Abstract
Myelodysplastic syndromes (MDSs) pose an important diagnostic and treatment challenge because of the genetic heterogeneity and poorly understood biology of the disease. To investigate initiating genomic alterations and the potential prognostic significance of cryptic genomic changes in low-risk MDS, we performed whole genome tiling path array comparative genomic hybridization (aCGH) on CD34+ cells from 44 patients with an International Prognostic Scoring System score less than or equal to 1.0. Clonal copy number differences were detected in cells from 36 of 44 patients. In contrast, cells from only 16 of the 44 patients displayed karyotypic abnormalities. Although most patients had normal karyotype, aCGH identified 21 recurring copy number alterations. Examples of frequent cryptic alterations included gains at 11q24.2-qter, 17q11.2, and 17q12 and losses at 2q33.1-q33.2, 5q13.1-q13.2, and 10q21.3. Maintenance of genomic integrity defined as less than 3 Mb total disruption of the genome correlated with better overall survival (P = .002) and was less frequently associated with transformation to acute myeloid leukemia (P = .033). This study suggests a potential role for the use of aCGH in the clinical workup of MDS patients.
Introduction
The myelodysplastic syndromes (MDSs) are a heterogeneous group of clonal hematologic malignancies characterized by peripheral cytopenias, a hypercellular marrow with ineffective hematopoiesis, and a propensity to progress to acute myeloid leukemia (AML).1,2 MDS is thought to arise from a primitive hematopoietic progenitor that has acquired genetic and/or epigenetic abnormalities.3 The cell of origin in MDS is thought to reside within the CD34+ population, based on multiparameter flow cytometry and fluorescence in situ hybridization (FISH) studies, global gene expression analysis, as well as on xenotransplantation of marrow cells from MDS patients into immunodeficient mice.4-7 However, in comparison with other hematologic malignancies, there is limited biologic understanding of the molecular abnormalities that contribute to the pathogenesis of MDS.
The current categorization of MDS according to the World Health Organization consists of 8 subtypes, based on biologic, genetic, and morphologic features.8,9 In addition, an International Prognostic Scoring System (IPSS), which takes into account the proportion of leukemic blast cells in the marrow, the number of cytopenias, and the karyotype, has been devised to assign patients to risk categories with respect to survival and the likelihood of AML transformation.2 The importance of cytogenetics in risk stratification has been verified in several studies, but this is of limited value in patients with lower-risk subtypes because approximately 50% of these patients do not have karyotypic abnormalities that are detectable using standard techniques.1-3 Greater than 50% of patients diagnosed with MDS are of the lower-risk groups, which generally correspond to the morphologic subtypes of refractory anemia (RA), refractory anemia with ringed sideroblasts (RARS), refractory cytopenia with multilineage dysplasia (RCMD), refractory cytopenia with multilineage dysplasia and ringed sideroblasts (RCMD-RS), and MDS with (del)5q.2,8
To further our biologic understanding, particularly of low-risk subtypes of MDS, we performed a high-resolution whole genome array comparative genomic hybridization (aCGH) analysis of CD34+ progenitor cells isolated from the marrow of MDS patients. Of note, the aCGH analyses in this study were all conducted on unamplified genomic DNA from the CD34+ fraction, thus eliminating biases secondary to PCR amplification. We also evaluated the effectiveness of high-resolution analysis of the structural integrity of the genome on providing additional prognostic information in patients with lower-risk MDS. Our whole genome aCGH analysis of CD34+ cells isolated from the marrow of a mixed group of karyotypically normal (n = 25) and abnormal (n = 15) MDS patients revealed that extensive genome alteration involving more than 3 Mb detectable by aCGH correlated with poorer overall survival and was more frequently associated with transformation to AML.
Methods
Sample collection
Bone marrow samples were obtained from 44 patients diagnosed with MDS, of which 38 were collected at the time of diagnosis. Of these, 40 patients with IPSS score less than or equal to 1.0 (Tables S1, S2, available on the Blood website; see the Supplemental Materials link at the top of the online article; median age, 76 years; range, 33-90 years; mean, 71.3 ± 1.9 years) were included in the survival analysis. Seven of the 44 patients received treatment during the course of the disease (3 received allogeneic bone marrow transplants, 2 received trials of erythropoietin, 1 received hydroxyurea, and 1 received antithymocyte globulin). Blood samples were obtained from 15 elderly subjects without evidence of marrow-derived malignancy after informed consent was obtained in accordance with the Declaration of Helsinki. Ten were healthy subjects with no evidence of cancer (other than 2 with nonmelanoma skin cancer) whose blood was collected as part of a healthy aging study. Five patients had marrow collected as part of the staging procedure for non-Hodgkin lymphoma but did not exhibit marrow or peripheral blood involvement by lymphoma. All protocols were approved by the Clinical Research Ethics Board of the British Columbia Cancer Agency/University of British Columbia. An MDS-derived CD34+ multipotent cell line (MDS-L) was used as an additional control.10,11
Cell isolation
CD34+ and CD3+ cells were purified from total marrow cells by magnetic cell separation (Miltenyi Biotec, Auburn, CA) as per the manufacturer's instructions (StemCell Technologies, Vancouver, BC). Flow-through (CD34−) cells were also collected for analysis. Purity after a single pass-through of the column of CD34+ and CD3+ cells was more than 70% as evaluated by flow cytometry (FACScan; BD Biosciences, San Jose, CA). Peripheral blood cells were prepared by Ficoll-Paque as suggested by the manufacturer (StemCell Technologies). Genomic DNA was extracted by AllPrep DNA/RNA Mini Kit (QIAGEN, Valencia, CA). Normal reference DNA was purchased as a pool of either male or female genomic DNA (Novagen, Madison, WI).
Whole genome tiling-path aCGH analysis
Details of whole genome array construction and probe labeling and hybridization have been described previously.12,13 The submegabase-resolution tiling set array contains 32 433 overlapping BAC-derived DNA segments that provide tiling coverage over the entire human genome with a theoretical resolution of 50 kb.12,14 All clones were spotted in duplicate. Sample (MDS), and reference (normal diploid) genomic DNA (50-200 ng each) were separately labeled using cyanine 3 and cyanine 5 dCTPs fluorescence markers, respectively. The images were captured with a charge-coupled device camera and analyzed using an ArrayWorx scanner and SoftWorx Tracker Spot Analysis software (Applied Precision, Issaquah, WA). SeeGH custom software (available by request to www.SeeGH@bccrc.ca) was used to visualize all data as log ratio plots.12,15 Clones with SD values between duplicate spots of more than 0.1 were filtered from the raw data. To avoid false-positives because of hybridization noise, a minimum of 2 overlapping consecutive clones showing change was required for a region to be considered altered. Breakpoints of genomic alterations were identified using a hidden Markov model algorithm and verified by visual inspection, as described.16 Copy number variants (CNVs) were excluded from analysis of the CD34+ population if the identical copy number alteration was found in matched CD3+ cells from the same patient. If matched CD3+ material was not available, copy number alterations identified in the normal controls and the Database of Genomic Variants were excluded from these patients. CNVs in the human population were identified by comparison to known polymorphic BAC clones according to the most current version (November 29, 2007) of the Database of Genomic Variants (www.projects.tcag.ca/variation).17
Fluorescent in situ hybridization
For FISH analysis, interphase chromosomes were prepared from the MDS-L cell line, and as a control, 3 normal BM samples to determine technical and biologic variation at these loci. For detection of gene amplification at chromosome band 17q12, BAC probes within the region of interest (RP11-072I20) and one outside the region of interest (RP11-686E5) were used. DNA from the BAC clones was isolated and labeled with either Spectrum Orange-dUTP or Spectrum Green-dUTP by nick translation (Abbott Laboratories, Abbott Park, IL). Interphase FISH using a modified formamide-free protocol (Abbott Laboratories) was followed. Image capture was performed with Ziess Axioplan II microscope equipped with the appropriate filters for DAPI, Spectrum Orange, and Spectrum Green, and the Metasystem ISIS imaging software. At least 100 interphase nuclei were analyzed. A locus was considered to be amplified when more than 2 FISH signals were present.
Statistical analysis
Spearman correlation was used to assess the relationship between age and total genomic alteration (TGA). Differences in TGA between normal controls and MDS patients were assessed using the Mann-Whitney test. Overall and AML-free survival was calculated by the Kaplan-Meier method, and differences between groups were determined by the log-rank test. AML-free survival was analyzed by censoring patients who died without progressing to AML. Analysis was carried out using GraphPad Prism4 (GraphPad Software, San Diego, CA) or JMP (JMP Software, Cary, NC).
Results
aCGH analysis reveals cryptic structural DNA alterations in CD34+ cells of patients with low-risk MDS that are not detectable by standard cytogenetic analysis
High-resolution genome analysis of cancer cell populations has proven to be a valuable tool to identify cryptic and submegabase alterations, define novel subtypes of cancer, and enhance prediction of overall survival in solid tumors.18-20 Unlike many leukemias, the bone marrow of MDS patients comprises a heterogeneous cell population with high rates of cell death.21-23 Thus, to identify early and potentially causal pathogenic changes in the genome that give rise to MDS, it is necessary to isolate the earliest stem/progenitor cell possible. Although blast cells in MDS express CD34, the proportion of CD34+ cells in the marrow remains low.4,24,25 Most commercially available aCGH platforms require a minimum of 1 μg of genomic DNA to perform analyses without amplification.14 Sample DNA amplification can introduce noise and bias by nonlinear amplification of sequences. We thus selected the submegabase-resolution tiling set BAC aCGH platform because the requirement for genomic DNA is much lower for efficient hybridization (50 ng without amplification), compared with other high-resolution platforms.14 Further, this platform can theoretically resolve genomic alterations as small as 50 kb with single-copy sensitivity.14
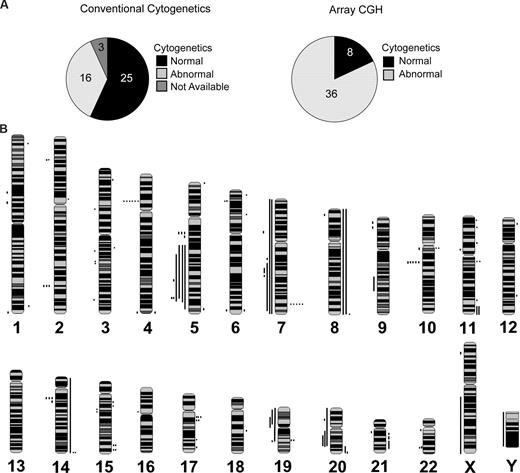
We analyzed 44 DNA samples of marrow CD34+ cells and matched CD3+ cells when available (n = 35) from low-risk MDS patients (24 RA/RCMD/5q-, 11 RARS/RCMD-RS, and 5 MDS/MPD), 4 samples from patients with RAEB-1, and 15 normal controls (Table S1). Copy number alterations in the CD34+ population of MDS patients were deemed “disease associated” (ie, clonal) if not present in matched normal CD3+ cells. If matched CD3+ cells were not available, the alterations were deemed “disease associated” when the alterations were absent in the cohortof age-matched normal controls and in the Database of Genomic Variants. At the time of diagnosis, conventional metaphase cytogenetic analysis was performed on 41 of 44 marrow specimens (Figure 1A). Results of the cytogenetic analysis are summarized in Tables 1 and S2. Twenty-five patients in this cohort had no karyotypic aberration. In contrast, only 8 of 44 patients had no detectable copy number alterations by aCGH (Figure 1A). Of the 16 patients with abnormal metaphase cytogenetic profiles, 11 were confirmed by aCGH to have the identical copy number alteration. In 2 cases (patients 5 and 25), the karyotype identified 2 independent clones. In both cases, the minor clone population was below the threshold of detection by aCGH (< 30%). The cytogenetic findings for patient 20 represent genomic alterations that do not involve a copy number change and would not be detectable by aCGH. For patients 11 and 22, deletion of the Y chromosome was not detectable by aCGH.
Array CGH detects cryptic alterations in MDS patients with normal cytogenetics. (A) Proportion of patients with abnormal cytogenetics as detected by conventional cytogenetics (left) and by aCGH (right). (B) Whole genome frequency distribution of chromosomal alterations in lower-risk patients (n = 44) as detected by aCGH and visualized by SeeGH. Lines to the right of the chromosome indicate gain of chromosomal material. Lines to the left of the chromosome indicate loss of chromosomal material.
Array CGH detects cryptic alterations in MDS patients with normal cytogenetics. (A) Proportion of patients with abnormal cytogenetics as detected by conventional cytogenetics (left) and by aCGH (right). (B) Whole genome frequency distribution of chromosomal alterations in lower-risk patients (n = 44) as detected by aCGH and visualized by SeeGH. Lines to the right of the chromosome indicate gain of chromosomal material. Lines to the left of the chromosome indicate loss of chromosomal material.
aCGH analysis of CD34+ bone marrow cells from 44 MDS patients
| Patient no. . | Diagnosis . | Karyotype . | Gain/loss . | Chromosome band . | Position, Mb* . | Change, Mb . |
|---|---|---|---|---|---|---|
| 1 | 5q- | 46,XX,del(5q) | L | 4q35.2 | 190.3-191.3 | 1 |
| L | 5q14.3-5q34 | 85.6-161.5 | 75.9 | |||
| G | 11q24.2-qter | 126.0-135.0 | 9 | |||
| L | 16p11.2 | 33.4-34.2 | 0.8 | |||
| 2 | RA | 47,XY,+14 | L | 7q22.1 | 99.6-101.4 | 1.8 |
| G | 14 | 17.4-105.4 | 88 | |||
| L | Xp22.13 | 14.9-15.6 | 0.7 | |||
| 3 | RA | 46,XX | G | 11q24.2-qter | 126.1-134.8 | 8.7 |
| G | 11p15.4 | 4.2-4.9 | 0.7 | |||
| G | 17q12 | 35.9-36.4 | 0.5 | |||
| G | 20q13.33 | 62.5-62.9 | 0.4 | |||
| 4 | RA | 46,XX,del(5q) | L | 5q14.3-5q35.1 | 86.3-168.5 | 82.2 |
| L | 9q21.33-q31.2 | 82.0-103.1 | 21.1 | |||
| 5 | RA | 47,XY,del(7),+8 | L | 7 | 0-158.1 | 158.1 |
| 6 | RA | 46,XX | G | 21q21.3 | 28.5-30.1 | 1.6 |
| G | 21q22.11-q22.12 | 33.9-36.0 | 2.1 | |||
| G | 21q22.13-21q22.2 | 38.2-40.8 | 2.6 | |||
| 7 | RA | 46,XX | G | 5q35.1 | 168.6-169.8 | 1.2 |
| L | 8q24.23 | 137.2-137.6 | 0.4 | |||
| G | 10q11.22 | 45.9-49.8 | 3.9 | |||
| G | 19q13.2-q13.31 | 47.8-48.2 | 0.4 | |||
| G | 21q21.3 | 27.9-28.1 | 0.2 | |||
| 8 | RA | 46,XY,del(20q) | L | 20q12-q13.2 | 40.2-51.9 | 11.7 |
| 9 | MDS/MPD (RARS-T) | 48,XY,del(20),+2mar | G | 6p25.1 | 6.4-6.8 | 0.4 |
| L | 20pter-q13.13 | 0-50.4 | 50.4 | |||
| G | 20q13.13-qter | 50.4-63.3 | 12.9 | |||
| 10 | 5q- | 46,XX,del(5q) | L | 5q14.3-q33.3 | 88.8-156.6 | 67.8 |
| L | 8p23.1 | 6.9-7.7 | 0.8 | |||
| 11 | RA | 46,XY/45,X,-Y | G | 6p21.2 | 39.2-40.2 | 1 |
| L | 7q21.11 | 83.4-83.6 | 0.2 | |||
| G | 7q34 | 141.6-142.0 | 0.4 | |||
| G | 15q24.1 | 71.6-71.7 | 0.1 | |||
| 12 | RCMD | 46,XY | G | 7q34 | 141.3-142.1 | 0.8 |
| L | 10q11.22 | 46.3-49.3 | 3 | |||
| L | 14q13.2 | 33.2-33.7 | 0.5 | |||
| L | 19q13.2-q13.31 | 47.8-48.3 | 0.5 | |||
| 13 | RA | 46,XX | G | 2p11.2 | 87.3-87.9 | 0.6 |
| L | 7q22.1 | 101.5-101.9 | 0.4 | |||
| G | 11q12.2 | 60.9-61.4 | 0.5 | |||
| 14 | RA | 46,XY | G | 8q21.2 | 86.1-86.7 | 0.6 |
| G | 17q12 | 35.9-36.3 | 0.4 | |||
| 15 | RAEB-1 | 46,XX | ND | |||
| 16 | RA | 46,XY | G | 6q27 | 167.8-168.2 | 0.4 |
| G | 15q13.3 | 30.0-30.4 | 0.4 | |||
| 17 | RA | 46,XX | L | 5q33.3 | 159.2-159.6 | 0.4 |
| 18 | MDS/MPD | 46,XX | L | 8p23.1 | 7.1-7.6 | 0.5 |
| 19 | RA | 46,XY | G | 11q12.2 | 61.2-61.4 | 0.2 |
| 20 | RA | 46,X,idic(X)(q13) | L | 6q26 | 160.5-161.0 | 0.5 |
| G | 10q11.22 | 46.2-47.2 | 1 | |||
| L | 20p12.1 | 14.6-15.1 | 0.5 | |||
| 21 | RA | 45,X,-Y | L | Y | 2.4-26.2 | 23.8 |
| 22 | RA | 45,X,-Y,del(7) | L | 4p14 | 39.1-40.6 | 1.5 |
| G | 5p15.33 | 0-2.3 | 2.3 | |||
| L | 5q13.2 | 68.9-70.1 | 0.2 | |||
| L | 7 | 0-158.1 | 158.1 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| G | 14q32.33 | 104.2-105.2 | 1 | |||
| L | 22q13.2 | 39.2-39.7 | 0.5 | |||
| 23 | RA | 46,XY,del(20q) | L | 20q11.21-q13.2 | 31.9-51.2 | 19.3 |
| 24 | 5q- | 46,XY,del(5q) | L | 5q21.1-q34 | 101.8-161.5 | 59.7 |
| G | 7q34 | 141.3-141.9 | 0.6 | |||
| 25 | RCMD | 47,XY,del(7),+8 | G | 2p11.2 | 87.3-87.9 | 0.6 |
| G | 7q34 | 141.4-141.9 | 0.5 | |||
| G | 8 | 0-143.9 | 143.9 | |||
| 26 | RARS | 46,XY | L | 1p22.1 | 91.6-93.1 | 1.5 |
| L | 1p31.1 | 77.3-77.9 | 0.6 | |||
| L | 2q33.1-q33.2 | 203-204 | 1 | |||
| L | 4p14 | 39.1-40.7 | 1.6 | |||
| G | 4q35.2 | 190.9-191.5 | 0.6 | |||
| L | 5q13.1-q13.2 | 68.1-69.0 | 0.9 | |||
| L | 6p23 | 14.9-15.5 | 0.6 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| L | 10q21.3 | 69.2-69.7 | 0.5 | |||
| L | 14q12 | 29.2-30.0 | 0.8 | |||
| G | 17q11.2 | 31.1-33.1 | 2 | |||
| 27 | RARS | 46,XX | ND | |||
| 28 | MDS/MPD (RARS-T) | 46,XY | ND | |||
| 29 | MDS/MPD (RARS-T) | 46,XX | L | 2q33.1-q33.2 | 203.0-204.3 | 1.3 |
| L | 4p14 | 39.1-40.0 | 0.9 | |||
| L | 9p24.1 | 6.6-6.8 | 0.2 | |||
| L | 10q21.3 | 64.6-65.2 | 0.6 | |||
| L | 14q12 | 29.4-29.9 | 0.5 | |||
| 30 | RAEB-1 | 46,XY | L | 4p14 | 39.0-39.8 | 0.8 |
| G | 12p13.31 | 10.8-11.5 | 0.7 | |||
| G | 15q25.3 | 86.0-86.5 | 0.5 | |||
| G | 15q24.1 | 71.2-71.8 | 0.6 | |||
| 31 | RARS | 46,XX | G | 15q14 | 33.6-33.9 | 0.3 |
| G | 18q21.1 | 44.3-44.5 | 0.2 | |||
| 32 | RARS | 46,XX | ND | |||
| 33 | RARS | 46,XY | L | 2p23.1 | 32.0-33.1 | 0.9 |
| L | 2q33.1-q33.2 | 202.9-204.8 | 1.9 | |||
| G | 3q13.12 | 108.7-109.2 | 0.5 | |||
| L | 3q26.2 | 170.8-171.6 | 0.8 | |||
| L | 4p14 | 39.0-39.8 | 0.8 | |||
| L | 5q13.1-q13.2 | 68.1-69.1 | 1 | |||
| L | 7p13 | 44.2-44.8 | 0.6 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| L | 14q12 | 29.3-30.1 | 0.8 | |||
| G | 17q11.2 | 31.1-32.9 | 1.8 | |||
| L | 19p | 0-24.3 | 24.3 | |||
| 34 | RARS | 46,XY | ND | |||
| 35 | RARS | 47,XY,+8 | G | 8 | 0-143.9 | 143.9 |
| 36 | RARS | 46,XY | ND | |||
| 37 | RARS | 46,XX | L | 22q11.21 | 169.5-173.0 | 0.5 |
| 38 | RARS | 46,XY | G | 1q42.3 | 231.0-231.4 | 0.4 |
| G | 7q34 | 141.2-141.8 | 0.6 | |||
| G | 14q32.33 | 104.2-105.3 | 1.1 | |||
| G | 15q25.3 | 85.9-86.5 | 0.6 | |||
| G | 17q24.1 | 63.1-63.4 | 0.3 | |||
| 39 | RAEB-1 | 46,XY | G | 3q21.3 | 127.7-128.5 | 0.8 |
| G | 3q21.3 | 129.1-129.5 | 0.4 | |||
| G | 11q13.2-q13.4 | 68.4-71.2 | 2.7 | |||
| 40 | RARS | 46,XY | G | 1q21.2 | 145.2-146.5 | 1.3 |
| L | 1q42.3 | 231.6-232.1 | 0.5 | |||
| L | 4p14 | 39.2-40.5 | 1.3 | |||
| L | 5q13.1-q13.2 | 68.3-68.9 | 0.6 | |||
| L | 5q31.1 | 133.5-134.3 | 0.8 | |||
| L | 6p23 | 14.9-15.4 | 0.5 | |||
| L | 9p22.2 | 16.6-17.3 | 0.7 | |||
| L | 10q21.3 | 64.6-65.2 | 0.6 | |||
| G | 17q11.2 | 31.2-33.1 | 1.9 | |||
| L | 19p12-13.3 | 1.1-24.2 | 23.1 | |||
| 41 | RA | NA | G | 8q21.2 | 86.2-86.6 | 0.4 |
| G | 11q23.2 | 114.4-115.0 | 0.6 | |||
| G | 12q23.3 | 106.0-106.6 | 0.6 | |||
| G | 17q25.1 | 72.0-72.6 | 0.6 | |||
| L | 21q21.1-q21.2 | 22.6-23.5 | 0.9 | |||
| L | Xq13.2-qter | 70.6-105.2 | 34.6 | |||
| 42 | RA | NA | ND | |||
| 43 | MDS/MPD (RARS-T) | NA | ND | |||
| 44 | RAEB | 46, XY, del (7q21-q31) | L | 2p23.1 | 32.0-33.0 | 1 |
| G | 3p25.3 | 10.3-10.9 | 0.6 | |||
| L | 3p14.3 | 56.4-57.8 | 1.4 | |||
| L | 3q13.2-q13.31 | 114.4-114.9 | 0.5 | |||
| L | 5q33.3 | 159.3-159.7 | 0.4 | |||
| L | 7q21.12-q36.2 | 87.6-152.6 | 65 | |||
| G | 11p15.2 | 14.9-16.0 | 1.1 | |||
| G | 11p12-p13 | 36.2-37.1 | 0.9 | |||
| L | 15q15.1 | 38.8-39.5 | 0.7 | |||
| L | 15q15.3-q21.1 | 41.9-42.5 | 0.6 | |||
| L | 19p-q13.2 | 0-46.3 | 46.3 | |||
| L | 22q13.1-q13.2 | 39.1-39.7 | 0.6 |
| Patient no. . | Diagnosis . | Karyotype . | Gain/loss . | Chromosome band . | Position, Mb* . | Change, Mb . |
|---|---|---|---|---|---|---|
| 1 | 5q- | 46,XX,del(5q) | L | 4q35.2 | 190.3-191.3 | 1 |
| L | 5q14.3-5q34 | 85.6-161.5 | 75.9 | |||
| G | 11q24.2-qter | 126.0-135.0 | 9 | |||
| L | 16p11.2 | 33.4-34.2 | 0.8 | |||
| 2 | RA | 47,XY,+14 | L | 7q22.1 | 99.6-101.4 | 1.8 |
| G | 14 | 17.4-105.4 | 88 | |||
| L | Xp22.13 | 14.9-15.6 | 0.7 | |||
| 3 | RA | 46,XX | G | 11q24.2-qter | 126.1-134.8 | 8.7 |
| G | 11p15.4 | 4.2-4.9 | 0.7 | |||
| G | 17q12 | 35.9-36.4 | 0.5 | |||
| G | 20q13.33 | 62.5-62.9 | 0.4 | |||
| 4 | RA | 46,XX,del(5q) | L | 5q14.3-5q35.1 | 86.3-168.5 | 82.2 |
| L | 9q21.33-q31.2 | 82.0-103.1 | 21.1 | |||
| 5 | RA | 47,XY,del(7),+8 | L | 7 | 0-158.1 | 158.1 |
| 6 | RA | 46,XX | G | 21q21.3 | 28.5-30.1 | 1.6 |
| G | 21q22.11-q22.12 | 33.9-36.0 | 2.1 | |||
| G | 21q22.13-21q22.2 | 38.2-40.8 | 2.6 | |||
| 7 | RA | 46,XX | G | 5q35.1 | 168.6-169.8 | 1.2 |
| L | 8q24.23 | 137.2-137.6 | 0.4 | |||
| G | 10q11.22 | 45.9-49.8 | 3.9 | |||
| G | 19q13.2-q13.31 | 47.8-48.2 | 0.4 | |||
| G | 21q21.3 | 27.9-28.1 | 0.2 | |||
| 8 | RA | 46,XY,del(20q) | L | 20q12-q13.2 | 40.2-51.9 | 11.7 |
| 9 | MDS/MPD (RARS-T) | 48,XY,del(20),+2mar | G | 6p25.1 | 6.4-6.8 | 0.4 |
| L | 20pter-q13.13 | 0-50.4 | 50.4 | |||
| G | 20q13.13-qter | 50.4-63.3 | 12.9 | |||
| 10 | 5q- | 46,XX,del(5q) | L | 5q14.3-q33.3 | 88.8-156.6 | 67.8 |
| L | 8p23.1 | 6.9-7.7 | 0.8 | |||
| 11 | RA | 46,XY/45,X,-Y | G | 6p21.2 | 39.2-40.2 | 1 |
| L | 7q21.11 | 83.4-83.6 | 0.2 | |||
| G | 7q34 | 141.6-142.0 | 0.4 | |||
| G | 15q24.1 | 71.6-71.7 | 0.1 | |||
| 12 | RCMD | 46,XY | G | 7q34 | 141.3-142.1 | 0.8 |
| L | 10q11.22 | 46.3-49.3 | 3 | |||
| L | 14q13.2 | 33.2-33.7 | 0.5 | |||
| L | 19q13.2-q13.31 | 47.8-48.3 | 0.5 | |||
| 13 | RA | 46,XX | G | 2p11.2 | 87.3-87.9 | 0.6 |
| L | 7q22.1 | 101.5-101.9 | 0.4 | |||
| G | 11q12.2 | 60.9-61.4 | 0.5 | |||
| 14 | RA | 46,XY | G | 8q21.2 | 86.1-86.7 | 0.6 |
| G | 17q12 | 35.9-36.3 | 0.4 | |||
| 15 | RAEB-1 | 46,XX | ND | |||
| 16 | RA | 46,XY | G | 6q27 | 167.8-168.2 | 0.4 |
| G | 15q13.3 | 30.0-30.4 | 0.4 | |||
| 17 | RA | 46,XX | L | 5q33.3 | 159.2-159.6 | 0.4 |
| 18 | MDS/MPD | 46,XX | L | 8p23.1 | 7.1-7.6 | 0.5 |
| 19 | RA | 46,XY | G | 11q12.2 | 61.2-61.4 | 0.2 |
| 20 | RA | 46,X,idic(X)(q13) | L | 6q26 | 160.5-161.0 | 0.5 |
| G | 10q11.22 | 46.2-47.2 | 1 | |||
| L | 20p12.1 | 14.6-15.1 | 0.5 | |||
| 21 | RA | 45,X,-Y | L | Y | 2.4-26.2 | 23.8 |
| 22 | RA | 45,X,-Y,del(7) | L | 4p14 | 39.1-40.6 | 1.5 |
| G | 5p15.33 | 0-2.3 | 2.3 | |||
| L | 5q13.2 | 68.9-70.1 | 0.2 | |||
| L | 7 | 0-158.1 | 158.1 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| G | 14q32.33 | 104.2-105.2 | 1 | |||
| L | 22q13.2 | 39.2-39.7 | 0.5 | |||
| 23 | RA | 46,XY,del(20q) | L | 20q11.21-q13.2 | 31.9-51.2 | 19.3 |
| 24 | 5q- | 46,XY,del(5q) | L | 5q21.1-q34 | 101.8-161.5 | 59.7 |
| G | 7q34 | 141.3-141.9 | 0.6 | |||
| 25 | RCMD | 47,XY,del(7),+8 | G | 2p11.2 | 87.3-87.9 | 0.6 |
| G | 7q34 | 141.4-141.9 | 0.5 | |||
| G | 8 | 0-143.9 | 143.9 | |||
| 26 | RARS | 46,XY | L | 1p22.1 | 91.6-93.1 | 1.5 |
| L | 1p31.1 | 77.3-77.9 | 0.6 | |||
| L | 2q33.1-q33.2 | 203-204 | 1 | |||
| L | 4p14 | 39.1-40.7 | 1.6 | |||
| G | 4q35.2 | 190.9-191.5 | 0.6 | |||
| L | 5q13.1-q13.2 | 68.1-69.0 | 0.9 | |||
| L | 6p23 | 14.9-15.5 | 0.6 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| L | 10q21.3 | 69.2-69.7 | 0.5 | |||
| L | 14q12 | 29.2-30.0 | 0.8 | |||
| G | 17q11.2 | 31.1-33.1 | 2 | |||
| 27 | RARS | 46,XX | ND | |||
| 28 | MDS/MPD (RARS-T) | 46,XY | ND | |||
| 29 | MDS/MPD (RARS-T) | 46,XX | L | 2q33.1-q33.2 | 203.0-204.3 | 1.3 |
| L | 4p14 | 39.1-40.0 | 0.9 | |||
| L | 9p24.1 | 6.6-6.8 | 0.2 | |||
| L | 10q21.3 | 64.6-65.2 | 0.6 | |||
| L | 14q12 | 29.4-29.9 | 0.5 | |||
| 30 | RAEB-1 | 46,XY | L | 4p14 | 39.0-39.8 | 0.8 |
| G | 12p13.31 | 10.8-11.5 | 0.7 | |||
| G | 15q25.3 | 86.0-86.5 | 0.5 | |||
| G | 15q24.1 | 71.2-71.8 | 0.6 | |||
| 31 | RARS | 46,XX | G | 15q14 | 33.6-33.9 | 0.3 |
| G | 18q21.1 | 44.3-44.5 | 0.2 | |||
| 32 | RARS | 46,XX | ND | |||
| 33 | RARS | 46,XY | L | 2p23.1 | 32.0-33.1 | 0.9 |
| L | 2q33.1-q33.2 | 202.9-204.8 | 1.9 | |||
| G | 3q13.12 | 108.7-109.2 | 0.5 | |||
| L | 3q26.2 | 170.8-171.6 | 0.8 | |||
| L | 4p14 | 39.0-39.8 | 0.8 | |||
| L | 5q13.1-q13.2 | 68.1-69.1 | 1 | |||
| L | 7p13 | 44.2-44.8 | 0.6 | |||
| L | 10q21.3 | 64.6-65.1 | 0.5 | |||
| L | 14q12 | 29.3-30.1 | 0.8 | |||
| G | 17q11.2 | 31.1-32.9 | 1.8 | |||
| L | 19p | 0-24.3 | 24.3 | |||
| 34 | RARS | 46,XY | ND | |||
| 35 | RARS | 47,XY,+8 | G | 8 | 0-143.9 | 143.9 |
| 36 | RARS | 46,XY | ND | |||
| 37 | RARS | 46,XX | L | 22q11.21 | 169.5-173.0 | 0.5 |
| 38 | RARS | 46,XY | G | 1q42.3 | 231.0-231.4 | 0.4 |
| G | 7q34 | 141.2-141.8 | 0.6 | |||
| G | 14q32.33 | 104.2-105.3 | 1.1 | |||
| G | 15q25.3 | 85.9-86.5 | 0.6 | |||
| G | 17q24.1 | 63.1-63.4 | 0.3 | |||
| 39 | RAEB-1 | 46,XY | G | 3q21.3 | 127.7-128.5 | 0.8 |
| G | 3q21.3 | 129.1-129.5 | 0.4 | |||
| G | 11q13.2-q13.4 | 68.4-71.2 | 2.7 | |||
| 40 | RARS | 46,XY | G | 1q21.2 | 145.2-146.5 | 1.3 |
| L | 1q42.3 | 231.6-232.1 | 0.5 | |||
| L | 4p14 | 39.2-40.5 | 1.3 | |||
| L | 5q13.1-q13.2 | 68.3-68.9 | 0.6 | |||
| L | 5q31.1 | 133.5-134.3 | 0.8 | |||
| L | 6p23 | 14.9-15.4 | 0.5 | |||
| L | 9p22.2 | 16.6-17.3 | 0.7 | |||
| L | 10q21.3 | 64.6-65.2 | 0.6 | |||
| G | 17q11.2 | 31.2-33.1 | 1.9 | |||
| L | 19p12-13.3 | 1.1-24.2 | 23.1 | |||
| 41 | RA | NA | G | 8q21.2 | 86.2-86.6 | 0.4 |
| G | 11q23.2 | 114.4-115.0 | 0.6 | |||
| G | 12q23.3 | 106.0-106.6 | 0.6 | |||
| G | 17q25.1 | 72.0-72.6 | 0.6 | |||
| L | 21q21.1-q21.2 | 22.6-23.5 | 0.9 | |||
| L | Xq13.2-qter | 70.6-105.2 | 34.6 | |||
| 42 | RA | NA | ND | |||
| 43 | MDS/MPD (RARS-T) | NA | ND | |||
| 44 | RAEB | 46, XY, del (7q21-q31) | L | 2p23.1 | 32.0-33.0 | 1 |
| G | 3p25.3 | 10.3-10.9 | 0.6 | |||
| L | 3p14.3 | 56.4-57.8 | 1.4 | |||
| L | 3q13.2-q13.31 | 114.4-114.9 | 0.5 | |||
| L | 5q33.3 | 159.3-159.7 | 0.4 | |||
| L | 7q21.12-q36.2 | 87.6-152.6 | 65 | |||
| G | 11p15.2 | 14.9-16.0 | 1.1 | |||
| G | 11p12-p13 | 36.2-37.1 | 0.9 | |||
| L | 15q15.1 | 38.8-39.5 | 0.7 | |||
| L | 15q15.3-q21.1 | 41.9-42.5 | 0.6 | |||
| L | 19p-q13.2 | 0-46.3 | 46.3 | |||
| L | 22q13.1-q13.2 | 39.1-39.7 | 0.6 |
ND indicates none detected; Mb, megabase; 5q-, 5q- syndrome; RA, refractory anemia; MPD, myeloproliferative disorder; RARS, refractory anemia with ringed sideroblasts; T, thrombocytosis; RAEB, refractory anemia with excess blasts; RCMD, refractory anemia with multilineage dysplasia; G, gain; L, loss; and NA, not applicable.
Mb position was determined using UCSC genome browser on Human Mar. 2003 assembly.
The total number of genomic copy number alterations detected by aCGH in 44 MDS samples was 135 (3.1 alterations per patient; range, 0-12 changes) (Figure 1B; Table 1). The median size of copy number alterations was 0.7 Mb with a mean of 10.5 Mb. In contrast, we detected copy number alterations in CD34+ cells from 15 nondiseased (“normal”) controls with a median size of 0.4 Mb and mean 0.5 Mb, all of which have been identified as common CNVs (Table S4). Most genomic amplifications displayed log2 ratios (∼ 0.4) indicative of low copy number gains (ie, duplications), which ranged in size from 0.1 to 143.9 Mb (Table 1; Figure 1B). In most cases, deletions represented loss of a single copy, and these deletions ranged in size from 0.2 to 158.1 Mb. Overall, we identified more than 85 distinct amplifications and deletions in CD34+ cells from 44 low-risk MDS patients by aCGH, of which 66 have not been previously described to our knowledge (Table 2). Twenty-one alterations involved recurring gains (n = 9) or deletions (n = 12) (Table S3). Of the 25 MDS patients with normal cytogenetic studies, 20 exhibited at least one disease-associated copy number alteration (Table 1).
Common regions of genomic alterations
| Chromosome . | Minimally altered region . | Size, Mb* . | Gain/loss . | Novel alteration . | Patient no. . |
|---|---|---|---|---|---|
| 1 | |||||
| p31.1 | 77.4-77.8 | 0.4 | L | Y | 26 |
| p22.1 | 91.6-93.1 | 1.5 | L | Y | 26 |
| q21.2 | 145.2-146.5 | 1.3 | G | Y | 40 |
| q42.3 | 231.0-231.4 | 0.4 | G | Y | 38 |
| q42.3 | 231.6-232.1 | 0.5 | L | Y | 40 |
| 2 | |||||
| p23.1 | 32.0-33.1 | 0.9 | L | Y | 33, 44 |
| p11.2 | 87.5-87.9 | 0.4 | G | Y | 13 |
| q33.1-q33.2 | 203-204 | 1 | L | Y | 26, 29, 33 |
| 3 | |||||
| p25.3 | 10.4-10.9 | 0.5 | G | Y | 44 |
| p14.3 | 56.4-57.8 | 1.4 | L | Y | 44 |
| q13.12 | 108.7-109.2 | 0.5 | G | Y | 33 |
| q13.2-q13.31 | 114.4-114.9 | 0.5 | L | Y | 44 |
| q21.3 | 127.7-128.5 | 0.8 | G | Y | 39 |
| q21.3 | 129.1-129.5 | 0.4 | G | Y | 39 |
| q26.2 | 170.8-171.6 | 0.8 | L | Y | 33 |
| 4 | |||||
| p14 | 39.2-39.8 | 0.6 | L | Y | 22,26,29,30,33,40 |
| q35.2 | 190.9-191.3 | 0.4 | G/L | Y | 1, 26 |
| 5 | |||||
| p15.33 | 0-2.3 | 2.3 | G | Y | 22 |
| q13.1-q13.2 | 68.3-68.9 | 0.6 | L | Y | 26, 33, 40 |
| q13.2 | 69.9-70.1 | 0.2 | L | Y | 22 |
| q14.3-q33 | 101.8-156.6 | 54.8 | L | Y | 1, 4, 10, 24 |
| q31.1 | 133.6-134.3 | 0.7 | L | Y | 40 |
| q33.3 | 159.3-159.6 | 0.3 | L | Y | 17, 44 |
| q35.1 | 168.9-169.6 | 0.7 | G | N | 7 |
| 6 | |||||
| p25.1 | 6.6-6.8 | 0.2 | G | Y | 9 |
| p23 | 14.9-15.4 | 0.5 | L | Y | 26, 40 |
| p21.2 | 39.2-40.2 | 1 | G | Y | 11 |
| q26 | 160.5-161.0 | 0.5 | L | Y | 20 |
| q27 | 167.8-168.2 | 0.4 | G | Y | 16 |
| 7 | |||||
| Entire chromosome | 0-158.1 | 158.1 | L | N | 5, 22 |
| p13 | 44.2-44.8 | 0.6 | L | Y | 33 |
| q21.11 | 83.4-83.6 | 0.2 | L | Y | 11 |
| q21.12-q36.2 | 87.6-152.6 | 65 | L | N | 44 |
| q22.1 | 99.6-101.4 | 1.8 | L | N | 2 |
| q22.1 | 101.5-101.9 | 0.4 | L | N | 13 |
| q34 | 141.6-141.8 | 0.2 | G | N | 11, 12, 24, 25, 38 |
| 8 | |||||
| Entire chromosome | 0-143.9 | 143.9 | G | N | 25, 35 |
| p23.1 | 7.2-7.6 | 0.4 | L | Y | 10 |
| q24.23 | 137.2-137.6 | 0.4 | L | Y | 7 |
| 9 | |||||
| p24.1 | 6.3-6.9 | 0.6 | L | Y | 29 |
| p22.2 | 16.6-17.3 | 0.7 | L | Y | 40 |
| q21.33-q31.2 | 82.0-103.1 | 21.1 | L | Y | 4 |
| 10 | |||||
| q11.22 | 46.3-46.9 | 0.6 | G/L | Y | 7,12,20 |
| q21.3 | 64.6-65.1 | 0.5 | L | Y | 22,26,29,33,40 |
| 11 | |||||
| p15.4 | 4.2-4.9 | 0.7 | G | Y | 3 |
| p15.2 | 14.9-16.0 | 1.1 | G | Y | 44 |
| p12-p13 | 36.2-37.1 | 0.9 | G | N | 44 |
| q12.2 | 61.2-61.4 | 0.2 | G | Y | 13, 19 |
| q13.2-q13.4 | 68.4-71.2 | 2.7 | G | Y | 39 |
| q23.2 | 114.4-115.0 | 0.6 | G | N | 41 |
| q24.2-qter | 126.1-135.0 | 9 | G | N | 1, 3 |
| 12 | |||||
| p13.31 | 10.8-11.5 | 0.7 | G | Y | 30 |
| q23.3 | 106.0-106.6 | 0.6 | G | N | 41 |
| 14 | |||||
| Entire chromosome | 17.4-105.4 | 88 | G | N | 2 |
| q12 | 29.4-29.9 | 0.5 | L | Y | 26, 29, 33 |
| q13.2 | 33.2-33.7 | 0.5 | L | Y | 12 |
| q32.33 | 104.2-105.0 | 0.8 | G/L | Y | 22, 38 |
| 15 | |||||
| q13.3 | 30.0-30.4 | 0.4 | G | Y | 16 |
| q14 | 33.6-33.9 | 0.3 | G | Y | 31 |
| q15.1 | 38.8-39.5 | 0.7 | L | Y | 44 |
| q15.3-q21.1 | 41.9-42.5 | 0.6 | L | Y | 44 |
| q24.1 | 71.6-71.7 | 0.1 | G | Y | 11, 30 |
| q25.3 | 86.0-86.5 | 0.5 | G | Y | 30, 38 |
| 16 | |||||
| p11.2 | 33.4-34.2 | 0.8 | L | Y | 1 |
| 17 | |||||
| q11.2 | 31.1-32.9 | 1.8 | G | Y | 26, 33, 40 |
| q12 | 35.9-36.3 | 0.4 | G | Y | 3, 14 |
| q24.1 | 63.1-63.4 | 0.3 | G | Y | 38 |
| q25.1 | 72.0-72.6 | 0.6 | G | N | 41 |
| 18 | |||||
| q21.1 | 44.3-44.5 | 0.2 | G | Y | 31 |
| 19 | |||||
| p-q13.2 | 0-46.3 | 46.3 | L | Y | 44 |
| p12-p13.2 | 9.0-24.1 | 15.1 | L | Y | 33, 40 |
| q13.2-q13.31 | 47.9-48.2 | 0.3 | G/L | Y | 7, 12, 30 |
| 20 | |||||
| p12.1 | 14.6-15.1 | 0.5 | L | N | 20 |
| q11.21-q13.13 | 40.2-50.4 | 10.2 | L | N | 8, 9, 23 |
| q13.13-qter | 50.4-63.3 | 12.9 | G | N | 9 |
| q13.33 | 62.5-62.9 | 0.4 | G | Y | 3 |
| 21 | |||||
| q21.1-q21.2 | 22.6-23.5 | 0.9 | L | Y | 41 |
| q21.3 | 27.9-28.1 | 0.2 | G | Y | 7 |
| q22.11-q22.12 | 33.9-36.0 | 2.1 | G | Y | 6 |
| q22.13-q22.2 | 38.2-40.8 | 2.6 | G | N | 6 |
| 22 | |||||
| q13.1-q13.2 | 39.2-39.7 | 0.5 | L | Y | 22, 44 |
| q11.21 | 16.9-17.3 | 0.4 | L | Y | 37 |
| X | |||||
| p22.13 | 14.9-15.6 | 0.7 | L | Y | 2 |
| q13.2-qter | 70.6-105.2 | 34.6 | L | N | 41 |
| Y | |||||
| Entire chromosome | 2.4-26.2 | 23.8 | L | N | 21 |
| Chromosome . | Minimally altered region . | Size, Mb* . | Gain/loss . | Novel alteration . | Patient no. . |
|---|---|---|---|---|---|
| 1 | |||||
| p31.1 | 77.4-77.8 | 0.4 | L | Y | 26 |
| p22.1 | 91.6-93.1 | 1.5 | L | Y | 26 |
| q21.2 | 145.2-146.5 | 1.3 | G | Y | 40 |
| q42.3 | 231.0-231.4 | 0.4 | G | Y | 38 |
| q42.3 | 231.6-232.1 | 0.5 | L | Y | 40 |
| 2 | |||||
| p23.1 | 32.0-33.1 | 0.9 | L | Y | 33, 44 |
| p11.2 | 87.5-87.9 | 0.4 | G | Y | 13 |
| q33.1-q33.2 | 203-204 | 1 | L | Y | 26, 29, 33 |
| 3 | |||||
| p25.3 | 10.4-10.9 | 0.5 | G | Y | 44 |
| p14.3 | 56.4-57.8 | 1.4 | L | Y | 44 |
| q13.12 | 108.7-109.2 | 0.5 | G | Y | 33 |
| q13.2-q13.31 | 114.4-114.9 | 0.5 | L | Y | 44 |
| q21.3 | 127.7-128.5 | 0.8 | G | Y | 39 |
| q21.3 | 129.1-129.5 | 0.4 | G | Y | 39 |
| q26.2 | 170.8-171.6 | 0.8 | L | Y | 33 |
| 4 | |||||
| p14 | 39.2-39.8 | 0.6 | L | Y | 22,26,29,30,33,40 |
| q35.2 | 190.9-191.3 | 0.4 | G/L | Y | 1, 26 |
| 5 | |||||
| p15.33 | 0-2.3 | 2.3 | G | Y | 22 |
| q13.1-q13.2 | 68.3-68.9 | 0.6 | L | Y | 26, 33, 40 |
| q13.2 | 69.9-70.1 | 0.2 | L | Y | 22 |
| q14.3-q33 | 101.8-156.6 | 54.8 | L | Y | 1, 4, 10, 24 |
| q31.1 | 133.6-134.3 | 0.7 | L | Y | 40 |
| q33.3 | 159.3-159.6 | 0.3 | L | Y | 17, 44 |
| q35.1 | 168.9-169.6 | 0.7 | G | N | 7 |
| 6 | |||||
| p25.1 | 6.6-6.8 | 0.2 | G | Y | 9 |
| p23 | 14.9-15.4 | 0.5 | L | Y | 26, 40 |
| p21.2 | 39.2-40.2 | 1 | G | Y | 11 |
| q26 | 160.5-161.0 | 0.5 | L | Y | 20 |
| q27 | 167.8-168.2 | 0.4 | G | Y | 16 |
| 7 | |||||
| Entire chromosome | 0-158.1 | 158.1 | L | N | 5, 22 |
| p13 | 44.2-44.8 | 0.6 | L | Y | 33 |
| q21.11 | 83.4-83.6 | 0.2 | L | Y | 11 |
| q21.12-q36.2 | 87.6-152.6 | 65 | L | N | 44 |
| q22.1 | 99.6-101.4 | 1.8 | L | N | 2 |
| q22.1 | 101.5-101.9 | 0.4 | L | N | 13 |
| q34 | 141.6-141.8 | 0.2 | G | N | 11, 12, 24, 25, 38 |
| 8 | |||||
| Entire chromosome | 0-143.9 | 143.9 | G | N | 25, 35 |
| p23.1 | 7.2-7.6 | 0.4 | L | Y | 10 |
| q24.23 | 137.2-137.6 | 0.4 | L | Y | 7 |
| 9 | |||||
| p24.1 | 6.3-6.9 | 0.6 | L | Y | 29 |
| p22.2 | 16.6-17.3 | 0.7 | L | Y | 40 |
| q21.33-q31.2 | 82.0-103.1 | 21.1 | L | Y | 4 |
| 10 | |||||
| q11.22 | 46.3-46.9 | 0.6 | G/L | Y | 7,12,20 |
| q21.3 | 64.6-65.1 | 0.5 | L | Y | 22,26,29,33,40 |
| 11 | |||||
| p15.4 | 4.2-4.9 | 0.7 | G | Y | 3 |
| p15.2 | 14.9-16.0 | 1.1 | G | Y | 44 |
| p12-p13 | 36.2-37.1 | 0.9 | G | N | 44 |
| q12.2 | 61.2-61.4 | 0.2 | G | Y | 13, 19 |
| q13.2-q13.4 | 68.4-71.2 | 2.7 | G | Y | 39 |
| q23.2 | 114.4-115.0 | 0.6 | G | N | 41 |
| q24.2-qter | 126.1-135.0 | 9 | G | N | 1, 3 |
| 12 | |||||
| p13.31 | 10.8-11.5 | 0.7 | G | Y | 30 |
| q23.3 | 106.0-106.6 | 0.6 | G | N | 41 |
| 14 | |||||
| Entire chromosome | 17.4-105.4 | 88 | G | N | 2 |
| q12 | 29.4-29.9 | 0.5 | L | Y | 26, 29, 33 |
| q13.2 | 33.2-33.7 | 0.5 | L | Y | 12 |
| q32.33 | 104.2-105.0 | 0.8 | G/L | Y | 22, 38 |
| 15 | |||||
| q13.3 | 30.0-30.4 | 0.4 | G | Y | 16 |
| q14 | 33.6-33.9 | 0.3 | G | Y | 31 |
| q15.1 | 38.8-39.5 | 0.7 | L | Y | 44 |
| q15.3-q21.1 | 41.9-42.5 | 0.6 | L | Y | 44 |
| q24.1 | 71.6-71.7 | 0.1 | G | Y | 11, 30 |
| q25.3 | 86.0-86.5 | 0.5 | G | Y | 30, 38 |
| 16 | |||||
| p11.2 | 33.4-34.2 | 0.8 | L | Y | 1 |
| 17 | |||||
| q11.2 | 31.1-32.9 | 1.8 | G | Y | 26, 33, 40 |
| q12 | 35.9-36.3 | 0.4 | G | Y | 3, 14 |
| q24.1 | 63.1-63.4 | 0.3 | G | Y | 38 |
| q25.1 | 72.0-72.6 | 0.6 | G | N | 41 |
| 18 | |||||
| q21.1 | 44.3-44.5 | 0.2 | G | Y | 31 |
| 19 | |||||
| p-q13.2 | 0-46.3 | 46.3 | L | Y | 44 |
| p12-p13.2 | 9.0-24.1 | 15.1 | L | Y | 33, 40 |
| q13.2-q13.31 | 47.9-48.2 | 0.3 | G/L | Y | 7, 12, 30 |
| 20 | |||||
| p12.1 | 14.6-15.1 | 0.5 | L | N | 20 |
| q11.21-q13.13 | 40.2-50.4 | 10.2 | L | N | 8, 9, 23 |
| q13.13-qter | 50.4-63.3 | 12.9 | G | N | 9 |
| q13.33 | 62.5-62.9 | 0.4 | G | Y | 3 |
| 21 | |||||
| q21.1-q21.2 | 22.6-23.5 | 0.9 | L | Y | 41 |
| q21.3 | 27.9-28.1 | 0.2 | G | Y | 7 |
| q22.11-q22.12 | 33.9-36.0 | 2.1 | G | Y | 6 |
| q22.13-q22.2 | 38.2-40.8 | 2.6 | G | N | 6 |
| 22 | |||||
| q13.1-q13.2 | 39.2-39.7 | 0.5 | L | Y | 22, 44 |
| q11.21 | 16.9-17.3 | 0.4 | L | Y | 37 |
| X | |||||
| p22.13 | 14.9-15.6 | 0.7 | L | Y | 2 |
| q13.2-qter | 70.6-105.2 | 34.6 | L | N | 41 |
| Y | |||||
| Entire chromosome | 2.4-26.2 | 23.8 | L | N | 21 |
G indicates gain; and L, loss.
Mb position was determined using UCSC genome browser on Human Mar. 2003 assembly.
Identification of genetic changes in peripheral blood using aCGH
To determine whether copy number changes identified were detectable in the peripheral blood, aCGH was performed on mononuclear cells obtained at the time of the diagnostic marrow specimen from select MDS patients. We detected copy number alterations in the circulating mononuclear cell population that had been identified in CD34+ cells, that were absent in matched CD3+ cells, in both cases examined. For patient 1, the amplification at chromosome band 11q24.2-qter detected in the CD34+ marrow cells was also found in the circulating mononuclear cells (Figure S1). Similarly, deletion of chromosome 7 in patient 22 was also detected in the peripheral blood mononuclear cells (Figure S1). These findings demonstrate that genomic changes are readily detectable in the peripheral blood and that aCGH of circulating nucleated cells may be a viable alternative to conventional cytogenetics in the diagnostic workup of MDS.
Amplification of the hominoid oncoprotein TBC1D3 is a novel and recurring genomic alteration in MDS
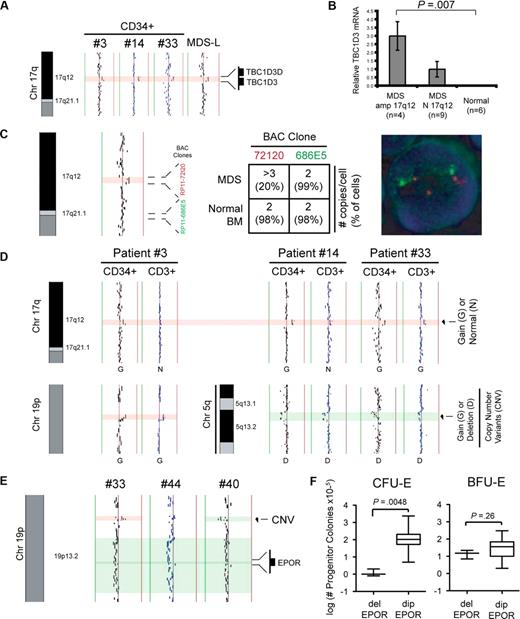
We identified a 0.5-Mb duplication at chromosome band 17q12 in 3 of the 44 patients and 3 of 6 MDS/AML cell lines (Figure 2A). The amplified locus at band 17q12 encompasses 2 of 8 paralogs of the hominoid oncoprotein TBC1D3.26 Because of limiting patient material, validation of the gain at band 17q12 by FISH analysis was performed on MDS-L cells using 2 BAC clones: one BAC clone (RP11-72I20) within the region of interest and one BAC clone (RP11-686E5) outside the region of interest. As shown in Figure 2C, FISH analysis confirmed an increase in copy number (∼ 20% of cells had > 3 copies per cell) at band 17q12, which was not detectable on 3 BM samples from healthy donors.
Detection of alterations at bands 19p13.2 and 17q12 by aCGH. (A) Alignment of aCGH profiles at chromosome 17q12 from 3 patients (3, 14, and 33) and 1 human MDS cell line (MDS-L). (B) Relative expression of TBC1D3 mRNA in MDS marrow samples from patients with (n = 4) and without (n = 9) amplification at band 17q12, as well as normal controls (n = 6) by quantitative reverse-transcribed polymerase chain reaction. (C) aCGH profile of the MDS cell line (MDS-L) at chromosome 17q12 showing position of the BAC clones used for FISH analysis of the MDS-L cell line (left). The average number of copies per cell (and proportion of total cells) as detected is tabulated (middle), and FISH analysis using BAC probes RP11-72I20 (red, within region of interest) and RP11-686E5 (green, outside region of interest) is shown on a representative cell (right). (D) Alignment at chromosome 17q, 19p, and 5q is shown to compare matched CD34+ and CD3+ profiles from 3 MDS patients (3, 14, and 33). Alterations at chromosomes 19p (patient 3) and 5q (patients 14 and 33) are within known polymorphic (CNV) regions and are used as an internal control. (E) Deletion encompassing the EPO receptor (EPOR) at chromosome 19p13.2 is seen in 3 patients (33, 40, and 44). Approximate boundaries of the EPO receptor locus and a CNV are highlighted. Vertical lines denote log2 signal ratios from −1 (green) to + 1 (red). Green shading delineates the borders of a deletion. Red shading delineates the borders of an amplification. (F) Erythroid progenitor colony assays were performed at the time of diagnosis. Comparison of CFU-E and BFU-E progenitor colony assays is shown for MDS patients with deletion of the EPOR locus (del EPOR; n = 3) and patients with a diploid EPOR locus (dip EPOR; n = 41).
Detection of alterations at bands 19p13.2 and 17q12 by aCGH. (A) Alignment of aCGH profiles at chromosome 17q12 from 3 patients (3, 14, and 33) and 1 human MDS cell line (MDS-L). (B) Relative expression of TBC1D3 mRNA in MDS marrow samples from patients with (n = 4) and without (n = 9) amplification at band 17q12, as well as normal controls (n = 6) by quantitative reverse-transcribed polymerase chain reaction. (C) aCGH profile of the MDS cell line (MDS-L) at chromosome 17q12 showing position of the BAC clones used for FISH analysis of the MDS-L cell line (left). The average number of copies per cell (and proportion of total cells) as detected is tabulated (middle), and FISH analysis using BAC probes RP11-72I20 (red, within region of interest) and RP11-686E5 (green, outside region of interest) is shown on a representative cell (right). (D) Alignment at chromosome 17q, 19p, and 5q is shown to compare matched CD34+ and CD3+ profiles from 3 MDS patients (3, 14, and 33). Alterations at chromosomes 19p (patient 3) and 5q (patients 14 and 33) are within known polymorphic (CNV) regions and are used as an internal control. (E) Deletion encompassing the EPO receptor (EPOR) at chromosome 19p13.2 is seen in 3 patients (33, 40, and 44). Approximate boundaries of the EPO receptor locus and a CNV are highlighted. Vertical lines denote log2 signal ratios from −1 (green) to + 1 (red). Green shading delineates the borders of a deletion. Red shading delineates the borders of an amplification. (F) Erythroid progenitor colony assays were performed at the time of diagnosis. Comparison of CFU-E and BFU-E progenitor colony assays is shown for MDS patients with deletion of the EPOR locus (del EPOR; n = 3) and patients with a diploid EPOR locus (dip EPOR; n = 41).
To determine whether the duplication at 17q12 results in increased mRNA levels of TBC1D3, we performed quantitative reverse-transcribed polymerase chain reaction on total RNA from MDS marrows exhibiting amplification at band 17q12 (n = 4), MDS marrows with diploid band 17q12 (n = 9), and normal controls (n = 6). MDS marrows with amplification at band 17q12 had increased mRNA expression of TBC1D3 compared with MDS patients or normal controls without the 17q12 amplification (Figure 2B).
Because the TBC1D3 locus lies within a region that is listed as a CNV (Variation_7155; frequency = 0.02), we determined whether the amplification at band 17q12 was a somatic or germ line mutation by performing aCGH on matched CD3+ T cells from the marrow of 3 patients from whom additional material was available. Although it is generally thought that MDS arises in a pluripotent stem progenitor cell, there is some debate as to whether the T-cell population is involved in this disorder.27 As shown in Figure 2D, the amplification at band 17q12 was absent in the CD3+ cells of 2 patients (patients 3 and 14) and clearly detectable in another (patient 33). Known high-frequency CNVs present in the CD34+ fractions at chromosome 5q (patients 14 and 33) and chromosome 19p (patient 3) were clearly detectable in the CD3+ cell populations and served as internal controls for the CD3+ aCGH analysis in each case (Figure 2D). These findings suggest that, at least in 2 cases (patients 3 and 14), the amplification at band 17q12 is not present as a constitutional variation.
Additional findings of interest include 3 patients (17, 33, and 40) with a deletion encompassing the erythropoietin receptor EPOR (band 19p31.2; Figure 2E). Reduced levels of EPOR have been reported in some MDS patients.28,29 To determine whether this deletion correlated with functional responses to erythropoietin, we examined hematopoietic progenitor assays performed under defined conditions (including exogenous erythropoietin) at the time of diagnosis in these patients. Loss of the region encompassing EPOR correlated with reduced colony formation of CFU-E, but not BFU-E, compared with MDS patients with a diploid EPOR locus (Figure 2F). These findings are consistent with the critical role of EPOR signaling in the proliferation and/or survival of relatively late erythroid progenitors (CFU-E), but not for early erythroid progenitors (BFU-E) as demonstrated in gene-targeted mice,30 and may explain the erythropoietin-resistance seen in some patients with MDS.
Comparison of CD34+ and CD34− cell populations by aCGH
It has been postulated that MDS arises from a genetically abnormal hematopoietic stem/progenitor cell that acquires additional genetic alterations during maturation.31 To determine whether there was an accumulation of additional changes in differentiated marrow cells (CD34−) compared with the stem/progenitor cell population (CD34+), we performed aCGH of CD34+ and CD34− cell fractions from the marrow of 5 matched patient cells and 2 normal controls. Although most of the genomic copy number alterations detected in CD34+ cells were present in the CD34− population, we also found that CD34− cells contained additional alterations that were not present in CD34+ cells (Table 3). Most strikingly, patient 17 had 1 change detected in the CD34+ cell population and an additional 13 unique alterations in the CD34− cell fraction, suggesting expansion of a specific clone favoring differentiation and/or proliferation or survival.
Comparison of alterations in matched CD34+ and CD34− population of cells from MDS patients
| Patient no. . | CD34+ . | CD34+ and CD34− . | CD34− . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| G/L . | Chromosome band . | Position, Mb . | G/L . | Chromosome band . | Position, Mb . | G/L . | Chromosome band . | Position, Mb . | |
| 13 | L | 7q22.1 | 101.5-101.9 | G | 11q12.2 | 60.9–61.4 | G | 1p31.1 | 77.3-78.0 |
| G | 4p14 | 38.7-40.7 | |||||||
| 17 | L | 5q33.3 | 159.2-159.6 | L | 2p21 | 42.2-42.9 | |||
| L | 2q33.1-q33.2 | 202.9-204.4 | |||||||
| G | 3q13.12 | 108.7-109.2 | |||||||
| L | 5q13.1-q13.2 | 68.2-68.7 | |||||||
| L | 6p23 | 14.9-15.4 | |||||||
| L | 7p13 | 44.2-45.1 | |||||||
| L | 9p13.3 | 33.7-34.3 | |||||||
| L | 10q21.3 | 64.6-65.2 | |||||||
| L | 10q21.3-q22.1 | 69.3-71.1 | |||||||
| L | 10q22.1-10q22.2 | 73.7-76.9 | |||||||
| L | 11p13 | 32.4-33.3 | |||||||
| L | 14q12 | 29.2-30.0 | |||||||
| L | 19 | 0-60.6 | |||||||
| 19 | G | 11q12.2 | 61.2-61.4 | L | 11q21 | 93.8-94.4 | |||
| G | 17q24.1 | 63.1-63.4 | |||||||
| 21 | L | Y | 2.4-26.2 | L | 6q26 | 160.4-161.0 | |||
| G | 17q12 | 35.9-36.3 | |||||||
| 31 | G | 15q14 | 33.6-33.9 | L | 1p36.11 | 24.5-25.1 | |||
| G | 22q11.21 | 17.1-17.3 | |||||||
| Normal-N1 | L | 7q35 | 143.2-143.5 | ||||||
| L | 16p11.2 | 32.0-34.1 | |||||||
| Normal-N2 | G | 8q21.2 | 86.2-86.6 | ||||||
| Patient no. . | CD34+ . | CD34+ and CD34− . | CD34− . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| G/L . | Chromosome band . | Position, Mb . | G/L . | Chromosome band . | Position, Mb . | G/L . | Chromosome band . | Position, Mb . | |
| 13 | L | 7q22.1 | 101.5-101.9 | G | 11q12.2 | 60.9–61.4 | G | 1p31.1 | 77.3-78.0 |
| G | 4p14 | 38.7-40.7 | |||||||
| 17 | L | 5q33.3 | 159.2-159.6 | L | 2p21 | 42.2-42.9 | |||
| L | 2q33.1-q33.2 | 202.9-204.4 | |||||||
| G | 3q13.12 | 108.7-109.2 | |||||||
| L | 5q13.1-q13.2 | 68.2-68.7 | |||||||
| L | 6p23 | 14.9-15.4 | |||||||
| L | 7p13 | 44.2-45.1 | |||||||
| L | 9p13.3 | 33.7-34.3 | |||||||
| L | 10q21.3 | 64.6-65.2 | |||||||
| L | 10q21.3-q22.1 | 69.3-71.1 | |||||||
| L | 10q22.1-10q22.2 | 73.7-76.9 | |||||||
| L | 11p13 | 32.4-33.3 | |||||||
| L | 14q12 | 29.2-30.0 | |||||||
| L | 19 | 0-60.6 | |||||||
| 19 | G | 11q12.2 | 61.2-61.4 | L | 11q21 | 93.8-94.4 | |||
| G | 17q24.1 | 63.1-63.4 | |||||||
| 21 | L | Y | 2.4-26.2 | L | 6q26 | 160.4-161.0 | |||
| G | 17q12 | 35.9-36.3 | |||||||
| 31 | G | 15q14 | 33.6-33.9 | L | 1p36.11 | 24.5-25.1 | |||
| G | 22q11.21 | 17.1-17.3 | |||||||
| Normal-N1 | L | 7q35 | 143.2-143.5 | ||||||
| L | 16p11.2 | 32.0-34.1 | |||||||
| Normal-N2 | G | 8q21.2 | 86.2-86.6 | ||||||
G indicates gain; and L, loss.
Total genomic alteration in CD34+ cells increases with age
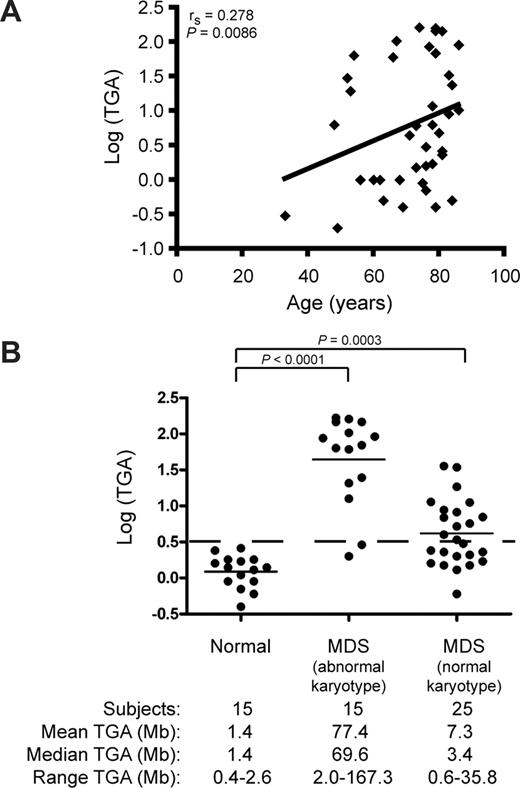
A related theory of the pathogenesis of MDS holds that the accumulation of genomic changes secondary to exposure to genotoxic damage over time gives rise to the abnormal hematopoietic stem cell clone.1 Indeed, increased age is a poor prognostic indicator in MDS.32 We thus examined whether the TGA, as measured by the sum of the sizes of amplifications and deletions, correlated with age. As demonstrated in Figure 3A, the TGA in CD34+ cells from 44 MDS patients increased with age (rS = 0.278 P = .009). This finding suggests that poorer outcome with increasing age may be related to greater disruption of the genome of hematopoietic stem/progenitor cells in the elderly. Alternatively, these findings may represent an epiphenomenon resulting from cumulative genotoxic exposure over time in older patients in general.
Impact of age on total genomic alteration (TGA) in CD34+ cells of lower-risk MDS patients. (A) Scatter-plot of age at diagnosis versus log (TGA). An increase in TGA is observed with increasing age in MDS patients (n = 44). (B) Distribution of TGA in CD34+ cells from normal controls (n = 15), MDS patients with an abnormal karyotype (n = 15), and MDS patients with a normal karyotype (n = 25). The dashed line represents a TGA of 3 Mb.
Impact of age on total genomic alteration (TGA) in CD34+ cells of lower-risk MDS patients. (A) Scatter-plot of age at diagnosis versus log (TGA). An increase in TGA is observed with increasing age in MDS patients (n = 44). (B) Distribution of TGA in CD34+ cells from normal controls (n = 15), MDS patients with an abnormal karyotype (n = 15), and MDS patients with a normal karyotype (n = 25). The dashed line represents a TGA of 3 Mb.
To distinguish between these possibilities, we examined CD34+ cells from peripheral blood or marrow from 15 elderly subjects with normal hematopoietic function (median age, 87 years; age range, 48-97 years). Controls showed minimal copy number alterations (median TGA, 1.4 Mb; range, 0.4-2.6 Mb) compared with the MDS patients with abnormal karyotype (median TGA, 69.6 Mb; range, 2.0-167.3 Mb; P < .001), or to patients with normal karyotype (median TGA, 3.4 Mb; range, 0.6-35.8 Mb; P < .001; Figure 3B). These findings suggest that the loss of genomic integrity seen in CD34+ cells from MDS patients was related to their disease, rather than their increased age.
Total genomic alteration predicts overall and leukemia-free survival
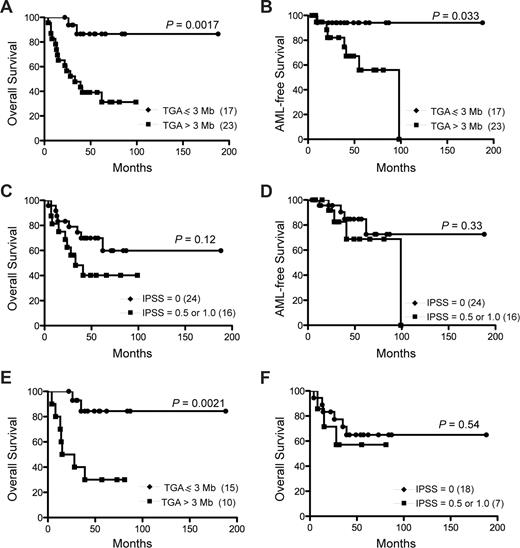
Cytogenetic findings are an established prognostic factor in MDS.33-35 However, although several recurrent genetic alterations were identified by aCGH (Table S3), the relatively small size of this cohort precluded assessment of prognosis based on specific genetic changes. We therefore asked whether TGA identified by aCGH could assign a prognostic category to patients with lower-risk MDS. The median duration of follow-up from time of diagnosis of the surviving study cohort was 54.5 months (range, 22-188 months). Because none of the age-matched controls had a TGA greater than 2.6 Mb, MDS patients were stratified into 2 groups based on whether the TGA in the CD34+ cells was less than or equal to 3 Mb (n = 17) or more than 3 Mb (n = 23). Comparison of the 2 groups revealed a median overall survival of 33 months for the group showing a TGA more than 3 Mb, whereas median overall survival was not reached in the group with a TGA less than or equal to 3 Mb (P = .002; Figure 4A). Similarly, AML-free survival was prolonged in the good-risk category (P = .033; Figure 4B). In contrast and as expected, differentiation by the IPSS score into low-risk (IPSS = 0) and the intermediate-1 groups (IPSS = 0.5 or 1.0) was not adequate to separate the 2 groups with respect to overall survival (P = .12; Figure 4C) or AML-free survival (P = .33; Figure 4D). When assessing only patients with a normal karyotype, the TGA cutoff of 3 Mb was still able to separate patients into a low-risk group (median survival not reached), and a higher-risk group (median survival, 39 months, P = .002; Figure 4E). Again, in this lower-risk group, the IPSS score was not able to substratify patients into prognostic groups (P = .54; Figure 4F). Analysis of only the patients whose diagnostic marrows were assessed by aCGH and who received only supportive treatment (n = 30) revealed that a TGA of 3 Mb still predicts overall survival (P = .001) and AML-free survival (P = .024) (Figure S2).
Overall survival of lower-risk MDS patients predicted by total genomic alterations (TGA). Overall (A,C,E,F) and AML-free (B,D) survival was determined when patients were stratified by TGA (A,B,E) or IPSS score (C,D,F). Overall survival and AML-free survival of all patients (n = 40) as stratified by TGA (A,B) and IPSS score (C,D). Overall survival of patients with normal karyotype (n = 25) as stratified by TGA (E) and IPSS (F). Numbers in parentheses represent the number of patients in each group.
Overall survival of lower-risk MDS patients predicted by total genomic alterations (TGA). Overall (A,C,E,F) and AML-free (B,D) survival was determined when patients were stratified by TGA (A,B,E) or IPSS score (C,D,F). Overall survival and AML-free survival of all patients (n = 40) as stratified by TGA (A,B) and IPSS score (C,D). Overall survival of patients with normal karyotype (n = 25) as stratified by TGA (E) and IPSS (F). Numbers in parentheses represent the number of patients in each group.
Discussion
The low-risk subtypes of MDS represent approximately 50% of all MDS patients. At least 50% of the low-risk MDS patients have a normal karyotype by conventional cytogenetics.2,8 To expand our understanding of lower-risk subtypes of MDS and identify MDS-initiating alterations, we performed submegabase BAC whole genome aCGH analysis of CD34+ cells of the marrow of 44 lower-risk MDS patients. Our findings revealed cryptic structural DNA alterations that were not detectable by standard cytogenetic analysis. Because of the heterogeneous nature of the marrow from MDS patients, acquisition of additional genetic abnormalities during malignant clone evolution, and the high propensity to undergo apoptosis, assaying the CD34+ cell population probably reveals genomic alterations that correspond to initiation of the disease. Although most patients tested had normal karyotype, our analysis identified 22 novel recurring copy number alterations by aCGH.
We also evaluated the effectiveness of high-resolution aCGH analysis in providing prognostic information for patients with lower-risk MDS. Our analysis of a mixed group of karyotypically normal (n = 25) and abnormal (n = 15) MDS patients revealed that minimal genomic alteration corresponding to less than 3 Mb detectable by aCGH correlated with significantly increased overall survival and was less frequently associated with transformation to AML, compared with IPSS stratification alone. Recently, it has been suggested that a pre-MDS phase, labeled idiopathic cytopenia of undetermined significance, may exist.36 It is possible that patients who do not meet the current criteria for MDS but still have unexplained cytopenias may fit into the group exhibiting minimal genomic alteration of CD34+ cells. However, further work will be needed to delineate these subgroups based on high-resolution aCGH.
Several groups have performed conventional and aCGH to identify genomic alteration in marrow cells from patients with MDS.37-43 In general, these studies used total marrow cells as the source of DNA and did not focus on the low-risk MDS subtypes. Nevertheless, single nucleotide polymorphism (SNP) genotyping by 2 independent groups revealed some copy number alterations in accordance with our findings43-45 (Table 2). The cell of origin in MDS is thought to reside within the CD34+ population. In addition, global gene expression profiling of highly purified (CD34+CD38−Thy1+) cells from MDS patients further support the hypothesis that the MDS clone originates from a hematopoietic stem cell.7 Because of the heterogeneous nature of the marrow from MDS patients, acquisition of additional genetic abnormalities during malignant clone evolution, and the high propensity to undergo apoptosis, use of purified CD34+ cells may reveal biologically significant copy number alterations that are present in the MDS-initiating clone. Our finding that there are more copy number alterations in the CD34− fraction compared with matched CD34+ cells lends credibility to the suggestion that the CD34+ fraction contains the MDS-initiating clonal population of cells. Of note, the platform we used allowed us to analyze genomic DNA at quantities as low as 50 ng, thereby permitting us to use unamplified DNA to eliminate biases generated by nonrandom polymerase chain reaction amplification.
Our data suggest that high-resolution aCGH will be a useful diagnostic and prognostic tool in the evaluation of patients with MDS falling into the low-risk group and that more extensive genome alterations will be a feature of older patients. In agreement with our findings, Mohamedali et al have shown that the presence of copy number changes in amplified DNA from CD34+ cells by SNP genotyping provides prognostic significance in MDS patients.44 Use of SNP genotyping on the total population of marrow cells from MDS patients of all risk groups did not substratify patients into prognostic groups.46 However, when only patients in the IPSS intermediate-1 subgroup were evaluated, additional SNP array copy number changes to those found by conventional cytogenetic analysis was predictive of poorer overall survival.46 Our findings suggest that measurement of TGA at high-resolution by aCGH provides prognostic information in lower-risk MDS patients (IPSS score ≤ 1) irrespective of conventional cytogenetic findings. Nevertheless, these aforementioned examples and our findings confirm that high-resolution genotyping improves prognostication in low-risk MDS patients independent of aCGH platform.
In our analysis, we did not take into consideration the favorable prognosis afforded by specific cytogenetic alterations, such as deletion of the long arm of chromosome 5.2 It is possible that substratification based on cytogenetic changes known to have prognostic implications could further improve the predictive capabilities of aCGH. Credence for this hypothesis is best exemplified by our aCGH finding on patient 1. This patient was originally diagnosed with 5q- syndrome, which has a favorable prognosis, yet died rapidly after the initial diagnosis. However, our aCGH findings of additional copy number alterations in this patient are in accordance with the less favorable prognosis of del(5q) in the presence of additional alterations. Thus, consideration should be given to incorporating aCGH findings into a prognostic scoring system for MDS, which could be achieved by replicating this study in a larger multicenter prospective trial.
There is minimal understanding of the specific molecular pathways that are involved in the pathogenesis of MDS. Copy number alterations in our analysis involving genes, such as EPO, EPOR, ETS1, FADD, SIRT1, TBC1D3, and Cyclin B1 (Table 1; Figure 2), are consistent with features of MDS, such as altered cytokine response, apoptosis, hematopoietic defects, and propensity to develop AML. Of interest, 2 patients demonstrated high-level amplification at chromosome band 11q24.2-qter, which encompasses several candidate genes, including HNT, BARX2, and ETS1. A novel and recurring copy number alteration found in MDS patients and cell lines was amplification (and corresponding increase in TBC1D3 mRNA) of the hominoid oncoprotein TBC1D3D (Figure 2). TBC1D3D (also called PRC17) was originally identified as a gene that was amplified in 15% of primary and 53% of metastatic prostate cancers.47 Furthermore, overexpression of TBC1D3D promoted anchorage-independent growth of NIH3T3 cells and tumor formation in nude mice.47 An extended analysis of a recurring duplication at band 17q12 by comparing CD34+ and CD3+ cell populations by aCGH revealed that, in 2 of 3 patients, the duplication was only present within the CD34+ cells. Interestingly, deletion of the TBC1D3 locus was noted in one normal subject (Table S4).
Some of the copy number alterations detected in CD34+ cells, but not in matched CD3+ cells, overlap with previously described regions known to harbor CNVs. These nonrandom segments of genomic DNA demonstrate variation in copy number, and this variation from person to person, or even within cell populations of a person, may have important functional consequences.48 As is the case with SNPs, some CNVs include potential oncogenes or tumor suppressor genes that have been implicated in other cancers, such as alterations at chromosome band 17q12 (TBC1D3)47 and band 5q13.1-q13.2 (RAD17)49 (Table S3). In both instances, the copy number change was not present in the CD3+ cells, suggesting a clonal copy number alteration in the CD34+ population. It has also recently been shown that a tumor suppressor gene (CHD5) on chromosome 1p is indeed within a known CNV.50 As such, it is conceivable that CNVs could contribute to the pathogenic phenotype observed in MDS.
Our finding of a high degree of genomic complexity and relatively few recurring alterations supports the view that MDS represents a heterogeneous disease manifested by the dysregulation of a large number of genes. The prognostic significance of a greater degree of genomic alteration suggests that DNA damage and aberrant DNA repair mechanisms may contribute to MDS and potentially offer an ancillary test for diagnosis and prognosis of low-risk MDS patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ron Deleeuw, Spencer Watson, and Emily Vucic for technical assistance and helpful discussions and Jennifer Collins for coordinating the collection of age-matched control samples.
This work was supported by Canadian Institutes of Health Research (CIHR), Leukemia and Lymphoma Society of Canada and Canadian Cancer Society grants (A.K.), Genome Canada, and CIHR grants (W.L.L., D.E.H.), a CIHR New Emerging Team grant (A.B.-W.), and CIHR and Michael Smith Foundation for Health Research (MSFHR) fellowships (D.T.S.). A.K. is a Senior Scholar of the MSFHR.
Authorship
Contribution: D.T.S. and A.K. participated in designing the research and drafting the manuscript; D.T.S, S.S., and A.T. performed experiments; S.V. and A.K. analyzed clinical data; J.J.S. performed statistical analysis; A.B.-W., C.J.E., and A.C.E. provided samples; C.J.E. assisted in drafting the manuscript; and K.T., W.L.L., and D.E.H. provided valuable reagents and expertise in aCGH and cytogenetics.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aly Karsan, Department of Medical Biophysics, British Columbia Cancer Research Centre, 675 West 10th Avenue, Vancouver, BC V5Z 1L3, Canada; e-mail: akarsan@bccrc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal