount of bcr/abl protein, the level of bcl-2 induction, and the degree of resistance to apoptosis.36 In addition, more frequent bcl-2 expression was observed in patients with advanced phase of CML.37 A recent study also demonstrated that the expression of apoptosis-related genes such as BCL2 or BAD is significantly reduced in the CML patients before imatinib therapy compared with healthy individuals.38
In the current study, the BCL2 SNP was suggested to be associated with susceptibility to CML. Single-marker and haplotype analyses consistently showed similar findings that the case with BCL2 risk allele had a 1.3- to 1.7-fold higher risk of developing CML. In addition, risk allele analysis for risk block of BCL2 showed that the group with 3 to 4 risk alleles showed 1.8-fold higher risk of increasing susceptibility to CML compared with those with 0 to 1 risk allele.
CML is a quite rare disease. Its annual incidence in Western countries is 1.8 per 100 000 population. With current technology, predicting CML is almost impossible. However, the current study showed that the SNP approach, using multiple candidate gene SNPs targeting multiple candidate pathways, could be useful in predicting the risk of CML in the general population.
Other candidate gene SNPs were suggested, such as VEGFA or CASP10, based on the current result. Further studies focusing on these SNPs are strongly warranted, especially in other ethnic groups and with larger numbers of cases.
The limitation of the present study is the selection of the control group. The impact of the covariates, including age and gender, was too strong. This control group had also served as a control group for another association study of the susceptibility to osteoarthritis in the white population. Accordingly, the group was old (median age, 72.4 years) and predominantly female. However, they were free of CML beyond the median age (49.1 years) of diagnosis of CML. Thus, the SNPs that showed a strong positive signal in the univariate analysis, but not in the multivariate analysis, should be reevaluated and analyzed in another study.
In conclusion, BCL2 SNP showed consistently significant association with increasing susceptibility to CML in both single-marker analysis and haplotype analysis. To reach a clear conclusion on this issue, further study will be needed to validate the current result with larger numbers of cases from different ethnic groups.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a grant from the Friends to Life Fund, Princess Margaret Hospital Foundation (Toronto, ON).
Authorship
Contribution: D.K. contributed to the design of the study, the supervision of data collection and interpretation, data analysis, and writing the manuscript; W.X. contributed to the design of the study, data interpretation, data analysis, and critical revision of the manuscript; C.M. was involved in the design of the study, data analysis, and critical revision of the manuscript; X.L. and K.S. were involved in the genotyping experiments and verification of the results; and H.M. and J.L. contributed to the design of the study, the supervision of data collection and interpretation, data analysis, and critical revision of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong Hwan (Dennis) Kim, Department of Hematology/Oncology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Irwon-dong 50, Gangnam-gu, Seoul, Korea, 135-710; e-mail: drkiim@medimail.co.kr; or Jeffrey H. Lipton, Chronic Myelogenous Leukemia Group, Princess Margaret Hospital, University of Toronto, 610 University Avenue, Toronto, ON M5G 2M9 Canada; e-mail: jeff.lipton@uhn.on.ca.
References
Author notes
*D.K. and W.X. contributed equally to the paper.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal