Abstract
Polymorphisms in folate pathway genes may influence the susceptibility to acute lymphoblastic leukemia (ALL). DNA was isolated from 245 pediatric ALL patients (cases) and from 500 blood bank donors (controls). Polymorphisms in methylene-tetrahydrofolate reductase (MTHFR 677C>T, 1298A>C), methionine synthase (MTR 2756A>G), methionine synthase reductase (MTRR 66A>G), methylenetetrahydrofolate dehydrogenase (MTHFD1 1958G>A), nicotinamide N-methyltransferase (NNMT IVS −151C>T), serine hydroxymethyl transferase (SHMT1 1420C>T), thymidylate synthase (TS 2R3R), and the reduced folate carrier (RFC1 80G>A) were detected. In ALL patients, an increased occurrence was observed of the RFC1 80AA variant (odds ratio [OR] = 2.1; 95% confidence interval [CI] = 1.3-3.2; P = .002) and the RFC1 80A allele (OR = 1.5; 95% CI, 1.1-2.1; P = .02). Likewise, the NNMT IVS −151TT genotype showed a 2.2-fold increased ALL risk (OR = 2.2; 95% CI, 1.1-4.6; P = .04). A 1.4-fold reduction in ALL risk was observed for (heterozygous or homozygous) carriers of the TS 2R allele and the MTHFR 677T allele (OR = 0.7; 95% CI, 0.5-1.0; P < .05). Furthermore, interactions between NNMT and MTHFR 677C>T and RFC1 were observed. NNMT IVS −151CC/MTHFR 677CT + TT patients exhibited a 2-fold reduction in ALL risk whereas RFC1 80AA/NNMT IVS −151CT + TT subjects had a 4.2-fold increase in ALL risk (P = .001). For the first time, we associate the RFC1 80G>A and NNMT IVS −151C>T variants to an increased ALL susceptibility.
Introduction
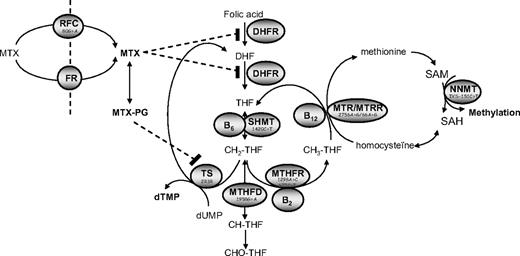
Folate metabolism is essential for cellular functioning because it provides one-carbon donors for the synthesis of de novo purines and pyrimidines necessary for RNA and DNA synthesis, the remethylation of homocysteine, and methylation reactions (eg, DNA methylation). Folate metabolism may become deranged by inadequate nutrition, altered cellular transport, and polymorphisms in folate-related genes. Several polymorphisms in folate-related genes have been described (Figure 1), although the biochemical consequences of most of these genes are unclear. However, 2 genetic variants in methylenetetrahydrofolate reductase (MTHFR 677 C>T and 1298 A>C) reduce enzyme activity, and the MTHFR 677TT variant also alters normal intracellular folate distribution: folates committed to purine and pyrimidine synthesis accumulate at the expense of 5-methyltetrahydrofolate (the most abundant form in wild-type MTHFR 677 CC subjects).1-3 The effects of other polymorphisms in folate-related genes on folate metabolism are unclear. However, the TS 3R/3R variant has been associated with an increased transcriptional activity in in vitro assays4 ; in tumor cells, no relation with enzyme activity was found5 in contrast to normal cells in which TS 3R/3R was associated with a higher activity than TS 2R/2R.
Simplified scheme depicting intracellular metabolism of folates. THF indicates tetrahydrofolate; DHF, dihydrofolate; CH2-THF, methylene-THF; CH3-THF, methyl-THF; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; dTMP, deoxy thymidine monophosphate; dUMP, deoxy uridine monophosphate; TS, thymidylate synthase; MTHFR, methylenetetrahydrofolate reductase; MTHFD, methylenetetrahydrofolate dehydrogenase; MTR, methionine synthase; MTRR, methionine synthase reductase; SHMT, serine-hydroxymethyltransferase; DHFR, dihydrofolate reductase; RFC, reduced folate carrier; FR, folate receptors, and MTG-PG, methotrexate-polyglutamate. Food folate (CH3-THF) and MTX enter the cell by way of FR or the RFC.
Simplified scheme depicting intracellular metabolism of folates. THF indicates tetrahydrofolate; DHF, dihydrofolate; CH2-THF, methylene-THF; CH3-THF, methyl-THF; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; dTMP, deoxy thymidine monophosphate; dUMP, deoxy uridine monophosphate; TS, thymidylate synthase; MTHFR, methylenetetrahydrofolate reductase; MTHFD, methylenetetrahydrofolate dehydrogenase; MTR, methionine synthase; MTRR, methionine synthase reductase; SHMT, serine-hydroxymethyltransferase; DHFR, dihydrofolate reductase; RFC, reduced folate carrier; FR, folate receptors, and MTG-PG, methotrexate-polyglutamate. Food folate (CH3-THF) and MTX enter the cell by way of FR or the RFC.
Low folate status or redistribution of folate vitamers may influence the risk of cancer6 through mechanisms of uracil misincorporation into DNA, possibly leading to double-strand breaks and chromosomal damage7 and DNA hypomethylation/dysmethylation8 of, for example, proto-oncogenes or tumor-suppressor genes. DNA hypomethylation and uracil misincorporation are considered important factors in carcinogenesis.9,10 The most studied gene variants in folate metabolism in relation to the risk of acute lymphoblastic leukemia (ALL) are MTHFR 677C>T and MTHFR 1298A>C (Figure 1). Both MTHFR variants reduce the susceptibility to adult and childhood lymphoid leukemia but not myeloid leukemia.11-18 However, studies investigating the effects of other polymorphisms within the folate pathway on risk of ALL are scarce and are relatively underpowered. Patients carrying variants in serine hydroxymethyl transferase (SHMT1 1420T allele) and thymidylate synthase gene (TS 3R variant) also appear to have a lower risk of ALL.19,20 Furthermore, both variants show interaction: in combination with SHMT1 1420CT/TT, patients with a variant in methionine synthase (MTR 2756AG genotype) showed a 5.6-fold reduction in adult ALL risk.19 MTR 2756G was a risk allele for ALL on itself but also in combination with the MTHFR 677T allele in adults in another study.18 Thus, the aim of this study is to investigate folate pathway polymorphisms other than those in MTHFR in a relatively large group of 245 ALL patients. The hypothesis of this study is that derangements in the patient's cellular folate status by polymorphisms in folate-related genes influence the susceptibility to ALL via reduced uptake or shifts in cellular distribution of folates.
Methods
Patients and samples
From 245 newly diagnosed West European pediatric ALL patients (< 18 years), DNA was obtained from unstained cytospin slides. Informed consent was obtained from all patients in accordance with the institutional guidelines and the Declaration of Helsinki. The ethical committees of all participating institutions approved the protocol. Leukemic blast samples were collected at the time of diagnosis. The mononuclear cell fraction was separated by density gradient centrifugation (Lymphoprep, 1.077 g/mL; Nycomed Pharma, Oslo, Norway). All samples contained more than 90% leukemic cells. Cytospins were made of 25 × 103 cells by centrifugation at 680 rpm for 7 minutes. Cytospins were wrapped in foil and stored at −20°C. To isolate DNA, bleached cotton was sterilized in an autoclave and cut into small pieces (0.5 × 0.5 cm). The pieces of cotton were first prewetted with H2O. Under sterile conditions, the leukemic blasts were then isolated by wiping the slides with the wetted cotton using forceps. Hereby the blasts became attached to the cotton. To be sure that as many blasts as possible were taken off the slide, a second piece of cotton was then used in an identical way. The 2 pieces of cotton were put in an Eppendorf vial, and 200 μL phosphate buffered saline was added. The vials were stored at 4°C until DNA isolation. Control DNA was obtained with informed consent from 500 white blood bank donors in Rotterdam, The Netherlands. DNA was obtained from ethylenediaminetetraacetic acid whole-blood in the control group. Genomic DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
Folate pathway polymorphisms
Polymorphisms in folate-related genes were determined as previously described (Figure 1).21 The genetic variants in methylenetetrahydrofolate reductase (MTHFR 677C>T, MTHFR 1298A>C), methionine synthase (MTR 2756A>G), serine hydroxymethyl transferase (SHMT1 1420C>T), and the reduced folate carrier (RFC1 80G>A) were determined with polymerase chain reaction (PCR)–restriction fragment length polymorphism (RFLP). Double (2R2R) or triple (3R3R) 28-bp tandem repeats in the promoter region of the thymidylate synthase gene (TS 2R3R) were visualized on agarose gel directly after the PCR reaction. The polymorphism in the methionine synthase reductase gene (MTRR 66A>G) was determined with real-time PCR using hybridization probes and Lightcycler Technology (Roche Diagnostics, Mannheim, Germany). The variant in the nicotinamide N-methyltransferase gene (NNMT IVS −151C>T) was determined with TaqMan technology (assay on demand).
