Abstract
MDS is characterized by ineffective hematopoiesis that leads to peripheral cytopenias. Development of effective treatments has been impeded by limited insight into pathogenic pathways governing dysplastic growth of hematopoietic progenitors. We demonstrate that smad2, a downstream mediator of transforming growth factor–β (TGF-β) receptor I kinase (TBRI) activation, is constitutively activated in MDS bone marrow (BM) precursors and is overexpressed in gene expression profiles of MDS CD34+ cells, providing direct evidence of overactivation of TGF-β pathway in this disease. Suppression of the TGF-β signaling by lentiviral shRNA-mediated down-regulation of TBRI leads to in vitro enhancement of hematopoiesis in MDS progenitors. Pharmacologic inhibition of TBRI (alk5) kinase by a small molecule inhibitor, SD-208, inhibits smad2 activation in hematopoietic progenitors, suppresses TGF-β–mediated gene activation in BM stromal cells, and reverses TGF-β–mediated cell-cycle arrest in BM CD34+ cells. Furthermore, SD-208 treatment alleviates anemia and stimulates hematopoiesis in vivo in a novel murine model of bone marrow failure generated by constitutive hepatic expression of TGF-β1. Moreover, in vitro pharmacologic inhibition of TBRI kinase leads to enhancement of hematopoiesis in varied morphologic MDS subtypes. These data directly implicate TGF-β signaling in the pathobiology of ineffective hematopoiesis and identify TBRI as a potential therapeutic target in low-risk MDS.
Introduction
The myelodysplastic syndromes (MDSs) are clonal stem cell disorders characterized by cytologic dysplasia and ineffective hematopoiesis.1-3 Although approximately one third of patients may progress to acute leukemia, refractory cytopenias are the principal cause of morbidity and mortality in patients with MDS.4 In fact, approximately two-thirds of patients present with lower risk disease characterized by hypercellular marrows with increased rates of apoptosis in the progenitor and differentiated cell compartments in the marrow.5-8 Ineffective hematopoiesis arising from abortive maturation leads to peripheral cytopenias. Higher grade or more advanced disease categories are associated with a significant risk of leukemia transformation, with a corresponding lower apoptotic index and higher percentage of marrow blasts.
Cytokines play important roles in the regulation of normal hematopoiesis, and a balance between the actions of hematopoietic growth factors and myelosuppressive factors is required for optimal production of different hematopoietic cell lineages. Excess production of inhibitory cytokines amplifies ineffective hematopoiesis inherent to the MDS clone. Transforming growth factor–β (TGF-β) is a myelosuppressive cytokine that has been implicated in the hematopoietic suppression in MDS. The plasma levels of TGF-β have been reported to be elevated in some9-13 but not all studies14-17 and are supported by greater TGF-β immunohistochemical staining in selected studies. In addition to direct myelosuppressive effects, TGF-β has also been implicated in the autocrine production of other myelosuppressive cytokines (TNF, IL-6, and IFNγ) in MDS.18 Conflicting data may arise from technical limitations of bone marrow immunohistochemical analyses of a secreted protein as well as the biologic heterogeneity of the disease itself. In addition, plasma levels of TGF-β may not be an accurate reflection of the biologic effects of this cytokine in the MDS bone marrow microenvironment. Thus we investigated the role of TGF-β in MDS by direct examination of receptor signal activation to conclusively determine its role in the pathogenesis of ineffective hematopoiesis in MDS.
Our previous studies have shown that signaling pathways activated by myelosuppressive cytokines can serve as therapeutic targets in low-risk MDS. We showed that interferons (IFNα, IFNβ, and IFNγ), TGF-β, and tumor necrosis factor α (TNFα) can all activate the p38 mitogen-activated protein kinase (MAPK) in primary human hematopoietic progenitors and that activation of p38 is required for myelosuppressive actions of these cytokines on hematopoiesis.19,20 We subsequently confirmed overactivation of p38 MAPK in the bone marrow progenitors of low-risk MDS patients. Our data showed that inhibition of this cytokine-stimulated p38 MAPK pathway partially rescues hematopoiesis in MDS progenitors. This led to a clinical trial of a p38 inhibitor, SCIO-469, in low-risk MDS; the preliminary results have shown modest clinical activity in some cases of lower-risk MDS.21 Having demonstrated that intracellular signaling pathways can serve as therapeutic targets in MDS, we decided to directly evaluate TGF-β signaling in MDS. We determined that the smad2 protein is heavily phosphorylated in MDS bone marrow progenitors and is found to be up-regulated in meta-analysis of MDS CD34+ cell gene expression studies, thereby demonstrating sustained TGF-β signal activation in this disease. We showed that inhibition of the TGF-β receptor I kinase (TBRI) by shRNA suppression or by small molecule inhibitors abrogates smad2 activation and reverses the suppressive effects of TGF-β on hematopoiesis. Most importantly, TBRI inhibition stimulated in vitro hematopoiesis from a variety of MDS progenitors, thus providing a therapeutic rationale for inhibiting TGF-β signaling in this disease.
Methods
Cells lines and reagents human
CD34+ cells were isolated from bone marrows of healthy or MDS patients, after obtaining informed consent approved by the institutional review boards of University of Texas (UT) Southwestern Medical School (Dallas, TX), the Dallas VAMC, the University of Arizona College of Medicine (Phoenix, AZ), and the University of South Florida (Tampa, FL), and in accordance with the Declaration of Helsinki. Some bone marrow CD34+ cells from various healthy donors were also obtained from Cambrex (Atlanta, GA). KG-1, K562, and HS-5 cell lines were purchased from ATCC (Manassas, VA). Erythroid progenitors at the colony-forming unit–erythroid (CFU-E) stage of differentiation were grown in cytokine-enriched Iscove modified Dulbecco medium (IMDM) media, similar to our previous studies.19,20,22 Briefly, bone marrow or peripheral blood mobilized CD34+ cells were immunomagnetically selected and cultured in 15% fetal bovine serum, 15% human serum, IMDM, 10 ng/mL IL-3, 2 units/mL erythropoietin (Epo), and 50 ng/mL stem cell factor (SCF). The cells were fed on days 3 and 6 to maintain an Epo concentration of 2 units/mL. The cells were sorted for glycophorin positivity on day 8 of culture and used for biochemical studies. SCF, thrombopoietin (TPO), Flt-3–ligand (FLT-3L), erythropoietin, and TGF-β1 were bought from R&D Systems (Minneapolis, MN). The phos-Smad2 (S465/467), Cdc2, and PCNA antibodies were from Cell Signaling Technology (Beverly, MA); cyclin E2 and Cdc6 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); and smad2 antibody was from Invitrogen (Carlsbad, CA). TBRI inhibitor SD-208 was provided by Scios (Fremont, CA). SD-208 is a selective 2,4-disubstituted pteridine-derived TGF-β receptor I (TGF-βI) kinase inhibitor23-28 that exhibits an in vitro specificity for TGF-βRI kinase of more than 100-fold compared with TGF-βRII kinase and at least 20-fold more than members of a panel of related protein kinases. SD-208 was diluted in dimethyl sulfoxide (DMSO; 20 mM stock solution) and kept at −20°C until use.
Cell lysis and immunoblotting
Immunohistochemistry
Paraffin-mounted bone marrow core biopsy sections from MDS patients and controls were obtained after informed consent. Controls had anemia from non–MDS-related causes. Slides were deparaffinized and hydrated. Mercury pigments from B5 fixative were removed by iodine-sodium thiosulfate sequences. After rinsing 3 times in PBS, all sections were immersed in 3% hydrogen peroxide for 20 minutes at room temperature to completely block endogenous peroxidases. Antigen retrieval (citrate buffer, pH 6.0) was used for all these antibodies. To prevent nonspecific binding with primary antibodies, sections were pretreated with 15% normal goat serum. After 3 washes with PBS, the sections were incubated with primary antibodies overnight at 4°C. The primary antibodies used in this study were rabbit phospho-smad2 polyclonal antibody diluted at 1:1000 dilution (Cell Signaling Technology). After 3 washes with PBS, the sections were then incubated with biotinylated goat antirabbit (Chemicon International, Temecula, CA) secondary antibody at 1:2000 dilution at room temperature for 30 minutes. Normal rabbit or mouse IgGs (Santa Cruz Biotechnology) were used as negative controls. All sections were then treated with avidin-biotin-horseradish peroxidase reagents (Vector, Burlingame, CA) and finally stained with diaminobenzidine (Research Genetic, Carlsbad, CA). After several more rinses, the sections were counterstained with hematoxylin and subsequently mounted with Permount (Fisher Scientific) mounting medium. The quantification of p-smad2 staining was analyzed by counting the total number of positively stained cells and by measuring the intensity of the positively stained cells in 5 hot fields (hot field is defined as area of high density of p-smad2 staining) for each patient sample under 400× magnification aided by Image Pro Plus (Media Cybernetics, Bethesda, MD) software. The results were expressed as mean numbers of positively stained cells per field and mean intensity per field for each individual patient sample.
ShRNA knockdown experiments
The human lentiviral shRNAmir (pCMV-GIN-Zeo) containing the hairpin sequence TGCTGTTGACAGTGAGCGAGGTACTACGTTGAAAGACTTATAGTGAAGCCACAGATGTATAAGTCTTTCAACGTAGTACCCTGCCTACTGCCTCGGA targeting TGF-βRI was obtained from Open Biosystems (Huntsville, AL). The scrambled control shRNAmir was cut out from pSM retroviral vector (Open Biosystems) by XhoI and EcoRI and ligated into lentiviral vector and used as a control. Recombinant lentiviruses were produced by transient transfection of the 293T cells with the transducing vector and 2 packaging vectors: pVSVg, a plasmid expressing the VSVg envelope gene, and pΔ8.9, a plasmid expressing the HIV-1 gag/pol gene. The supernatants were collected 48 hours after transfection. Cell lines HS-5 and K562 were transduced with the virus supernatant in the presence of 8 μg/mL polybrene, and transduced cells were identified by expression of green fluorescent protein (GFP) and sorted by fluorescence-activated cell sorting (FACS). Nucleofection of CD34+ cells or MDS BM mononuclear cells (MNCs) was performed according to the manufacturer's instructions, using the Nucleofector machine (Amaxa, Cologne, Germany). The CD34+ cells were thawed and cultured for 2 hours; 2 × 106 cells were resuspended in 100 μL human CD34+ nucleofection solution (Amaxa). Samples were transferred into cuvettes and transfected using program U08. CD34+ cells were collected, dispensed in the wells of a 24-well plate containing 1 mL prewarmed StemSpan (StemCell Technologies, Vancouver, BC), and supplemented with 100 ng/mL human FLt-3, SCF, and TPO. The lentiviral transfected CD34+ cells were sorted 24 hours later according to the GFP intensity using Moflow (Becton Dickinson, San Jose, CA). Quantitative reverse-transcription–polymerase chain reaction (RT-PCR) for TBRI expression was done on total RNA from HS-5 and K562 cells obtained by RNAeasy mini kit (Qiagen, Valencia, CA). cDNA was synthesized with SuperScript Reverse Transcriptase III (Invitrogen). Real-time PCR was performed with SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA), and TBRI and GAPDH (control) were simultaneously amplified with specific primers.
Primers for GAPDH were forward, 5′-cgaccactttgtcaagctca-3′ and reverse, 5′-ccctgttgctgtagccaaat-3′. Primers for TBRI were forward, 5′-tcgtctgcatctcactcat and reverse, 5′-gataaatctctgcctcacg-3′.
Meta-analysis
National Center for Biotechnology Information's (NCBI, Bethesda, MD) GEO database29 was searched for gene expression studies on MDS and normal CD34 cells. A total of 2 MDS datasets with 69 unique MDS CD34+ studies30,31 and 57 normal CD34+ datasets from 6 studies30-34 were identified and downloaded. Fifty-nine MDS samples belonged to low-grade risk group, whereas 10 belonged to high-grade International Prognostic Scoring System (IPSS) risk group. The data were integrated using UniGene IDs.35 Studies were done using Affymetrix U133A/B and Plus 2.0 platforms (Santa Clara, CA). After interarray quantile normalization, smad2 expression was assessed and visually represented as a heat map using R statistical software (http://www.r-project.org/).
CD34+ cell proliferation and cell-cycle assays
CD34+ cells, cultured in IMDM medium (Mediatech, Manassas, VA) containing 15% fetal bovine serum (FBS; HyClone, Logan, UT) and cytokines (50 ng/mL each of SCF, TPO, and FLT-3L), were pretreated with DMSO or 0.5 μM SD-208 for 1 hour before TGF-β1 (0.5 ng/mL final concentration) was added. Cells were cultured for 7 days at 37°C/5% CO2. On days 3, 5, and 7, cell aliquots were taken and viable cell concentration was determined using Guava ViaCount (Guava Technologies, Hayward, CA). The experiment was repeated at least 3 times (using multiple donors of CD34+ cells) with similar results. Cell-cycle distribution of CD34+ cells (gated with PE-conjugated CD34 antibody [BD Biosciences, San Jose, CA]) was determined on day 7 using the APC BrdU Flow Kit and the LSR-II flow cytometer (BD Biosciences) according to the manufacturer's instructions.
cDNA microarray analysis
CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1. After 24 hours, total CD34+ RNA was extracted using Qiagen's RNeasy kit. Details of microarray and data analysis have been described previously.36 The data were lowess normalized37 using the maNorm function in microarray package of Bioconductor version 1.5.8 (Fred Hutchinson Cancer Research Center, Seattle, WA). Differential expression values were expressed as the ratio of the median of background-subtracted fluorescent intensity of the experimental RNA to the median of background-subtracted fluorescent intensity of the control RNA.
Murine experiments
Transgenic mice expressing a fusion gene (Alb/TGF) consisting of modified porcine TGF-β1 cDNA under the control of the regulatory elements of the mouse albumin gene38 were used under animal institute–approved protocol. Mice were given SD-208 in 1% methylcellulose solution by gastric lavage using a curved 14-G needle. Blood counts were analyzed by Advia (Bayer) machine. Mice femurs were flushed and bone marrows cells were used for clonogenic assays. Methocult GF M3534 (StemCell Technologies) was used to assess myeloid (granulocyte/macrophage colony-forming unit [CFU-GM]) colony formation, and Methocult SF M3436 was used for erythroid colony (erythroid burst-forming unit [BFU-E]) assessment.
TGF-β gene reporter assay
HS-5 cells were plated in 6-well plates 24 hours before transfection. Cells were transfected in triplicate with SuperFect (Qiagen) according to the manufacturer's instruction using the reporter plasmid (pGL2-3TP-Lux); plasmids that contain kinase-null TBRI (pRK5-TBRI-KR) were used as positive control.39 pSV-B-Gal was used to normalize the transfection efficiency. After an overnight incubation, the medium was replaced with serum-free medium with or without SD-208 as necessary for the experiment. After 48 hours, cells were treated with 10 ng/mL TGF-β1. Cells were harvested 24 hours later in reporter lysis buffer (Promega, Madison, WI), and luciferase and β-galactosidase activities were determined using Dual Luciferase System (Promega).
Hematopoietic progenitor cell assays
Hematopoietic progenitor colony formation was determined by clonogenic assays in methylcellulose, as in our previous studies.19,20 All participants in the study signed informed consent, approved by the institutional review board of UT Southwestern and Dallas VA Medical Center. Mononuclear cells were isolated from MDS patient bone marrow aspirates by Ficoll-Hypaque density gradient separation. These cells were cultured in methylcellulose media (Methocult GF H4434; StemCell Technologies) containing recombinant human stem cell factor, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-3 and erythropoietin. Granulocyte/macrophage colony-forming units (CFU-GMs) and erythroid burst-forming units (BFU-Es) were scored on day 14 of culture.
Results
smad2 is activated in MDS bone marrow precursors
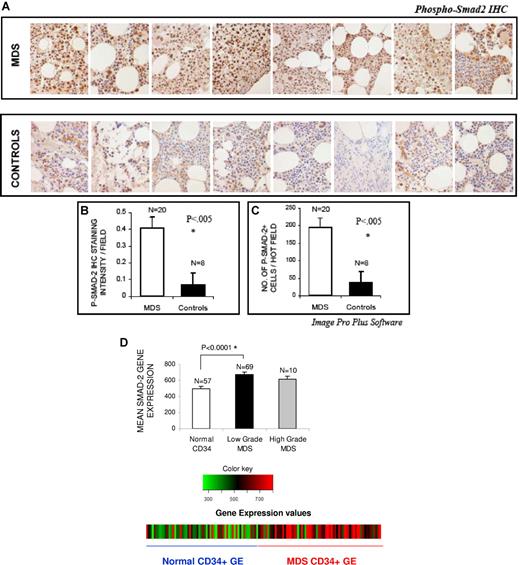
Bone marrows of 20 patients with MDS were assessed for the activation/phosphorylation state of smad2 by immunohistochemistry. Most (16 of 20) patients belonged to lower (low and Int-1 IPSS) risk category of MDS. MDS bone marrow samples were compared with age-matched controls with non-MDS causes of cytopenias (iron deficiency anemia [1], chemotherapy-related anemia [1], hyperplenism [1], drug-induced marrow suppression [1], autoimmune anemia [1], myelofibrosis [1], and unexplained cytopenias in the absence of any dysplasia [2]). Notable activation of smad2 was seen in bone marrow cells of all patients with MDS, (Figure 1A) with a greater number of positively staining cells (Figure 1B) and significantly higher intensity of nuclear staining (Figure 1C) compared with controls. Activation of smad2 was seen in all subtypes of MDS examined (7 with refractory anemia [RA]), 11 with refractory cytopenias with multilineage dysplasia [RCMD], and 2 with refractory anemia with excess blasts [RAEB]). Because smad2 is ubiquitously expressed, we also investigated the phenotypes of bone marrow cells that expressed the activated kinase. Histologic examination revealed that smad2 was activated in hematopoietic progenitors of all lineages including erythroid and myeloid precursors and even megakaryocytes.
smad2 is activated in MDS. Bone marrow (BM) biopsies from patients with MDS and controls with non-MDS causes of cytopenias were fixed and immunostained with antibody against phospho-smad2 (A). Histologic examination reveals more intense staining in MDS samples. Eight representative samples of MDS and controls are shown in panel A. The quantification of p-smad2 staining was analyzed by counting the total number of positively stained cells (B) and by measuring intensity of the positively stained cells (C) in 5 hot fields (which is defined as area of high density of p-p38 staining) and aided by Image Pro Plus (Nikon, 400×). Two-tailed t test shows significantly higher smad2 activation/field in MDS samples. Differences in smad2 expression were also evaluated in normalized meta-analysis of MDS CD34+ (69 cases)– and normal CD34+ (57 cases)–cell–derived gene expression microarray studies. Smad2 gene expression was significantly up-regulated in low-grade MDS bone marrow CD34+ cells (2-tailed t test) (D). Error bars represent SEM.
smad2 is activated in MDS. Bone marrow (BM) biopsies from patients with MDS and controls with non-MDS causes of cytopenias were fixed and immunostained with antibody against phospho-smad2 (A). Histologic examination reveals more intense staining in MDS samples. Eight representative samples of MDS and controls are shown in panel A. The quantification of p-smad2 staining was analyzed by counting the total number of positively stained cells (B) and by measuring intensity of the positively stained cells (C) in 5 hot fields (which is defined as area of high density of p-p38 staining) and aided by Image Pro Plus (Nikon, 400×). Two-tailed t test shows significantly higher smad2 activation/field in MDS samples. Differences in smad2 expression were also evaluated in normalized meta-analysis of MDS CD34+ (69 cases)– and normal CD34+ (57 cases)–cell–derived gene expression microarray studies. Smad2 gene expression was significantly up-regulated in low-grade MDS bone marrow CD34+ cells (2-tailed t test) (D). Error bars represent SEM.
Because sustained TGF-β signaling can also lead to up-regulation of smad2 protein, we decided to analyze its expression in a meta-analysis of publicly available microarray studies in MDS. A total of 69 unique MDS patient CD34+ gene expression profiles were obtained from published studies,30,31 integrated using UniGene protein IDs,35 subjected to interarray normalization, and then used for analysis (Figures S1,S2 and Table S1 showing schema of meta-analysis, available on the Blood website; see the Supplemental Materials link at the top of the online article). Fifty-seven normal CD34+ gene expression profiles were also obtained from a total of 6 studies30-34,40 and used as controls. This strategy was shown to be a biologically valid strategy for analysis as it was able to cluster datasets based on their cell of origin and phenotype (D.S. and A.V., manuscript submitted). Our analysis of this integrated dataset demonstrated that smad2 gene was greatly up-regulated in low-grade MDS CD34+ cells compared with normal cells (Figure 1D, P < .001). This result is based on other independent studies and confirms our histologic findings, while offering further evidence of overactivated TGF-β signaling in a large number of MDS samples.
Down-regulation of TGF β receptor I can inhibit smad2 activation in hematopoietic cells and stimulate MDS hematopoiesis
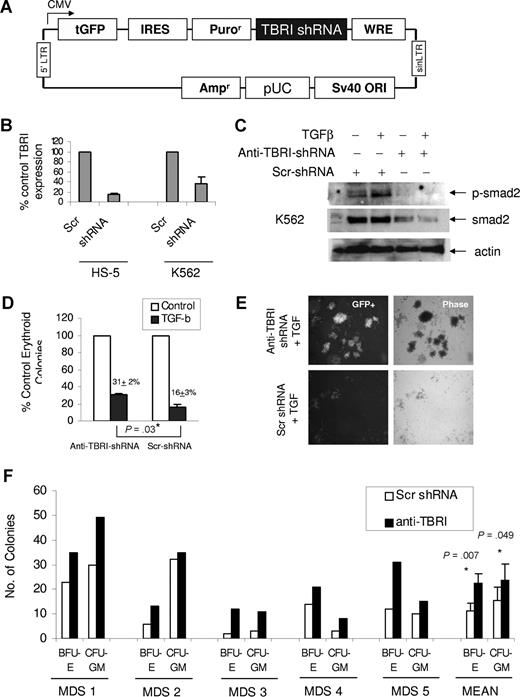
Because smad2 is phosphorylated by TGF-beta receptor I kinase (alk5 kinase, TBRI), lentiviral vectors containing shRNA targeting TBRI were developed (Figure 2A) to determine the biologic role of TGF-smad2 pathway activation in MDS hematopoiesis. The anti–TBRI-shRNA was successfully able to inhibit TBRI expression compared with scrambled control in hematopoietic (K562) and bone marrow stromal (HS-5) cell lines with stable expression of GFP+ lentiviral construct (Figure 2B). Down-regulation of TBRI also led to inhibition of TGF-mediated smad2 activation (Figure 2C), demonstrating a functional level of TBRI inhibition with the shRNA. Further functional validation showed that introduction of anti–TBRI-shRNA in primary CD34+ hematopoietic progenitors led to their resistance to TGF-β–mediated suppression of erythroid colony formation (Figure 2D). Hematopoietic progenitors with GFP+ TBRI-shRNA expression also formed bigger colonies, again demonstrating resistance to TGF-β–mediated antiproliferative signals (Figure 2E).
Down-regulation of TGF beta receptor I (TBRI) can inhibit smad2 activation in hematopoietic cells and stimulate MDS hematopoiesis. GFP-expressing lentiviral-based shRNA against TBRI (A) was used to knock down TBRI in hematopoietic cells. qPCR shows decrease in TBRI mRNA expression after lentiviral shRNA-TBRI infection in bone marrow stromal (HS-5) and leukemia cells (K562) compared with scrambled control (B). K562 cells with stable expression of TBRI-shRNA lentivirus show decreased smad2 phosphorylation after TGF stimulation (C). Primary CD34+ progenitors were electroporated with GFP coexpressing anti–TBRI-shRNA construct and sorted after 48 hours. GFP-positive cells were grown in methylcellulose with cytokines, and erythroid colonies were counted after 14 days. TBRI-shRNA–transfected progenitors were less inhibited by TGF-β compared with cells transfected with scrambled control shRNA (31% colonies/control vs 16% colonies/control). Expressed as means (± SEM) of 4 independent experiments (P = .03, 2-tailed t test) (D). CD34+ cells transfected with anti–TBRI-shRNA also formed bigger colonies in the presence of TGF-β1 compared with controls (E; Nikon, 40×). Primary bone marrow–derived mononuclear cells from 5 patients with MDS were transfected with shRNA targeting TBRI and control, and equal numbers of cells (for each individual patient) were grown in methylcellulose with cytokines. Erythroid (BFU-E) and myeloid (CFU-GM) colonies were counted after 14 days of culture and demonstrated an increase after anti–TBRI-shRNA transfection (significance between means calculated by 2-tailed t test) (F).
Down-regulation of TGF beta receptor I (TBRI) can inhibit smad2 activation in hematopoietic cells and stimulate MDS hematopoiesis. GFP-expressing lentiviral-based shRNA against TBRI (A) was used to knock down TBRI in hematopoietic cells. qPCR shows decrease in TBRI mRNA expression after lentiviral shRNA-TBRI infection in bone marrow stromal (HS-5) and leukemia cells (K562) compared with scrambled control (B). K562 cells with stable expression of TBRI-shRNA lentivirus show decreased smad2 phosphorylation after TGF stimulation (C). Primary CD34+ progenitors were electroporated with GFP coexpressing anti–TBRI-shRNA construct and sorted after 48 hours. GFP-positive cells were grown in methylcellulose with cytokines, and erythroid colonies were counted after 14 days. TBRI-shRNA–transfected progenitors were less inhibited by TGF-β compared with cells transfected with scrambled control shRNA (31% colonies/control vs 16% colonies/control). Expressed as means (± SEM) of 4 independent experiments (P = .03, 2-tailed t test) (D). CD34+ cells transfected with anti–TBRI-shRNA also formed bigger colonies in the presence of TGF-β1 compared with controls (E; Nikon, 40×). Primary bone marrow–derived mononuclear cells from 5 patients with MDS were transfected with shRNA targeting TBRI and control, and equal numbers of cells (for each individual patient) were grown in methylcellulose with cytokines. Erythroid (BFU-E) and myeloid (CFU-GM) colonies were counted after 14 days of culture and demonstrated an increase after anti–TBRI-shRNA transfection (significance between means calculated by 2-tailed t test) (F).
After demonstrating TBRI as a functionally important mediator of TGF-smad2 signaling, we expressed anti–TBRI-shRNA in 5 primary MDS bone marrow patient samples and performed clonogenic assays in methycellulose. Three patients belonged to RCMD subtype, and 1 each belonged to RA and RAEB subtype. The patient with RAEB had moderate marrow fibrosis. All MDS samples demonstrated an increase in erythroid and myeloid colony formation after anti–TBRI-shRNA expression compared with scrambled control, demonstrating an important functional role of this kinase in hematopoietic suppression seen in MDS (Figure 2F).
SD-208 is an effective and functionally active inhibitor of TGF-β signaling in hematopoietic cells
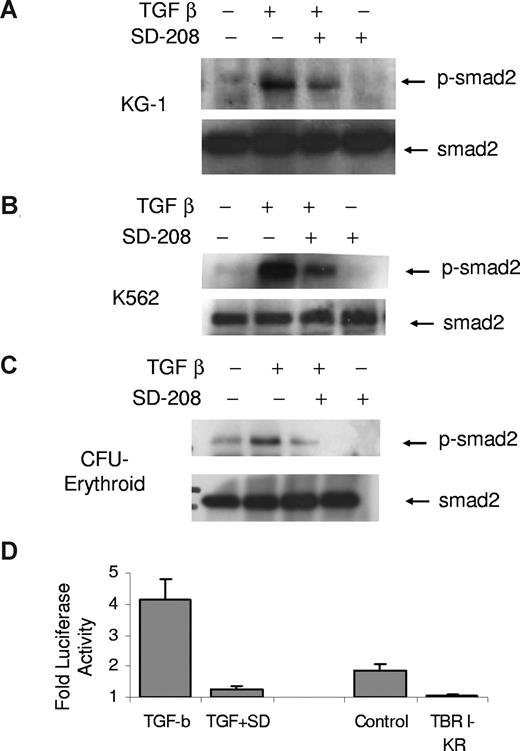
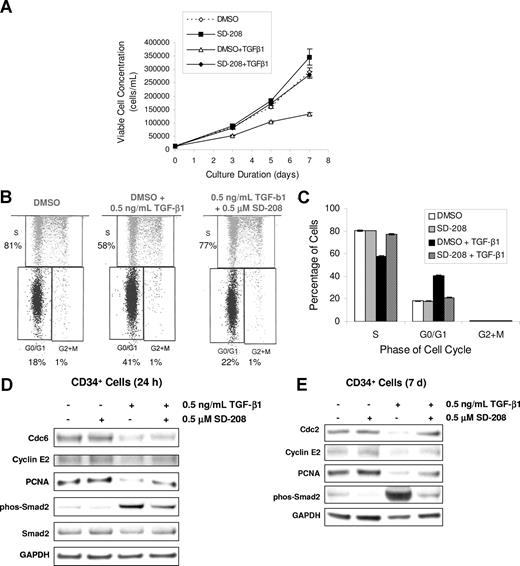
Having demonstrated an important role of TBRI in TGF-smad signaling in hematopoiesis, we next wanted to test the efficacy of SD-208, a novel small molecule pyridopyrimidine inhibitor of TBRI. We first demonstrated that SD-208 was able to potently inhibit smad2 activation in myeloid hematopoietic cell lines KG-1 and K562 as well as in erythroid progenitors derived from primary BM CD34+ cells (Figure 3A-C). Because the bone marrow microenvironment plays an important role in cytokine signaling and production in bone marrow failure states, we next tested the efficacy of SD-208 in human bone marrow stromal cells. SD-208 was also able to effectively inhibit TGF-smad2/3–mediated gene transcription in the bone marrow stromal cell line, HS-5. (Figure 3D) Furthermore, functional testing revealed that SD-208 was also able to abrogate the suppressive affects of TGF-β on primary human CD34 proliferation (Figure 4A). These effects were dependent on its ability to block TGF-mediated G0/G1 cell-cycle arrest in these hematopoietic progenitors (Figure 4B,C). Gene expression analysis also supported these findings by demonstrating that cell- cycle progression proteins were not down-regulated by TGF-β in the presence of SD-208 (Table 1). Immunoblotting validated these microarray finings (Figure 4E), thus demonstrating the functional ability of TBRI inhibitor in reversing TGF-mediated actions in hematopoiesis.
SD-208 is an inhibitor of TGF-β signaling in hematopoietic cells. Leukemic cells (K562 and KG-1) and primary hematopoietic progenitors at the colony-forming unit–erythroid stage of maturation (CFU-E) were treated with TGF-β1 (20 ng/mL) in the presence and absence of SD-208 (.5 μM) and assessed for smad2 phosphorylation by immunoblotting. SD-208 pretreament (1 hour) led to attenuation of activation/phosphorylation of smad2 (A-C). Bone marrow stroma–derived cells (HS-5) were transfected with plasmids expressing smad binding 3TP-luciferase and β-galactisidose (transfection control) and stimulated with TGF-β1 in the presence and absence of SD-208 (dose .5 μM). TGF-β1–induced control-normalized luciferase activity was potently inhibited by SD-208. A kinase-null mutant of TGF-β receptor I (TBRI-KR) was used a positive control (D). Error bars represent SEM.
SD-208 is an inhibitor of TGF-β signaling in hematopoietic cells. Leukemic cells (K562 and KG-1) and primary hematopoietic progenitors at the colony-forming unit–erythroid stage of maturation (CFU-E) were treated with TGF-β1 (20 ng/mL) in the presence and absence of SD-208 (.5 μM) and assessed for smad2 phosphorylation by immunoblotting. SD-208 pretreament (1 hour) led to attenuation of activation/phosphorylation of smad2 (A-C). Bone marrow stroma–derived cells (HS-5) were transfected with plasmids expressing smad binding 3TP-luciferase and β-galactisidose (transfection control) and stimulated with TGF-β1 in the presence and absence of SD-208 (dose .5 μM). TGF-β1–induced control-normalized luciferase activity was potently inhibited by SD-208. A kinase-null mutant of TGF-β receptor I (TBRI-KR) was used a positive control (D). Error bars represent SEM.
TBRI inhibition can inhibit TGF-β–mediated cell-cycle arrest of CD34+ cells. Equal numbers of BM CD34+ cells were grown in the presence of SCF, TPO, and FLT-3L and were pretreated with DMSO or 0.5 μM SD-208 for 1 hour before TGF-β1 (0.5 ng/mL final concentration) was added. On days 3, 5, and 7, cell aliquots were taken and viable cell concentration was determined using Guava ViaCount. The experiment was repeated at least 3 times (using multiple donors of CD34+ cells) and means plus or minus SEM is shown (A). CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 for 7 days. Cell-cycle distribution of CD34+ cells (gated with PE-conjugated CD34 antibody) was determined on day 7 using the APC BrdU Flow Kit and the LSR-II flow cytometer (BD Biosciences; B; representative sample, C). Error bars represent SEM. CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 as described above. After 24 hours, cDNA was prepared and hybridized on a cDNA microarray. Selected cell-cycle progression genes that were down-regulated by TGF-β by 2-fold were validated at the protein level by Western blotting. CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 as described above. Cells were collected at the indicated time points and lysed in radioimmunoprecipitation assay (RIPA) buffer. Equal protein was separated on a 10% Bis-Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to nitrocellulose membrane and immunoblotted with the antibodies (D). GAPDH levels were used as protein loading controls, and p-smad2 was used as positive control for TGF-β1 stimulation.
TBRI inhibition can inhibit TGF-β–mediated cell-cycle arrest of CD34+ cells. Equal numbers of BM CD34+ cells were grown in the presence of SCF, TPO, and FLT-3L and were pretreated with DMSO or 0.5 μM SD-208 for 1 hour before TGF-β1 (0.5 ng/mL final concentration) was added. On days 3, 5, and 7, cell aliquots were taken and viable cell concentration was determined using Guava ViaCount. The experiment was repeated at least 3 times (using multiple donors of CD34+ cells) and means plus or minus SEM is shown (A). CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 for 7 days. Cell-cycle distribution of CD34+ cells (gated with PE-conjugated CD34 antibody) was determined on day 7 using the APC BrdU Flow Kit and the LSR-II flow cytometer (BD Biosciences; B; representative sample, C). Error bars represent SEM. CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 as described above. After 24 hours, cDNA was prepared and hybridized on a cDNA microarray. Selected cell-cycle progression genes that were down-regulated by TGF-β by 2-fold were validated at the protein level by Western blotting. CD34+ cells were treated with DMSO or SD-208 in the presence or absence of TGF-β1 as described above. Cells were collected at the indicated time points and lysed in radioimmunoprecipitation assay (RIPA) buffer. Equal protein was separated on a 10% Bis-Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to nitrocellulose membrane and immunoblotted with the antibodies (D). GAPDH levels were used as protein loading controls, and p-smad2 was used as positive control for TGF-β1 stimulation.
Cell-cycle progression genes down-regulated by TGF-β
| Clone ID . | TGF-β1 (24 h)* . | SD-208 plus TGF-β1 (24 h) . | Symbol . | Name . |
|---|---|---|---|---|
| P01125_G01 | −2.3 | 1.8 | CDC2 | Cell division cycle 2 |
| P01066_F04 | −2.6 | 2.0 | CDC6 | Cell division cycle 6 |
| P01091_D07 | −2.0 | 2.2 | CDC7 | Cell division cycle 7 |
| P01108_D07 | −2.0 | 1.9 | CDC45L | Cell division cycle 45-like |
| P01077_E02 | −3.0 | 2.2 | CCNA2 | Cyclin A2 |
| P01093_D10 | −1.9 | 1.9 | CCNE1 | Cyclin E1 |
| P01113_A07 | −2.1 | 2.0 | CCNE2 | Cyclin E2 |
| P01071_H02 | −2.3 | 2.1 | PCNA | Proliferating cell nuclear antigen |
| P01071_E06 | −1.9 | 2.0 | RFC3 | Replication factor C3 |
| P01102_D08 | −2.1 | 1.8 | E2F1 | E2F transcription factor 1 |
| Clone ID . | TGF-β1 (24 h)* . | SD-208 plus TGF-β1 (24 h) . | Symbol . | Name . |
|---|---|---|---|---|
| P01125_G01 | −2.3 | 1.8 | CDC2 | Cell division cycle 2 |
| P01066_F04 | −2.6 | 2.0 | CDC6 | Cell division cycle 6 |
| P01091_D07 | −2.0 | 2.2 | CDC7 | Cell division cycle 7 |
| P01108_D07 | −2.0 | 1.9 | CDC45L | Cell division cycle 45-like |
| P01077_E02 | −3.0 | 2.2 | CCNA2 | Cyclin A2 |
| P01093_D10 | −1.9 | 1.9 | CCNE1 | Cyclin E1 |
| P01113_A07 | −2.1 | 2.0 | CCNE2 | Cyclin E2 |
| P01071_H02 | −2.3 | 2.1 | PCNA | Proliferating cell nuclear antigen |
| P01071_E06 | −1.9 | 2.0 | RFC3 | Replication factor C3 |
| P01102_D08 | −2.1 | 1.8 | E2F1 | E2F transcription factor 1 |
Fold change of gene expression over control unstimulated CD34+ cells.
TBRI inhibition can improve anemia in a model of bone marrow failure
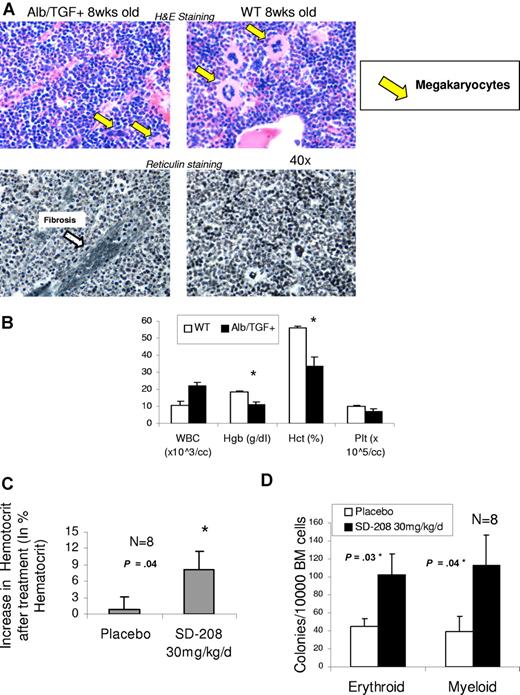
To further examine the effects of TGF-β on bone marrow function, we evaluated the hematopoietic effects of constitutive TGF secretion in a transgenic mouse expressing a fusion gene (Alb/TGF) consisting of modified porcine TGF-β cDNA under the control of the regulatory elements of the mouse albumin gene.38 We determined that these mice become anemic at an early age (3 weeks), and histologic examination of their bone marrows reveals dysplastic megakaryocytes and focal marrow reticulin fibrosis, features commonly seen in human bone marrow failure states such as MDS (Figure 5A,B). These mice were used to determine the specificity and efficacy of SD-208 in an in vivo setting. Mice were randomized into treatment or placebo groups on the basis of pretreatment hematocrits. Blood counts were measured after 14 days of oral administration of SD-208 or vehicle. TBRI inhibitor treatment led to significant increase in hematocrit in these mice, demonstrating the specificity of SD-208 in inhibiting TGF-β signaling. (Figure 5C). Furthermore, bone marrow progenitors of treated mice showed increased erythroid and myeloid colony forming ability compared with placebo control, thus demonstrating the efficacy of the TBRI inhibitor in rescuing hematopoietic activity in vivo (Figure 5D).
SD-208 can improve anemia in a murine model of TGF-β1–driven bone marrow failure. Mice transgenic for alb/TGF-β were killed at 8 weeks of age and their bone marrows were stained for histology (hematoxylin and eosin and reticulin stain). Transgenic mice demonstrated dysplastic micromegakaryocytes and patchy fibrosis. (A) Blood counts were analyzed at 3 weeks by Advia (Bayer) counter, and alb/TGF+ transgenic mice were found to be significantly anemic compared with WT controls (n = 4; means ± SEM; Nikon, 40×) (B). alb/TGF+ mice were treated with either SD-208 (30 mg/kg per day) or vehicle (placebo, daily) by gastric lavage for 14 days. Blood counts were done on the 14th day and revealed a significant rise in hematocrit after SD-208 treatment (C). The mice were killed and bone marrow cells were plated in methylcellulose with Epo (for BFU-E colonies) and IL-3, IL-6, and SCF (for CFU-GM colonies). SD-208 treatment led to a significant increase in both erythroid and myeloid colony-forming potential compared with placebo (n = 8; means ± SEM; 2-tailed t test; D).
SD-208 can improve anemia in a murine model of TGF-β1–driven bone marrow failure. Mice transgenic for alb/TGF-β were killed at 8 weeks of age and their bone marrows were stained for histology (hematoxylin and eosin and reticulin stain). Transgenic mice demonstrated dysplastic micromegakaryocytes and patchy fibrosis. (A) Blood counts were analyzed at 3 weeks by Advia (Bayer) counter, and alb/TGF+ transgenic mice were found to be significantly anemic compared with WT controls (n = 4; means ± SEM; Nikon, 40×) (B). alb/TGF+ mice were treated with either SD-208 (30 mg/kg per day) or vehicle (placebo, daily) by gastric lavage for 14 days. Blood counts were done on the 14th day and revealed a significant rise in hematocrit after SD-208 treatment (C). The mice were killed and bone marrow cells were plated in methylcellulose with Epo (for BFU-E colonies) and IL-3, IL-6, and SCF (for CFU-GM colonies). SD-208 treatment led to a significant increase in both erythroid and myeloid colony-forming potential compared with placebo (n = 8; means ± SEM; 2-tailed t test; D).
Pharmacologic inhibition of TBRI kinase can stimulate MDS hematopoiesis
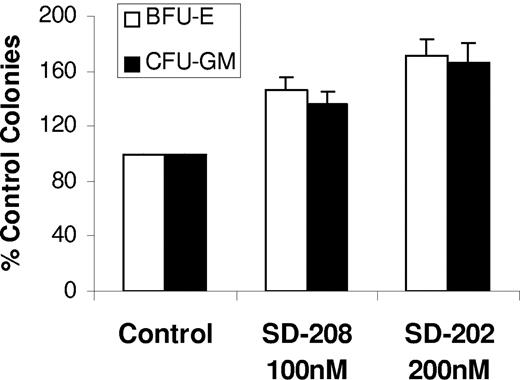
Finally, we tested the ability of SD-208 in vitro in 15 MDS bone marrow samples. Most of the patients had low-grade MDS and did not have increased blast counts (Table 2). Mononuclear cells were grown in methylcellulose with cytokines in the presence and absence of various doses of SD-208. Consistent with results seen with siRNAs, treatment with the TBRI inhibitor (Figure 6) resulted in a significant increase in erythroid (BFU-E) and myeloid (CFU–granulocytic monocytic) colony numbers (Figure 6A) These results point to a high therapeutic potential of TBRI inhibition in low-grade MDS.
Patient characteristics
| Age, y/sex . | WBC count, ×109/L . | Hgb level, g/dL . | Plt count, ×109/L . | Cytogenetics . | Path . | IPSS . | Risk . |
|---|---|---|---|---|---|---|---|
| 89/M | 4.6 | 9.2 | 169 | N | CMML | 0 | Low |
| 71/M | 3.9 | 8.1 | 154 | N | RA | 0 | Low |
| 76/M | 6 | 7 | 137 | N | RA | 0 | Low |
| 55/M | 3.4 | 10 | 146 | N | RA | 0 | Low |
| 69/M | 2.1 | 9.6 | 120 | N | RCMD | 0.5 | Int-1 |
| 65/M | 2.4 | 10.8 | 90 | N | RCMD | 0.5 | Int-1 |
| 58/M | 3 | 12 | 106 | N | RA | 0.5 | Int-1 |
| 88/M | 2.4 | 8.3 | 155 | N | RCMD | 0.5 | Int-1 |
| 77/M | 2 | 12 | 174 | N | RCMD | 0.5 | Int-1 |
| 76/M | 5.2 | 9.4 | 41 | t(11:14) | RCMD | 1 | Int-1 |
| 70/M | 4.7 | 12 | 20 | del 20q | RAEB | 1 | Int-1 |
| 78/M | 0.6 | 6 | 30 | del 16 (q22) | RCMD | 1.5 | Int-2 |
| 48/F | 5.2 | 8.4 | 95 | −1q, −11q | RAEB | 1.5 | Int-2 |
| 73/M | 1.2 | 9.4 | 88 | −20q, iso 17q | RCMD | 2 | Int-2 |
| 65/F | 3.1 | 8.3 | 54 | N | RAEB | 1.5 | Int-2 |
| Age, y/sex . | WBC count, ×109/L . | Hgb level, g/dL . | Plt count, ×109/L . | Cytogenetics . | Path . | IPSS . | Risk . |
|---|---|---|---|---|---|---|---|
| 89/M | 4.6 | 9.2 | 169 | N | CMML | 0 | Low |
| 71/M | 3.9 | 8.1 | 154 | N | RA | 0 | Low |
| 76/M | 6 | 7 | 137 | N | RA | 0 | Low |
| 55/M | 3.4 | 10 | 146 | N | RA | 0 | Low |
| 69/M | 2.1 | 9.6 | 120 | N | RCMD | 0.5 | Int-1 |
| 65/M | 2.4 | 10.8 | 90 | N | RCMD | 0.5 | Int-1 |
| 58/M | 3 | 12 | 106 | N | RA | 0.5 | Int-1 |
| 88/M | 2.4 | 8.3 | 155 | N | RCMD | 0.5 | Int-1 |
| 77/M | 2 | 12 | 174 | N | RCMD | 0.5 | Int-1 |
| 76/M | 5.2 | 9.4 | 41 | t(11:14) | RCMD | 1 | Int-1 |
| 70/M | 4.7 | 12 | 20 | del 20q | RAEB | 1 | Int-1 |
| 78/M | 0.6 | 6 | 30 | del 16 (q22) | RCMD | 1.5 | Int-2 |
| 48/F | 5.2 | 8.4 | 95 | −1q, −11q | RAEB | 1.5 | Int-2 |
| 73/M | 1.2 | 9.4 | 88 | −20q, iso 17q | RCMD | 2 | Int-2 |
| 65/F | 3.1 | 8.3 | 54 | N | RAEB | 1.5 | Int-2 |
WBC indicates white blood cell; Hgb, hemmoglobin; Plt, platelet; Path, pathological condition; and CMML, chronic myelomonocytic leukemia
TBRI inhibition stimulates hematopoiesis in MDS. MDS bone marrow–derived MNCs from 15 patients were plated in methylcellulose and cytokines in the presence and absence of TBRI inhibitor SD-208 (100 nM and 200 nM). Colonies were scored at day 14 and results were expressed as means (± SEM) of 15 independent experiments.
TBRI inhibition stimulates hematopoiesis in MDS. MDS bone marrow–derived MNCs from 15 patients were plated in methylcellulose and cytokines in the presence and absence of TBRI inhibitor SD-208 (100 nM and 200 nM). Colonies were scored at day 14 and results were expressed as means (± SEM) of 15 independent experiments.
Discussion
Progress in the discovery of disease-selective treatments for MDS has been challenged by the limited insight into molecular pathogenesis of the ineffective hematopoiesis seen in these disorders. Our work has shown that TGF-smad2 pathway is commonly overactivated in a variety of subtypes of MDS. Inhibition of the TGF-β receptor I kinase (TBRI) blocks smad2 activation and promotes MDS hematopoiesis in vitro, suggesting that TGF-β–smad2 signaling pathway is a functionally important inhibitory pathway in MDS.
Transforming growth factor–beta (TGF-β) is an important physiologic regulator of cell proliferation and differentiation.41 TGF-β has been shown to affect both early and late stages of hematopoiesis and generally leads to suppressive effects on erythroid and myeloid cell proliferation41 Deregulation of TGF-β signaling has been implicated in a variety of carcinogenesis and metastasis models, illustrating an important biologic role in oncogenesis. Because MDS is a preleukemic condition characterized by dysplastic hematopoiesis, earlier studies have also examined the role of TGF-β in this disease. Most reports to date have relied upon immunohistochemical detection of TGF-β in patient bone marrows or measurement of TGF-β levels in patient plasma. Some of these studies showed increased TGF-β in MDS,10-13 whereas some did not observe any correlation.14-17 As local concentrations of cytokines can be more informative than serum concentrations and immunohistochemical detection of cell surface proteins in bone marrow sections has technical limitations, we focused our studies on direct evaluation of nuclear smad2 phosphorylation in MDS bone marrows. Smad2 activation and nuclear translocation are specific and accurate markers of TGF-β signaling, and Smad2 up-regulation occurs with sustained TGF-β signaling. Thus, our results are the first to directly show evidence that TGF-β signaling is a common pathway activated in MDS and indicate that a downstream signaling biomarker can identify a valid therapeutic target.
Because MDS is a heterogeneous disease, we validated our findings in a larger number of MDS samples. We devised a meta-analytic approach using publicly available gene expression data to test our hypothesis. Recent publications have concluded that microarray data generated in different laboratories and different platforms can be compared after adequate integration and normalization.42-47 Our database covered 6 independent studies30-34,40 and provides further validation of our findings that is hard to achieve with single-center studies in a complex disease such as MDS. We hope that creation of this database will also help other MDS investigators.
Our findings are consistent with other work that has been done in defining molecular pathways in MDS. Our earlier data demonstrated that cytokine signaling cascades such as p38 MAPK can be functionally important in regulating hematopoietic failure in MDS.48 It has also been shown that TGF-β can activate p38 and other MAP kinase pathways.19,41,49 Thus our earlier results can in part be explained by overactivated TGF-β signaling in MDS. A recent clinical trial evaluating thalidomide in MDS observed that patients with increased red cell counts after treatment had a significant decrease in TGF-β levels.9 Even though this was a small study it reinforces our findings, suggesting an important role of TGF-β signaling in ineffective hematopoiesis. In addition, our data point to a greater role of TGF-β–suppressive signaling in low-grade MDS. The bone marrow at this stage of the disease is hypercellular and is characterized by increased rates of apoptosis. At this stage of the disease, both normal and cytogenetically abnormal hematopoietic clones are found to exist in the marrow.50 With disease progression toward high-risk stages, normal progenitors gradually diminish, resulting in a bone marrow composed mainly of the resistant abnormal clones with an increased percentage of myeloblasts. These observations in combination with our data may allude to a pathogenic role of TGF-β signaling in suppressing hematopoiesis in normal clonal progenitors in low-grade MDS. As preponderance of leukemic clones seen in higher grade MDS is characterized by relative resistance to suppressive effects of TGF-β,41,51-53 the therapeutic efficacy of TBRI inhibitors may be greater in low-grade disease subtypes.
Ineffective hematopoiesis is the cause of most of morbidity in patients with low-risk MDS. Two-thirds of all MDS cases are at the low-risk stage, have a lower chance of progressing to leukemia, and suffer problems mainly associated with low blood counts. Thus strategies aimed at raising blood counts are needed at this stage. The erythroid lineage is the most commonly affected hematopoietic lineage in MDS. TGF-β has recently been shown to regulate both primitive and late stages of erythropoiesis.54 by distinct molecular processes. After smad2/3 phosphorylation by the TBRI kinase, these proteins associate with partner smads and other proteins to bind distinct DNA sequences and regulate different biologic functions. It has recently been shown that smad2-smad4 complex enters the nucleus and negatively regulates suppression of primitive erythropoiesis, whereas the smad2-TIFγ complex can regulate late stages of erythroid differentiation.54 Thus TBRI-dependent smad2 overactivation in MDS can affect different facets of erythropoiesis and can be abrogated by inhibition of the alk5 kinase.
In addition to MDS, TGF-β has also been implicated in the pathobiology of idiopathic myelofibrosis (IMF). Due to the fibrogenic actions of TGF-β, oversecretion of this cytokine has been correlated with marrow fibrosis seen in IMF.55,56 In addition, myelofibrosis seen after thrombopoietin overexpression in mice can be abrogated by depletion of TGF-β1 by a soluble receptor, suggesting that TGF-β secretion is an important downstream effector of marrow fibrosis in various cytokine-induced models of hematopoietic failure.57 These data coupled with our findings provide a rationale for using inhibitors of TGF-β signaling (including TBRI inhibitors) in other bone marrow failure syndromes. TBRI selectively participates in TGF-β signaling, whereas other activin-like and TGF-β receptors can also participate in BMP and activin ligand signaling49,58 and thus provide selective therapeutic efficacy. SD-208 is a selective 2,4-disubstituted pteridine-derived TGF-βRI kinase inhibitor23-28 that has demonstrated specificity for this kinase. In addition, other TBRI inhibitors are in development27,59 for various other indications such as chronic renal diseases. Our findings provide a preclinical rationale for bringing these agents into clinical trials for MDS.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH, Bethesda, MD) 1R01HL082946-01 and Community Foundation of Southeastern Michigan (Detroit) J. P. McCarthy fund award (A.V.); by NIH RO1 AG029138 (L.C.P.); and by Immunooncology Training Program T32 CA009173 (L.Z.).
National Institutes of Health
Authorship
Contribution: L.Z. and A.N.N. designed and performed experiments and wrote the paper; D.S., J.Y.M., P.P., K.G., J.H., A.C., Y.M., T.D.B., B.D., A.M.K., T.A.N., and A.P. performed experiments; S.P., S.K., I.B., Y.Z., A.W., J.B., A.F.L., and M.B. contributed samples; S.M., L.C.P, and L.S.H. participated in design of studies; and A.V. designed experiments and wrote the paper.
Conflict-of-interest disclosure: A.N.N., J.Y.M., A.M.K., T.A.N., S.M., and L.S.H. are employees of Scios, Inc. The remaining authors declare no competing financial interests.
Correspondence: Amit Verma, Chanin 302B, Albert Einstein Cancer Center, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: averma@aecom.yu.edu.