Statistical analysis
Univariate logistic regression analysis was performed to obtain odds ratios (ORs) and 95% confidence intervals (CIs) to estimate relative risks for each polymorphism independently. Then, potential bivariate interaction between polymorphisms was explored using multiplicative interaction terms in a logistic regression model. Interactions with a P value less than .2 were further explored. To increase power, sometimes the heterozygous and homozygous mutant groups were combined. All analyses were carried out using SPSS, version 11.0, to calculate ORs and CIs. Stratification of the cases according to immunophenotype was not performed because smaller numbers gave insufficient power to detect significant differences.
Results
Of the 245 patients, 60% were male and 40% female. The phenotype distribution was 75% (precursor-) B-ALL (61.8% common-ALL, 33.7% pre–B-ALL, 3.9% pro–B-ALL, and 0.6% B-ALL) and 25% T-ALL. The genotype distribution of the folate pathway gene polymorphisms and the univariate risk estimates compared with the wild-type genotype are depicted in Table 1. All genotype distributions in the controls and the patients were in Hardy-Weinberg equilibrium, except for NNMT in ALL patients (P = .04)
Distribution of genotypes in childhood ALL patients and controls
| VariantGenotype . | ALL cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | P . |
|---|---|---|---|---|
| MTHFR 677C>T | ||||
| CC | 130 (53.1) | 219 (44.2) | 1 | |
| CT | 93 (38.0) | 223 (45.0) | 0.7 (0.5-1.0) | .03* |
| TT | 22 (9.0) | 54 (10.9) | 0.7 (0.4-1.2) | .17 |
| CT + TT | 115 (46.9) | 277 (55.8) | 0.7 (0.5-1.0) | .02* |
| MTHFR 1298A>C | ||||
| AA | 110 (44.9) | 229 (47.0) | 1 | |
| AC | 100 (40.8) | 213 (43.7) | 1.0 (0.7-1.4) | .89 |
| CC | 35 (14.3) | 45 (9.2) | 1.6 (1.0-2.7) | .06 |
| AC + CC | 135 (55.1) | 258 (52.9) | 1.1 (0.8-1.5) | .57 |
| MTR 2756A>G | ||||
| AA | 162 (66.1) | 340 (69.5) | 1 | |
| AG | 74 (30.2) | 137 (28.0) | 1.1 (0.8-1.6) | .47 |
| GG | 9 (3.7) | 12 (2.5) | 1.6 (0.7-3.8) | .32 |
| AG + GG | 83 (33.9) | 149 (30.5) | 1.2 (0.8-1.6) | .35 |
| MTRR 66A>G | ||||
| AA | 59 (24.4) | 101 (20.2) | 1 | |
| AG | 117 (48.3) | 245 (49.1) | 0.8 (0.6-1.2) | .31 |
| GG | 66 (27.3) | 153 (30.7) | 0.7 (0.5-1.1) | .17 |
| AG + GG | 183 (75.6) | 397 (79.6) | 0.8 (0.5-1.1) | .20 |
| SHMT 1420C>T | ||||
| CC | 130 (53.3) | 232 (46.7) | 1 | |
| CT | 86 (35.2) | 213 (42.9) | 0.7 (0.5-1.0) | .05 |
| TT | 28 (11.5) | 52 (10.5) | 1.0 (0.6-1.6) | .88 |
| CT + TT | 114 (46.7) | 265 (53.3) | 0.8 (0.6-1.0) | .09 |
| RFC 80G>A | ||||
| GG | 69 (28.6) | 186 (37.6) | 1 | |
| GA | 120 (49.8) | 241 (48.7) | 1.3 (0.9-1.9) | .10 |
| AA | 52 (21.6) | 68 (13.7) | 2.1 (1.3-3.2) | .002* |
| GA + AA | 172 (71.4) | 309 (62.4) | 1.5 (1.1-2.1) | .02* |
| TS 2R3R | ||||
| 3R3R | 80 (32.8) | 123 (25.1) | 1 | |
| 3R2R | 113 (46.3) | 252 (51.3) | 0.7 (0.5-1.0) | .04* |
| 2R2R | 51 (20.9) | 116 (23.6) | 0.7 (0.4-1.0) | .08 |
| 3R2R + 2R2R | 164 (67.2) | 368 (74.9) | 0.7 (0.5-1.0) | .03* |
| NNMT IVS-151C>T | ||||
| CC | 161 (66.0) | 354 (71.2) | 1 | |
| CT | 68 (27.9) | 128 (25.8) | 1.2 (0.8-1.7) | .40 |
| TT | 15 (6.1) | 15 (3.0) | 2.2 (1.1-4.6) | .04* |
| CT + TT | 83 (34.0) | 143 (28.8) | 1.3 (0.9-1.8) | .15 |
| VariantGenotype . | ALL cases, no. (%) . | Controls, no. (%) . | OR (95% CI) . | P . |
|---|---|---|---|---|
| MTHFR 677C>T | ||||
| CC | 130 (53.1) | 219 (44.2) | 1 | |
| CT | 93 (38.0) | 223 (45.0) | 0.7 (0.5-1.0) | .03* |
| TT | 22 (9.0) | 54 (10.9) | 0.7 (0.4-1.2) | .17 |
| CT + TT | 115 (46.9) | 277 (55.8) | 0.7 (0.5-1.0) | .02* |
| MTHFR 1298A>C | ||||
| AA | 110 (44.9) | 229 (47.0) | 1 | |
| AC | 100 (40.8) | 213 (43.7) | 1.0 (0.7-1.4) | .89 |
| CC | 35 (14.3) | 45 (9.2) | 1.6 (1.0-2.7) | .06 |
| AC + CC | 135 (55.1) | 258 (52.9) | 1.1 (0.8-1.5) | .57 |
| MTR 2756A>G | ||||
| AA | 162 (66.1) | 340 (69.5) | 1 | |
| AG | 74 (30.2) | 137 (28.0) | 1.1 (0.8-1.6) | .47 |
| GG | 9 (3.7) | 12 (2.5) | 1.6 (0.7-3.8) | .32 |
| AG + GG | 83 (33.9) | 149 (30.5) | 1.2 (0.8-1.6) | .35 |
| MTRR 66A>G | ||||
| AA | 59 (24.4) | 101 (20.2) | 1 | |
| AG | 117 (48.3) | 245 (49.1) | 0.8 (0.6-1.2) | .31 |
| GG | 66 (27.3) | 153 (30.7) | 0.7 (0.5-1.1) | .17 |
| AG + GG | 183 (75.6) | 397 (79.6) | 0.8 (0.5-1.1) | .20 |
| SHMT 1420C>T | ||||
| CC | 130 (53.3) | 232 (46.7) | 1 | |
| CT | 86 (35.2) | 213 (42.9) | 0.7 (0.5-1.0) | .05 |
| TT | 28 (11.5) | 52 (10.5) | 1.0 (0.6-1.6) | .88 |
| CT + TT | 114 (46.7) | 265 (53.3) | 0.8 (0.6-1.0) | .09 |
| RFC 80G>A | ||||
| GG | 69 (28.6) | 186 (37.6) | 1 | |
| GA | 120 (49.8) | 241 (48.7) | 1.3 (0.9-1.9) | .10 |
| AA | 52 (21.6) | 68 (13.7) | 2.1 (1.3-3.2) | .002* |
| GA + AA | 172 (71.4) | 309 (62.4) | 1.5 (1.1-2.1) | .02* |
| TS 2R3R | ||||
| 3R3R | 80 (32.8) | 123 (25.1) | 1 | |
| 3R2R | 113 (46.3) | 252 (51.3) | 0.7 (0.5-1.0) | .04* |
| 2R2R | 51 (20.9) | 116 (23.6) | 0.7 (0.4-1.0) | .08 |
| 3R2R + 2R2R | 164 (67.2) | 368 (74.9) | 0.7 (0.5-1.0) | .03* |
| NNMT IVS-151C>T | ||||
| CC | 161 (66.0) | 354 (71.2) | 1 | |
| CT | 68 (27.9) | 128 (25.8) | 1.2 (0.8-1.7) | .40 |
| TT | 15 (6.1) | 15 (3.0) | 2.2 (1.1-4.6) | .04* |
| CT + TT | 83 (34.0) | 143 (28.8) | 1.3 (0.9-1.8) | .15 |
OR indicates odds ratio; and CI, confidence interval.
Significant value.
The MTHFR 677CT variant was underrepresented in ALL patients (38%) compared with healthy controls (45%; OR = 0.7; 95% CI, 0.5-1.0; P = .03); T-allele carriers were also significantly lower in ALL cases (47%) than healthy controls (56%), indicating that the T allele is related to a reduced leukemia risk (OR = 0.7; 95% CI, 0.5-1.0; P = .02). Reduced leukemia risk was also observed for the TS 3R2R variant (OR = 0.7; 95% CI, 0.4-1.0; P = .04) and TS 2R allelic carriers (OR = 0.7; 95% CI, 0.5-1.0; P = .03). RFC1 80AA homozygotes and A-allelic carriers have an increased susceptibility to ALL. Risk was increased 2.1 times (95% CI, 1.3-3.2; P = .002) with the RFC1 80AA variant and 1.5 times (95% CI, 1.1-2.1; P = .02) for A-allelic carriers. Similarly, the NNMT IVS −151 TT variant also is associated with increased susceptibility to ALL (OR = 2.2; 95% CI, 1.1-4.6; P = .04). When the T-ALL patients (n = 59) were left out from the univariate analysis, this did not lead to significantly different point estimates of the ORs or the CIs. For example, the OR of the RFC 80 A-allele carriers was 1.4 (95% CI, 0.9-2.0; P = .09); the OR of the MTHFR T-allele carriers was identical to the combined analysis (OR = 0.7; 95% CI, 0.9-2.0; P = .09; Table 1). However, because numbers are lower, P values generally increase.
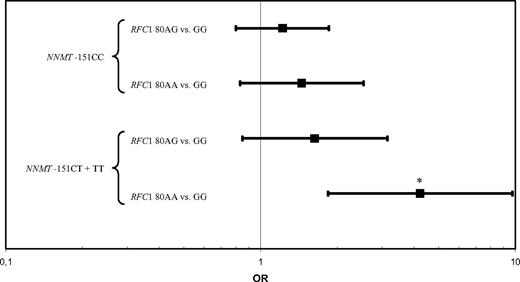
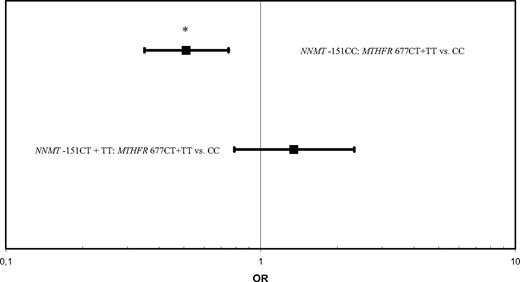
Then, we tested several gene × gene interactions. There was an interaction between the RFC1 80G>A and NNMT IVS −151 C>T polymorphisms (P = .1). To gain power in the analysis, NNMT −151CT and TT variants were combined because only one case and one control were present in the NNMT IVS −151TT/RFC 80AA genotype. Specifically, among persons with the RFC1 80AA variant, the OR of NNMT IVS −151CT + TT vs −151CC was 4.2 (95% CI, 1.8-9.7; P = .001; Figure 2). Furthermore, the MTHFR 677C>T and NNMT IVS −151C>T variants interacted (P = .004). In carriers of the wild-type NNMT IVS −151CC variant, a significantly reduced risk for ALL was observed for the MTHFR 677CT + TT genotype (OR = 0.5; 95% CI, 0.4-0.8; P = .001; Figure 3). To increase power, NNMT −151CT and TT and MTHFR 677 CT + TT variants were combined in the analyses.
RFC1 80AA and the NNMT IVS −151T allele interact to increase the risk of pediatric acute lymphoblastic leukemia. Odds ratios and 95% confidence intervals for NNMT IVS −151 CT + TT versus CC genotypes at levels of RFC1 80G>A. *P = .001.
RFC1 80AA and the NNMT IVS −151T allele interact to increase the risk of pediatric acute lymphoblastic leukemia. Odds ratios and 95% confidence intervals for NNMT IVS −151 CT + TT versus CC genotypes at levels of RFC1 80G>A. *P = .001.
The MTHFR 677T allele and NNMT IVS −151CC interact to decrease the risk of pediatric acute lymphoblastic leukemia. Odds ratios and 95% confidence intervals for NNMT IVS −151 CT + TT versus CC genotypes at levels of MTHFR 677C>T. *P = .001.
The MTHFR 677T allele and NNMT IVS −151CC interact to decrease the risk of pediatric acute lymphoblastic leukemia. Odds ratios and 95% confidence intervals for NNMT IVS −151 CT + TT versus CC genotypes at levels of MTHFR 677C>T. *P = .001.
Discussion
Polymorphisms in folate-related genes may influence cellular folate metabolism by lowering folate status or shifts in cellular folate distribution. This may influence the risk of cancer6 through several mechanisms. First, low availability of folates or redistribution of folates toward homocysteine remethylation reduces availability of folates for the TS reaction and DNA synthesis (Figure 1) and results in uracil misincorporation into DNA, possibly leading to double-strand breaks and chromosomal damage.7 On the other side, reduced availability of folate for the methionine cycle results in reduced transmethylation capacity (low S-adenosylmethionine/S-adenosylhomocysteine ratio, Figure 1) and DNA hypomethylation/dysmethylation8 of, for example, proto-oncogenes or tumor suppressor genes. DNA hypomethylation and uracil misincorporation are considered important factors in carcinogenesis.9,10
This is the first report that identifies an association between the RFC1 80G>A polymorphism and ALL risk. The RFC1 80G>A variant was the strongest modulator of leukemia risk: risk increased 1.5 times and 2.1 times in A-allelic carriers and 80AA homozygotes, respectively. In addition, a 4.2-fold increased risk of ALL was observed in carriers of the RFC1 80GG/NNMT IVS −151CT + TT genotype. The RFC1 80G>A allele frequency of the 500 blood-bank donors (38%) was similar to earlier reports in 169 healthy French adults (36%),22 602 healthy English children (41%),23 and 1041 healthy English elderly people (44%).24 The RFC1 80G>A polymorphism results in a change of arginine-27 to histidine-27. However, the functional effect of the 80G>A variant is scarcely investigated. Whetstine et al25 investigated the transport properties of this genetic polymorphism in K562 cells, and these in vitro experiments suggest that the RFC1 80G>A polymorphism has marginal functional effects. Cellular uptake of most oxidized folates, such as folic acid, is predominantly mediated by the folate receptor and not via the RFC. Thus, whether these in vitro results can be translated to the human in vivo situation is questionable and hardly investigated. Interestingly, patients carrying the polymorphic RFC1 80A allele showed an increased percentage of extracellular folate relative to intracellular folate.26 This observation suggests that the RFC1 80G>A polymorphism renders a reduced efficiency in the cellular uptake of methyl-THF. Similarly, mean plasma folate was slightly higher in healthy subjects carrying the RFC1 80AA variant: 19 nmol/L versus 15 nmol/L in G-allelic carriers.22 Whether the RFC1 80G>A variant induces shifts in intracellular folate redistribution is not known. In addition, other regulating factors in cellular folate metabolism might counteract the diminished influx. Thus, patients carrying the RFC1 80A allele may have lower folate or methotrexate (MTX) uptake, which might increase ALL risk and decrease MTX efficacy. Interestingly, the RFC1 gene is located on chromosome 21. Extra copies of this chromosome in favorable subgroups of ALL are associated with enhanced RFC1 mRNA expression and MTX-PG accumulation27,28 and, hence, may be a selective advantage. Earlier studies did not show a relation between the RFC1 80G>A polymorphism and the risk of non–Hodgkin lymphoma,29,30 neural tube defects,23,31 and colon cancer.32 However, the RFC1 A allele is associated with an increased risk of distal gastric cancer,33 whereas the maternal RFC1 G allele increases the risk of offspring with Down syndrome, especially in combination with the MTHFR 1298C allele.34-36 Interestingly, the RFC1 80A allele has been associated with antifolate toxicity in children with ALL,37 although we did not find altered MTX sensitivity21 or toxicity38 associated with this polymorphism.
The most studied gene variants in folate metabolism in relation to cancer risk are the 677C>T and the 1298A>C polymorphisms in the MTHFR gene. The 677TT and the 1298CC variants are associated with moderately reduced risks for colorectal cancer.39 Both MTHFR variants also reduce the susceptibility to adult and childhood lymphoid leukemia but not myeloid leukemia.11-18 A recent meta-analysis concluded that MTHFR 677TT reduces risk of adult ALL but not childhood ALL; the MTHFR 1298A>C polymorphism did not influence susceptibility to childhood or adult ALL.40 In line with this recent meta-analysis,40 we observed no protective effect of the MTHFR 1298A>C variant on ALL risk. In the present study, MTHFR 677CT and T-allelic carriers exhibited decreased risk of ALL. Moreover, MTHFR 677C>T interacted with NNMT IVS −151C>T: a 49% reduction in ALL risk was observed for patients carrying the MTHFR CT + TT/NNMT IVS −151CC genotype. Discrepant literature results for the MTHFR variants may have to do with the folate status of the patients. Few studies investigated the role of gene-environment interactions on the risk of childhood ALL. Both increased folate intake,41,42 and the MTHFR 677C>T and 1298A>C polymorphisms12,13,16,17 separately are related to a reduced susceptibility to ALL. Krajinovic et al16 investigated gene-environment interaction by grouping the children into those born before or after 1996, the time of the recommendation of folic acid supplementation during pregnancy in Canada. They observed that the protective effect of the MTHFR polymorphisms was only present in those born before 1996 and that the protective effect disappeared in (folate replete) subjects born after this date. Because the year of birth is a surrogate marker of the folate status, more studies are necessary that measure plasma and erythrocyte folate levels together with polymorphisms in folate-related genes to study the influence of gene-environment interactions on ALL risk. Similarly, MTHFR 677TT increases gastric cancer risk in subjects with low folate status but not in folate replete subjects.43 Another possibility that is hardly explored is that pediatric ALL risk is modulated by the folate status of the mother during pregnancy. Against this hypothesis is the observation that the effects of the MTHFR 677C>T and 1298A>C genotypes on ALL risk were abolished when parental genotypes were compared with controls.16 The authors concluded that it is the patient's genotype that determines leukemia risk. The biochemical consequences of both the MTHFR 677C>T and 1298A>C variants are best characterized. The MTHFR 677C>T polymorphism yields a thermolabile enzyme with approximately 30% and 60% residual activity for the TT and CT variants, respectively.44 The variant lies at the base of the binding site for the MTHFR cofactor, flavin adenine dinucleotide. The 677TT variant increases total homocysteine by approximately 25%,44,45 especially when folate status is low,46 and influences folate redistribution and TS flux.1-3 The other variant in the MTHFR gene, 1298A>C, also decreases enzyme activity,47-49 although to a lesser extent than the 677C>T variant.50 The 1298A>C variant lies in the S-adenosylmethionine-regulatory domain of the enzyme and seems not to affect total homocysteine.48-50
Polymorphisms other than those in the MTHFR gene are scarcely studied, and results are contradictory. Patients carrying the SHMT1 1420T allele and the TS 3R variant have a lower risk of ALL19,20 ; furthermore, both variants show interaction. In our study, we could not confirm the protective effect of the TS 3R variant. Instead, we observed a protective effect of the TS 2R variant. Thus, because of the relatively small sample sizes of our study (n = 245) and that of Skibola et al19 (n-71) and Hishida et al20 (n = 108), the reported and contrasting effects may be the result of chance. More studies are needed to elucidate the effect of the TS 2R3R polymorphism on leukemia risk. In line with previous studies,19,20 we also observed trends to protective effects of the SHMT1 1420CT variant (P = .05) and T-allelic carriers (P = .09).
Both in our study and in other studies,13,19 the MTR 2756A>G variant does not modulate ALL risk in univariate analysis. However, in combination with SHMT1 1420CT/TT, patients with the MTR 2756AG variant showed a 5.6-fold reduction in ALL risk.19 In contrast, MTR 2756G was a risk allele for ALL on itself but also in combination with the MTHFR 677T allele in adults.18 We could not confirm these observed interactions between the MTR 2756A>G and SHMT1 1420C>T or MTHFR 677C>T polymorphisms. The MTRR 66A>G variant was unrelated to ALL risk in our study and in the study of Gemmati et al,18 although only in the latter study were significant interaction effects on ALL risk with the MTR 2756AG/GG and MTHFR 677TT genotypes observed.
A genome-wide study51 identified NNMT IVS −151C>T as the major determinant of plasma homocysteine. Little is known about the functional effects of this polymorphism. We describe, for the first time, that this NNMT gene variant increases ALL risk and interacts with the MTHFR 677C>T and RFC1 80G>A variants. More studies on this polymorphism are needed. NNMT serum levels are increased in patients with colorectal cancer,52 and NNMT is up-regulated in tumor cells of favorable subgroups of oral squamous cell carcinoma.53
We are well aware of the risk of spurious findings when using multiple comparisons. However, we controlled chance findings by (1) carefully selecting a priori 8 single nucleotide polymorphisms (SNPs) with documented biologic and/or clinical effects and (2) the selection of SNPs was hypothesis driven. Therefore, we did not adjust for multiple comparisons. A major disadvantage of adjusting for multiple comparisons is that it becomes very hard to detect a possible effect of a promising SNP in a relatively small patient population. Hence, we would increase the risk of throwing out a useful SNP, although indeed there is an effect (type 2 error). When correcting for multiple comparisons, the effect of the homozygous RFC1 80AA variant remained significant. In earlier studies using a subgroup of patients, we demonstrated that polymorphisms in folate-related genes are associated with MTX sensitivity21 and toxicity.38
MTX uptake/metabolism is different between T-ALL and B-ALL patients, as we have shown.54 MTX polyglutamylation is lower in T-ALL because of lower folylpolyglutamate synthetase activity compared with c/pre–B-ALL. However, whether T-ALL and B-ALL patients also differ in folate kinetics (eg, because of folate polymorphisms) is hardly explored. When we left out the T-ALL patients (n = 59) from the analysis, this did not lead to significantly different results; therefore, T-ALL and B-ALL patients are presented as one group. Future studies with larger numbers of patient will allow better investigation of whether T-ALL and B-ALL patients differ in folate metabolism and how this influences their susceptibility to ALL. Furthermore, we did not assess nutritional status (eg, serum or erythrocyte folate). Future studies should investigate the influence of nutritional status because folate intake most probably interacts with polymorphisms in one-carbon metabolism, as has been shown for MTHFR. Our study shows, in line with several other studies, that folate status should be considered a risk factor for ALL and warrants further research. For comparison of the risk estimates we observed (eg, RFC 80 A allele: OR = 1.5; 95% CI, 1.1-2.1), a recent meta-analysis (n = 10 282) showed that a birth weight more than 4000 g is associated with a higher risk of ALL (OR = 1.26; 95% CI, 1.17-1.37).
In conclusion, the RFC1 and NNMT gene variants are associated with increased ALL risk, whereas the MTHFR and TS gene variants are associated with decreased ALL risk. The underlying mechanisms of the increased ALL risk associated with the RFC1 80G>A and NNMT IVS −151C>T polymorphisms may be reduced cellular uptake of folate and changed methylation status, respectively; the reduced risk associated with the MTHFR 677C>T and TS 2R/3R polymorphisms may be the result of changed intracellular folate redistribution.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.d.J. designed research, performed SNP and statistical analysis, and wrote the paper; W.J.E.T. provided patient samples and clinical data and designed research; J.H.H. provided patient samples and helped in setting up DNA analysis and interpretation of data; G.J. provided patient samples and helped in discussion and interpretation; G.J.L.K. provided patient samples; J.L. designed research; G.J.P. helped in setting up DNA analysis, interpretation of data, and discussion; and R.P. designed research and provided patient samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert de Jonge, Department of Clinical Chemistry, Erasmus MC (L-137), POB 2040, 3000 CA Rotterdam, The Netherlands; e-mail: r.dejonge@erasmusmc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal