Abstract
Recent studies of retroviral-mediated gene transfer have shown that retroviral integrations themselves may trigger nonmalignant clonal expansion of hematopoietic stem cells (HSCs) in transplant recipients. These observations suggested that previous conclusions of HSC dynamics based on gamma-retroviral gene marking should be confirmed with improved vectors having a more limited capacity to transactivate endogenous genes. Because of the low trans-activation activity of self-inactivating lentiviral vectors (LVs), we have investigated whether the LV marking of mouse HSCs induces a competitive repopulation advantage in recipients of serially transplants. As deduced from analyses conducted in primary and secondary recipients, we concluded that lentivirally transduced HSCs have no competitive repopulation advantages over untransduced HSCs. By linear amplification-mediated polymerase chain reaction (LAM-PCR) analysis, we characterized LV-targeted genes in HSC clones that engrafted up to quaternary recipients. Although 9 clones harbored integrations close to defined retroviral insertion sites, none was characterized as a common integration site, and none was present in HSC clones repopulating quaternary recipients. Taken together, our results show unaltered repopulation properties of HSCs transduced with LVs, and confirm early studies suggesting the natural capacity of a few HSC clones to generate a monoclonal or oligoclonal hematopoiesis in transplant recipients.
Introduction
Retrovirus-derived vectors currently constitute the most efficient tool for the stable genetic modification of the hematopoietic stem cells (HSCs). Early studies demonstrated the efficacy of gamma-retroviral vectors (gRVs) for stably transducing the mouse and also the human HSCs1-5 and for the treatment of monogenic diseases.6-9 Since then, some difficulties have been found in the application of these vectors to human gene therapy. These include the necessity of stimulating the HSCs prior to transduction with gRVs,4,5,10-14 the in vivo silencing of LTR promoter,4,15-17 and, principally, the risks of insertional oncogenesis. In this respect, previous studies have shown the generation of leukemias in experimental animals18-20 as well as in X1-linked severe combined immunodeficiency (X1-SCID) patients21-23 who received a transplant of gRV-transduced BM grafts. Significantly, in all these cases, trans-activation of cellular oncogenes by the strong LTR promoters present in these vectors was demonstrated. In addition, in the gene therapy trial of chronic granulomatous disease both treated patients developed myelodysplastic syndrome (MDS) and showed expanded clones with insertions in the MDS1/EVI1 gene.24 Besides insertional mutagenesis, proviral inactivation through methylation of the LTR promoter has been recently reported in that trial.9,24
Most of the limitations of gRVs have been essentially overcome by self-inactivating lentiviral vectors (LVs). In contrast to gRVs, LVs can transduce both dividing and nondividing cells, thus facilitating the transduction of unstimulated HSCs.25-32 In addition, stable transgene expression has been observed after transplantation of hematopoietic grafts previously transduced with sinLVs harboring endogenous promoters.32,33 Concerning the risks of insertional oncogenesis, sinLVs have a lower preference for integration in regions close to the transcription start site of genes (TSS), compared with gRVs.34-37 This fact, together with the advanced self-inactivating design of sinLVs, accounts for the reduced risks of insertional oncogenesis associated with these vectors.38
Although early gene marking studies with gRVs allowed investigators to propose that hematopoiesis in the long term is oligoclonal or even monoclonal,1,2 recent gene marking studies of Kustikova et al showed that dominant long-term repopulating clones had gRVs insertions proximal to genes with a role in self-renewal or cell survival.39,40 Consequently, observations made by this group could limit previous conclusions of HSC gene marking with gRVs, suggesting the convenience of conducting further studies of HSC repopulation using gene marking strategies with improved vectors having a more limited capacity to transactivate endogenous genes.
Because of the low trans-activation activity of LVs,34-38 here we have investigated whether the LV marking of mouse HSCs induces a repopulation advantage of the transduced population in patients who underwent serial transplantations. In addition, we have investigated whether genetically marked clones capable of extensive repopulation contain LV insertions in the vicinity of genes involved in cell survival or self-renewal. To determine the repopulating properties of LV-transduced samples, we conducted BM competition experiments in which chimeric grafts consisting of BM cells exposed to sinLVs and BM cells sham-exposed to LVs were serially transplanted into irradiated recipient mice. To facilitate the detection of clonal expansions and even leukemias that could appear after a proliferative stress on the HSCs, up to quaternary BM transplantation rounds have been conducted. These serial transplantations would enable us to trace and detect by linear amplification-mediated polymerase chain reaction (LAM-PCR) the insertion sites in LV-transduced HSCs with longevities exceeding several times the life span of a mouse.
Methods
Lentiviral vector production
The LV used in these experiments was the pCCL-CMV-EGFP-Wpre that expressed the enhanced green fluorescent protein (EGFP) under the control of the immediate-early human cytomegalovirus (CMV) promoter. This vector also contains a central polypurine tract (cPPT) and the woodchuck hepatitis virus postregulatory element (Wpre; Figure 1). Vector stocks of VSV-G pseudotyped LVs were prepared by 4-plasmid calcium phosphate-mediated transfection in 293T cells, essentially as described.33 Constructs were kindly provided by Dr L. Naldini (San Raffaele Telethon Institute for Gene Therapy–Vita-Salute San Raffaele University Medical School, Milano, Italy). High-titer viral vector stocks were prepared by 2 rounds of ultracentrifugation before freezing and storing at −70°C. Functional titers of LVs were determined by transduction of HeLa cells with serial dilutions of the vector stocks, followed by flow cytometry analysis of EGFP+ cells. Viral titer of 1.2 × 108 transduction units (TU)/mL were routinely obtained.
Mice and irradiation procedures
All mice used in this study were P3D2F1 (B6.SJL-Ptprca Pepcb/BoyJ × DBA/2J) and B6D2F1 (C57BL/6J × DBA/2J), bred at the CIEMAT Laboratory Animals Facility (registration number 28079-21 A). Breeding pairs were originally obtained from The Jackson Laboratory (Bar Harbor, ME). Animals were routinely screened for pathogens according to FELASA (Federation of European Laboratory Animal Science Associations, Tamworth, United Kingdom) procedures and no pathogens were found. Mice aged 8 to 10 weeks old were used as donors and recipients of bone marrow transplant. All experimental procedures were carried out according to European and Spanish laws and regulations (European convention ETS 1 2 3, about the use and protection of vertebrate mammals used in experimentation and other scientific purposes; Spanish RD 1201/2005, about the protection and use of animals in scientific research). Procedures were approved by the Animal Experimentation Ethical Committee of the CIEMAT according to all external and internal biosafety and bioethics guidelines.
P3D2F1 and B6D2F1 mice were congenic for the Ly5 locus, expressing the Ptprc protein (protein tyrosine phosphatase receptor type C). Hematopoietic cells derived from these mice can be distinguished by Ly5.1 isoform, since P3D2F1 mice are Ly5.1/Ly5.2 and B6D2F1 are Ly5.2/Ly5.2. Throughout the paper, mice will be referred as Ly5.1 (P3D2F1) and Ly5.2 (B6D2F1). Recipient mice (Ly5.2 females) were total body irradiated with a fractionated dose of 11 Gy (2 doses of 5.5 Gy spaced 4 hours apart). Animals were irradiated with a Philips MG 324 X-ray equipment (Philips, Hamburg, Germany), set at 300 kV, 10 mA, and a dose rate of 1.03 Gy/min. Earlier studies from our laboratory showed the myeloablative properties of this irradiation protocol.41
Lin−Sca1+ purification
BM cells were flushed from the femurs and tibias of donor male mice (Ly5.1 and Ly5.2) with Iscove modified Dulbecco medium (IMDM) with 25 mM Hepes and l-glutamine (BioWhittaker Europe, Verviers, Belgium). To purify Lin−Sca1+ cells, lineage-positive cells from BM samples were removed using a Lineage Cell Depletion Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) following the manufacturer's recommendations. Lin− cells were then stained with anti-Sca1 MicroBeads (Miltenyi Biotech) for 30 minutes at 4°C, and washed with purification buffer (PBS− 1 × + 0.5% BSA). Cells were subjected to a positive immunomagnetic selection. A third column was finally applied to enrich Sca1+ cells. Lin−Sca1+ cells were washed with purification buffer and resuspended in IMDM. On average, 89% pure populations of Lin−Sca1+ were obtained, with recovery of 30% to 60% of the input number of Lin−Sca1+ cells.
Transduction of Lin−Sca1+ cells
Purified Lin−Sca1+ cells, from both Ly5.1 and Ly5.2 male mice, were prestimulated for 24 hours with 100 ng/mL human interleukin-11 (hIL-11; kindly provided by Genetics Institute, Cambridge, MA) and 100 ng/mL murine stem cell factor (mSCF, Chemicon International, Temecula, CA) in IMDM medium and 20% of FBS. Prestimulated Lin−Sca1+ Ly5.1 cells were exposed to LVs for 24 hours in IMDM supplemented with 20% of FBS, 100 ng/mL hrIL-11, and 100 ng/mL mrSCF. Incubations were conducted in P24 plates, at a density of 5 × 105 cells/mL and multiplicity of infection (MOI) of 20 TU/mL (Figure 1). At the end of the incubation period cells were washed and counted. In parallel, BM Lin−Sca1+ cells from Ly5.2 mice were manipulated under identical conditions, but not exposed to LVs (Figure 1).
Competitive repopulation assays
A chimeric population consisting of 2 × 105 cells exposed to LVs (Ly5.1+ cells) and the same number of cells not exposed to the LVs (Ly5.2+ cells) was transplanted into irradiated female Ly5.2 mice. At periodic intervals after transplantation (up to 9 months after transplantation), samples of 200 μL PB were obtained from the tail vein and mixed with 20 μL 0.5 M EDTA (pH 8.0) to avoid coagulation. Samples were analyzed by fluorescence-activated cell sorting (FACS) to determine the proportion of Ly5.1+ and EGFP+ cells. In addition, the proportion of granulocytes and of T- and B-lymphoid cells was determined. When mice were killed, samples from BM, spleen, and thymus were also analyzed by FACS. For the transplantation of secondary and tertiary recipients, 10 million BM cells from primary and secondary recipients were infused in the respective recipients, on an individual mouse-to-mouse basis. Quaternary recipients received a transplant of 3 million BM cells from tertiary recipients. In this case, 2 quaternary recipients received a transplant from one tertiary recipient.
To determine the level of exogenous hematopoiesis in recipient mice, the presence of donor cells (all of them from male animals) was determined by real-time quantitative PCR (qPCR) analyses using primers and a probe for the SRY gene, specific to the Y chromosome: FSRY (5′-TGTTCAGCCCTACAGCCACA-3′), RSRY (5′-CCTCTCACCACGGGACCAC-3′), and the probe P1SRY (5′ 6-FAM-ACAATTGTCTAGAGAGCATGGAGGGCCA-3′). Amplification of the mouse genomic β-actin sequence was achieved using the primers F1β-actin (5′-ACGGCCAGGTCATCACTATTG-3′) and R1β-actin (5′-ACTATGGCCTCAAGGAGTTTTGTCA-3′) and detected with the taqman probe: P1β-actin 5′ TR-AACGAGCGGTTCCGATGCCCT-3′. qPCR analyses were conducted in a Rotor Gene RG-3000 (Corbett Research, Sydney, Australia). The amplification was performed using one cycle of 10 minutes at 95°C, followed by 45 cycles of 30 seconds at 95°C, and 30 seconds at 58°C. To determine the proportion of exogenous cells, a standard curve of SRY/β-actin amplification ratio was made using genomic DNA extracts from samples containing 0% to 100% of BM cells from male/female mice. The contribution of donor hematopoietic repopulation in primary, secondary, and tertiary recipients was in most cases higher than 90%. Only in quaternary recipients was a low level of exogenous reconstitution observed in some animals due to the mild conditioning of these animals (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Flow cytometric analysis
To analyze the phenotype of PB, BM, spleen, and thymus cells, erythrocytes were first lysed for 10 minutes at room temperature in ammonium chloride lysis solution (0.155 mM NH4Cl + 0.01 mM KHCO3 + 10−4 mM EDTA) and washed with PBA (PBS− 1 × + 0.1% BSA + 0.02% NaN3). For conducting differentiation analyses, PB, BM, spleen, and thymus cells were stained with B220-biotinylated, CD3-biotinylated, and MAC-biotinylated plus GR1-biotinylated monoclonal antibodies (MoAbs; all of them from Pharmingen, Palo Alto, CA). After washing the cells with PBA, samples were stained with streptavidin-tricolor (Caltag, Burlingame, CA) and PE-conjugated anti–mouse CD45.1 (clone A20; Pharmingen, San Diego, CA) for 30 minutes at 4°C. Finally, cells were washed, resuspended in PBA with 2 μg/mL propidium iodide (Sigma-Aldrich, Bergisch Gladbach, Germany), and analyzed in an EPICS XL flow cytometer (Coulter Electronics, Hialeah, FL). A minimum number of 104 viable cells was acquired. Offline analysis was done with WinMDI (http://www.cyto.purdue.edu/flowcyt/software/Winmdi.htm) free software package (kindly provided by Dr J. Trotter, The Scripps Research Institute, La Jolla, CA).
Provirus quantification
Detection and quantification of integrated provirus was accomplished using complementary primers to the EGFP sequence. PCR product was amplified by the primers F1EGFP (5′-GTAAACGGCCACAAGTTCAGC-3′) and R1EGFP (5′-TGGTGCAGATGAACTTCAGGG-3′) and detected with the taqman MGB probe PEGFP (5′-6-FAM-CTTGCCGTAGGTGGC-MGB-3′). Amplification of the murine genomic β-actin was conducted as previously described. For negative controls, 5 μL H2O and 5 μL genomic DNA from untransduced cells were used. The amplification was performed using one cycle of 95°C for 10 minutes, followed by 45 cycles of 95°C for 30 seconds and 58°C for 30 seconds. To measure the average of proviral DNA per transduced cell, a standard curve of EGFP and murine DNA amplification ratio was made. Next, the average of proviral number per cell was estimated by interpolation of the EGFP/β-actin ratio from each DNA sample in the standard curve.42
Clonogenic assays
To determine the number of granulocyte-macrophage colony-forming unit (CFU-GM) progenitors present in purified Lin−Sca1+ cells, samples were plated in MethoCult GF M3534 culture media (StemCell Technologies, Vancouver, BC) at a concentration of 1 to 2 × 103 cells/plate. Samples were cultured at 37°C in 5% CO2 and fully humidified air, and 7 days later, colonies of at least 50 cells were scored under inverted microscope. To determine the gene transfer efficiency into CFU-GM progenitors, EGFP+ colonies were also scored under fluorescence inverted microscope.
LAM-PCR
In one experiment (experiment no. 2 in Figures 4,5) the pattern of LV integration sites was determined in BM cells from primary, secondary, and also quaternary recipients. For detection of lentiviral integration sites, DNA was extracted and preamplified by repeated primer extension using vector-specific 5′-biotinylated primers, SK-LTR1 (5′-GAGCTCTCTGGCTAACTAGG-3′) and SK-LTR2 (5′-GAACCCACTGCTTAAGCCTCA-3′) from 50 ng of each DNA sample. Selection of biotinylated extension products was performed with 200 μg magnetic beads according to the manufacturer's instructions (Dynal, Oslo, Norway). The samples were incubated with Klenow polymerase (2 U; Roche, Mannheim, Germany), dNTPs (250 μM), and random hexanucleotide mixture (Roche) in a volume of 20 μL for 1 hour at 37°C. Samples were washed on the magnetic particle concentrator (Dynal) and incubated with Tsp509I endonuclease (4 U in 20 μL; New England Biolabs, Beverly, MA) for 1 hour at 65°C. After an additional wash step, 100 pmol of a double-stranded asymmetric linker cassette and Fast Link Ligation Kit (2 U; Epicentre Technologies, Madison, WI) were incubated with the beads in a volume of 10 μL at room temperature for 5 minutes. Denaturing was performed with 5 μL of 0.1 N NaOH for 10 minutes at room temperature. Each ligation product was amplified with AmpliTaq gold polymerase (5 U; Applied Biosystems, Weiterstadt, Germany), vector-specific primer SK-LTR3 (5′-AGCTTGCCTTGAGTGCTTCA-3′) (0.5 μM), and linker cassette primer LCP1 (5′-GACCCGGGAGATCTGAATTC-3′) (0.5 μM). Of each PCR product, 2% served as a template for a second, nested PCR with internal primers SK-LTR4 (5′-AGTAGTGTGTGCCCGTCTGT-3′) and LCP2 (5′-GATCTGAATTCAGTGGCACAG-3′) at identical conditions. Of this final product, one-fifth was separated on a Spreadex high-resolution gel EL800 (Elchrom Scientific, Cham, Switzerland). PCR products were cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and sequenced (GATC Biotech, Konstanz, Germany). We obtained the sequences by LAM-PCR database from CONSERT project (https://consert.gatc-biotech.com/lampcr).43
Sequence analysis
Sequences were trimmed to remove all vector elements, and were considered valid when they contained the 5′LTR sequence and the linker sequence intact, and when they showed at least 90% identity in the alignment. The junction sequence was mapped to the mouse genome using the BLAT tool with the UCSC Mouse genome Project (Assembly February 2006; http://genome.ucsc.edu).44 The analysis of cluster of genes involved in Biological Process or Molecular Function was made by DAVID easy tool (GO: http://david.abcc.ncifcrf.gov).45 We also use Retroviral Tagged Cancer Database (http://rtcgd.abcc.ncifcrf.gov)46 for identification of genes involved in tumorigenesis.
Analysis of leukemias (hematopathology)
Cytometry analysis performed to detect multilineage reconstitution of transduced cells was also informative to detect leukemias. In addition, killed mice were examined for pathological abnormalities and blood cells counts were done using an automatic analyzer (Technicon H-1E System of Bayer) equipped with a multispecies software version 3.0 with capacity to measure differential white blood cells, including atypical cells with characteristics of blastic cells. Leukocyte morphology was analyzed in May-Grunwald-Giemsa–stained blood smears and cytospins of BM and spleen cells.
Statistics
Data are presented as mean plus or minus standard error. The significances of differences between groups were determined using Student t test.
Results
Preserved competitive repopulation ability of hematopoietic stem cells transduced with lentiviral vectors
In these experiments, a population of Lin−Sca1+ (LS) BM cells from Ly5.1+ male mice was first transduced with EGFP-LVs as described in “Methods.” Similarly, another population of LS cells from Ly5.2+ male mice was subjected to mock transduction, under identical in vitro conditions, but in the absence of LVs. Thereafter, the same number of both populations was mixed together, and a total number of 2 × 105 cells was transplanted into irradiated Ly5.2 female mice. Periodically, PB samples from recipient mice were analyzed by FACS to determine the proportion of Ly5.1+ and EGFP+ cells (Figure 1).
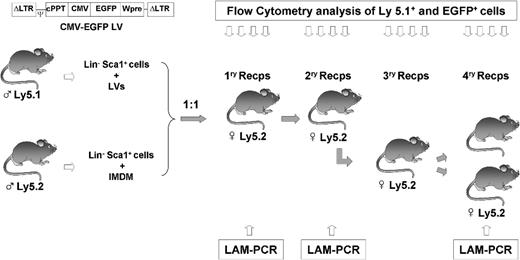
Illustration of the experimental protocol used to investigate the competitive repopulating ability (CRA) of mouse HSCs transduced with lentiviral vectors related to HSCs not exposed to lentiviral particles. Prestimulated Lin−Sca1+ BM cells from male Ly5.1 mice were exposed to LVs, as described in “Transduction of Lin−Sca1+ cells.” Another Lin−Sca1+ population from male Ly5.2 mice was treated under identical conditions without LVs. After a total incubation period of 48 hours, cells were mixed 1:1 and then transplanted into irradiated recipients. Nine months after transplantation, BM from primary recipients was retransplanted into secondary recipients. At periodic intervals, peripheral blood cells from primary and secondary recipients were sampled to determine the proportion of Ly5.1+ and EGFP+ cells (Figures 2,3). In one experiment, BM from secondary recipients was retransplanted into tertiary and quaternary recipients to conduct further studies of hematopoietic repopulation (Figure 4). In this experiment, BM cells from primary and quaternary recipients were analyzed by LAM-PCR to determine the LV integration sites in the corresponding repopulating cells (Figure 5).
Illustration of the experimental protocol used to investigate the competitive repopulating ability (CRA) of mouse HSCs transduced with lentiviral vectors related to HSCs not exposed to lentiviral particles. Prestimulated Lin−Sca1+ BM cells from male Ly5.1 mice were exposed to LVs, as described in “Transduction of Lin−Sca1+ cells.” Another Lin−Sca1+ population from male Ly5.2 mice was treated under identical conditions without LVs. After a total incubation period of 48 hours, cells were mixed 1:1 and then transplanted into irradiated recipients. Nine months after transplantation, BM from primary recipients was retransplanted into secondary recipients. At periodic intervals, peripheral blood cells from primary and secondary recipients were sampled to determine the proportion of Ly5.1+ and EGFP+ cells (Figures 2,3). In one experiment, BM from secondary recipients was retransplanted into tertiary and quaternary recipients to conduct further studies of hematopoietic repopulation (Figure 4). In this experiment, BM cells from primary and quaternary recipients were analyzed by LAM-PCR to determine the LV integration sites in the corresponding repopulating cells (Figure 5).
Two independent experiments were conducted. In the first one (data shown in Figure 2), 69% of colony forming cells (CFCs) seeded after LV transduction were EGFP+ (not shown). After transplantation of the chimeric BM, the proportion of Ly5.1 cells (donor cells exposed to LVs) was maintained both in primary and secondary recipients (Figure 2A). When the expression of the EGFP marker gene was analyzed, we observed that 77.1% plus or minus 3.9% of Ly5.1 cells were EGFP+ at 1 month after transplantation. This value was maintained for the whole observation period in primary recipients, and also in 6 of 8 secondary recipients (Figure 2B), reflecting the efficient transduction and stable expression of the transgene in a high proportion of the primitive HSCs.
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 1). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they underwent transplantation on an individual basis (n = 10). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells [ ]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [
]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [ ]; non–LV-exposed cells: Ly5.2+ cells [▭]). Because of the efficacy of the LV used in these experiments to express the EGFP reporter gene, LV-transduced cells are defined as EGFP+ cells (“Results”).
]; non–LV-exposed cells: Ly5.2+ cells [▭]). Because of the efficacy of the LV used in these experiments to express the EGFP reporter gene, LV-transduced cells are defined as EGFP+ cells (“Results”).
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 1). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they underwent transplantation on an individual basis (n = 10). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells [ ]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [
]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [ ]; non–LV-exposed cells: Ly5.2+ cells [▭]). Because of the efficacy of the LV used in these experiments to express the EGFP reporter gene, LV-transduced cells are defined as EGFP+ cells (“Results”).
]; non–LV-exposed cells: Ly5.2+ cells [▭]). Because of the efficacy of the LV used in these experiments to express the EGFP reporter gene, LV-transduced cells are defined as EGFP+ cells (“Results”).
Based on our observations, and on previous studies showing the stable expression properties of LVs used in these experiments,28-32 the chimeric graft that was transplanted into irradiated mice can be divided in 3 different types of populations: (1) cells transduced with the LVs, defined as Ly5.1+/EGFP+ cells, (2) cells exposed—but not transduced—to the LVs, defined as Ly5.1+/EGFP− cells, and (3) cells subjected to identical in vitro manipulation conditions, but not exposed to the LV particles. These cells are Ly5.2 cells, and always were EGFP− (not shown). The average competitive repopulating properties corresponding to each of these populations in primary and secondary recipients is shown in Figure 2C. Although the engraftment observed in individually analyzed recipients with the 3 types of donor-derived cells (Ly5.1+/EGFP+, Ly5.1+/EGFP−, and Ly5.2+) was somewhat heterogeneous (Figure 2B), average data showed the absence of a generalized repopulation advantage or disadvantage in any of the 3 groups of transplanted HSCs (Figure 2C).
To demonstrate that transduction with LVs was not affecting the differentiation potential of the HSCs, the expression of the transgene in cells of different lymphohematopoietic lineages was routinely investigated in PB at different times after transplantation. As expected, a normal distribution of cell lineages was observed in recipient mice. Also as expected, EGFP was expressed in the myeloid (GR-1/MAC), B-lymphoid (B220), and T-lymphoid (CD3) cells (see representative analyses in Figure S1A). When mice were killed, cells from the BM, spleen, and thymus were also analyzed by flow cytometry. Data obtained in BM and spleen were highly consistent with results obtained in PB. In the thymus, however, the proportion of Ly5.1+/EGFP+ cells was generally lower, suggesting a modest preferential repopulation of this organ with untransduced Ly5.1+ cells (Figure S1B,C).
Figure 3 shows data obtained in a duplicate experiment as that shown in Figure 2. In this case, 53% of colonies generated after LV transduction were EGFP+ (not shown). Compared with the previous experiment, a more heterogeneous repopulation of Ly5.1+ cells and of EGFP+ cells was observed in individual recipient mice, although again a sustained repopulation of Ly5.1+ (Figure 3A) and of EGFP+ (Figure 3B) cells was observed in primary and secondary recipients. As happened in the previous experiment, the average repopulation of the 3 HSC populations present in the transplanted graft was maintained along the posttransplantation period, in both primary and secondary recipients (Figure 3C). Taken together, the results of these 2 experiments show the sustained competitive repopulating ability of the different HSC populations infused into recipient mice: LV-transduced HSCs; HSCs exposed, but not transduced, with the LVs; and HSCs not exposed to the LVs.
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 2). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they were underwent transplantation on an individual basis (n = 9). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells [ ]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [
]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [ ]; non–LV-exposed cells: Ly5.2+ cells [▭]). In this experiment, 2 secondary recipients died on days 9 and 12 after transplantation.
]; non–LV-exposed cells: Ly5.2+ cells [▭]). In this experiment, 2 secondary recipients died on days 9 and 12 after transplantation.
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 2). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they were underwent transplantation on an individual basis (n = 9). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells [ ]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [
]; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells [ ]; non–LV-exposed cells: Ly5.2+ cells [▭]). In this experiment, 2 secondary recipients died on days 9 and 12 after transplantation.
]; non–LV-exposed cells: Ly5.2+ cells [▭]). In this experiment, 2 secondary recipients died on days 9 and 12 after transplantation.
To investigate more deeply the repopulation potential of LV-transduced HSC clones capable of extensive repopulation potential, 2 additional rounds of BM transplantation were conducted with BM from 2 secondary recipients (mouse B2 and mouse F2 in Figure 3). Data in Figure 4 show the individual repopulation kinetics of donor Ly5.1+ cells and of donor transduced cells (represented as EGFP+ cells in the Ly5.1+ population from primary to quaternary recipients). In the first 3 rounds of transplantation, BM from each recipient was transplanted into one recipient subjected to standard conditioning (“Mice and irradiation procedures”). In the case of quaternary recipients, BM from each tertiary recipient was transplanted into 2 quaternary recipients subjected to submyeloablative conditioning (7 Gy), to prevent mouse mortality. In the case of recipients from mouse F (Figure 4A open triangles), a progressive increase in the repopulation of donor cells exposed to LVs (Ly5.1+) was observed between secondary (F2) to quaternary (F4.1 and F4.2) recipients (Figure 4A). In the case of recipients from mouse B (Figure 4A, mice B4.1 and B4.2), the proportion of Ly5.1+ cells went down at the expense of a high level of endogenous reconstitution after 4 rounds of BM transplantation (Ly5.2+ cells of female origin; data were confirmed by qPCR analyses of Y-chromosome sequences; Figure 4A; Table S1).
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary to quaternary recipients. (A) CRA of HSCs exposed to LVs relative to the CRA of non–LV-exposed HSCs (percentage of Ly5.1 positive cells with respect to the whole population) in primary to quaternary recipients corresponding to experiment no. 2. (B) CRA of HSCs transduced with LVs relative to the CRA of HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary to quaternary recipients. ■ correspond to retransplantation data obtained from primary recipient B1, whereas △ correspond to retransplantation data from recipient F1.
Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary to quaternary recipients. (A) CRA of HSCs exposed to LVs relative to the CRA of non–LV-exposed HSCs (percentage of Ly5.1 positive cells with respect to the whole population) in primary to quaternary recipients corresponding to experiment no. 2. (B) CRA of HSCs transduced with LVs relative to the CRA of HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary to quaternary recipients. ■ correspond to retransplantation data obtained from primary recipient B1, whereas △ correspond to retransplantation data from recipient F1.
Concerning the kinetics of the genetically marked cells (EGFP+ cells), evident differences were observed when recipients of the B and F groups were analyzed. Whereas a progressive decrease in the repopulation of EGFP+ cells—at the expense of an increase in Ly5.1+/EGFP− cells—was observed from primary B1 to quaternary B4 recipients, a progressive increase in the repopulation of EGFP+ cells was observed in recipients of the F group. In this case, most repopulating cells exposed to LVs were transduced and expressed the marker transgene, not only in primary and secondary but also in tertiary and quaternary recipients (Figure 4B).
In no instance was evidence of myelodysplasia or leukemia observed in any of the mice that underwent transplantation. Nevertheless, to investigate whether insertional mutagenesis had a role in the repopulating properties of clones engrafting quaternary recipients (particularly in mouse F, whose PB cells were almost all EGFP+), LAM-PCR analyses were conducted with BM samples from primary (B1 and F1), secondary (B2 and F2), and quaternary (B4.1 and B4.2, and F4.1 and F4.2) recipients of groups B and F (Figure 5A).
Integration site analyses in the bone marrow of primary and quaternary recipients infused with LV-transduced Lin−Sca1+ samples. (A) Representative LAM-PCR analysis of BM samples from primary and quaternary recipients that received a transplant of the chimeric BM graft corresponding to experiment no. 2. M indicates size markers; 5′IC, internal control band amplified in LV-transduced samples. (B) Distribution of LV integrations in genes, or in repetitive sequences (LINES, SINES, LTR, and RNA) of the mouse genome. (C) Distance of 24 LV integration sites to the transcription start sites of targeted genes in the mouse genome. The distance is represented by intervals of plus or minus 10 Kb from 0 to 100 Kb around TSSs, or by intervals of plus or minus 50 Kb for distances higher than 100 Kb.
Integration site analyses in the bone marrow of primary and quaternary recipients infused with LV-transduced Lin−Sca1+ samples. (A) Representative LAM-PCR analysis of BM samples from primary and quaternary recipients that received a transplant of the chimeric BM graft corresponding to experiment no. 2. M indicates size markers; 5′IC, internal control band amplified in LV-transduced samples. (B) Distribution of LV integrations in genes, or in repetitive sequences (LINES, SINES, LTR, and RNA) of the mouse genome. (C) Distance of 24 LV integration sites to the transcription start sites of targeted genes in the mouse genome. The distance is represented by intervals of plus or minus 10 Kb from 0 to 100 Kb around TSSs, or by intervals of plus or minus 50 Kb for distances higher than 100 Kb.
Hematopoietic stem cell clones characterized by extensive repopulating capacity have no lentiviral integrations in genes with repopulation potential
In the LAM-PCR, a total of 960 TopoTA clones were sequenced and 327 sequences fulfilled the quality criterion. After the comparison of the sequences to remove the repetitive ones, a total of 44 sequences were identified (Figure 5B). Integration sites were then mapped to investigate whether proviruses were close to genes involved in cell survival or repopulation. As expected, a high number of integrations (19 of 44) occurred within genes (all of them within introns), although 15 integrations occurred out of genes. The remaining 10 integrations occurred in repetitive elements (LINE, SINE, LTR, and RNA repeat sequences; Figure 5B; Table 1). Concerning the distribution of insertions around the transcription start site (TSS) of targeted genes, 16.6% of integrations were found within plus or minus 10 kb of the TSS, and 75% within plus or minus 100 kb of the TSS (Figure 5C; Table 1).
Chromosomal localization of HIV-derived vector integration sites with corresponding hit-RefSeq gene
| Integration . | Frequencies* . | Gene name . | Gene symbol . | Gene ID . | Distance to TSS, Kb . | |||
|---|---|---|---|---|---|---|---|---|
| B1 . | B2 . | B4.1 . | B4.2 . | |||||
| b-01 | 15 (112) | 11 (60) | 2 (3) | 14 (15) | Similar to nsfl1 (p97) cofactor (p47) | LOC433885 | 433885 | −281 |
| b-02 | 6 (112) | - | - | - | Topoisomerase (dna) ii beta | Top2b | 21974 | 34 |
| b-03 | 3 (112) | - | - | - | Activated leukocyte cell adhesion molecule | Alcam (RIS) | 11658 | −493 |
| b-04 | 1 (112) | - | - | - | Similar to nsfl1 (p97) cofactor (p47) | LOC433885 | 433885 | −280 |
| b-05 | 4 (112) | - | - | - | Repeat seq.LINE | |||
| b-06 | 2 (112) | - | - | - | Similar to zinc finger, cchc domain containing 9 (predicted) | LOC383743 | 383743 | −144 |
| b-07 | 25 (112) | - | - | 1 (15) | Nudc domain containing 1 | Nudcd1 | 67429 | 6 |
| b-08 | 2 (112) | - | - | - | Similar to heterogeneous nuclear ribonucleoprotein c isoform b | LOC329893 | 329893 | −177 |
| b-09 | 5 (112) | - | - | - | Tbc1 domain family, member 15 | Tbc1d15 | 66687 | 20 |
| b-10 | 1 (112) | - | - | - | Septin 7 | Sept7 | 235072 | 49 |
| b-11 | 1 (112) | - | - | - | Repeat seq. SINE | |||
| b-12 | 1 (112) | - | - | - | Repeat seq. LTR | |||
| b-13 | 1 (112) | - | - | - | Repeat seq. LINE | |||
| b-14 | 1 (112) | - | - | - | Atpase, class vi, type 11b | Atp11b (RIS) | 76295 | 31 |
| b-15 | 2 (112) | - | - | - | Thymoma viral proto-oncogene 3 | Akt3 (RIS) | 23797 | 69 |
| b-16 | 4 (112) | - | - | - | Transient receptor potential cation channel, subfamily m, member 3 | Trpm3 | 226025 | 24 |
| b-17 | 2 (112) | - | - | - | Repeat seq. LINE | |||
| b-18 | 4 (112) | - | - | - | Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 | RassF9 | 237504 | −235 |
| b-19 | 12 (112) | - | - | - | Transmembrane and tetratricopeptide repeat containing 3 | Tmtc3 | 237500 | 23 |
| b-20 | 5 (112) | 9 (60) | - | - | Zinc finger protein 804A | Zfp804a (RIS) | 241514 | −535 |
| b-21 | 8 (112) | 13 (60) | - | - | Repeat seq. LINE | |||
| b-22 | 5 (112) | 25 (60) | - | - | Cell adhesion molecule with homology to L1CAM | Chl1 | 12661 | −348 |
| b-23 | 1 (112) | - | - | - | Protection of telomeres 1A | Pot1a | 101185 | −1110 |
| b-24 | 1 (112) | 2 (60) | - | - | Zinc finger protein 804A | Zfp804a (RIS) | 241514 | −319 |
| b-25 | - | - | 1 (3) | - | Similar to svh protein | LOC382626 (RIS) | 382626 | −193 |
| Integration . | Frequencies* . | Gene name . | Gene symbol . | Gene ID . | Distance to TSS, Kb . | |||
|---|---|---|---|---|---|---|---|---|
| B1 . | B2 . | B4.1 . | B4.2 . | |||||
| b-01 | 15 (112) | 11 (60) | 2 (3) | 14 (15) | Similar to nsfl1 (p97) cofactor (p47) | LOC433885 | 433885 | −281 |
| b-02 | 6 (112) | - | - | - | Topoisomerase (dna) ii beta | Top2b | 21974 | 34 |
| b-03 | 3 (112) | - | - | - | Activated leukocyte cell adhesion molecule | Alcam (RIS) | 11658 | −493 |
| b-04 | 1 (112) | - | - | - | Similar to nsfl1 (p97) cofactor (p47) | LOC433885 | 433885 | −280 |
| b-05 | 4 (112) | - | - | - | Repeat seq.LINE | |||
| b-06 | 2 (112) | - | - | - | Similar to zinc finger, cchc domain containing 9 (predicted) | LOC383743 | 383743 | −144 |
| b-07 | 25 (112) | - | - | 1 (15) | Nudc domain containing 1 | Nudcd1 | 67429 | 6 |
| b-08 | 2 (112) | - | - | - | Similar to heterogeneous nuclear ribonucleoprotein c isoform b | LOC329893 | 329893 | −177 |
| b-09 | 5 (112) | - | - | - | Tbc1 domain family, member 15 | Tbc1d15 | 66687 | 20 |
| b-10 | 1 (112) | - | - | - | Septin 7 | Sept7 | 235072 | 49 |
| b-11 | 1 (112) | - | - | - | Repeat seq. SINE | |||
| b-12 | 1 (112) | - | - | - | Repeat seq. LTR | |||
| b-13 | 1 (112) | - | - | - | Repeat seq. LINE | |||
| b-14 | 1 (112) | - | - | - | Atpase, class vi, type 11b | Atp11b (RIS) | 76295 | 31 |
| b-15 | 2 (112) | - | - | - | Thymoma viral proto-oncogene 3 | Akt3 (RIS) | 23797 | 69 |
| b-16 | 4 (112) | - | - | - | Transient receptor potential cation channel, subfamily m, member 3 | Trpm3 | 226025 | 24 |
| b-17 | 2 (112) | - | - | - | Repeat seq. LINE | |||
| b-18 | 4 (112) | - | - | - | Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 | RassF9 | 237504 | −235 |
| b-19 | 12 (112) | - | - | - | Transmembrane and tetratricopeptide repeat containing 3 | Tmtc3 | 237500 | 23 |
| b-20 | 5 (112) | 9 (60) | - | - | Zinc finger protein 804A | Zfp804a (RIS) | 241514 | −535 |
| b-21 | 8 (112) | 13 (60) | - | - | Repeat seq. LINE | |||
| b-22 | 5 (112) | 25 (60) | - | - | Cell adhesion molecule with homology to L1CAM | Chl1 | 12661 | −348 |
| b-23 | 1 (112) | - | - | - | Protection of telomeres 1A | Pot1a | 101185 | −1110 |
| b-24 | 1 (112) | 2 (60) | - | - | Zinc finger protein 804A | Zfp804a (RIS) | 241514 | −319 |
| b-25 | - | - | 1 (3) | - | Similar to svh protein | LOC382626 (RIS) | 382626 | −193 |
| . | F1 . | F2 . | F4.1 . | F4.2 . | . | . | . | . |
|---|---|---|---|---|---|---|---|---|
| f-01 | 6 (59) | - | - | - | Riken cdna c730024g19 gene | C730024G19Rik | 232566 | −98 |
| f-02 | 1 (59) | - | 5 (16) | 3 (13) | Cerebellum postnatal development associated protein 2 | Cadps2 | 320405 | 488 |
| f-03 | 1 (59) | 39 (49) | 6 (16) | 9 (13) | Repeat seq. LINE | |||
| f-04 | 1 (59) | - | - | - | Repeat seq. SINE | |||
| f-05 | 1 (59) | - | - | - | Repeat seq. RNA | |||
| f-06 | 1 (59) | - | - | - | Yth domain family 3 | Ythdf3 | 229096 | 12 |
| f-07 | 1 (59) | - | - | - | Riken cdna 3830405g04 gene | 3830405G04Rik | 70681 | 2 |
| f-08 | 18 (59) | - | - | - | Riken cdna 1300010f03 gene | 1300010F03Rik | 219189 | 178 |
| f-09 | 1 (59) | - | - | - | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | Slc4a7 (RIS) | 218756 | 79 |
| f-10 | 3 (59) | - | - | - | Riken cdna 2610316d01 gene | 2610316D01Rik | 72511 | 14 |
| f-11 | 1 (59) | - | - | - | N-deacetylase/n-sulfotransferase 4-like | LOC545976 | 545976 | 37 |
| f-12 | 10 (59) | - | - | - | Myo3a myosin IIIA | Myo3a (RIS) | 667663 | −285 |
| f-13 | 13 (59) | - | - | - | Sorting nexin 13 | Snx13 | 217463 | 90 |
| f-14 | 1 (59) | - | - | - | Nuclear receptor coactivator 3 | Ncoa3 (RIS) | 17979 | 17 |
| f-15 | - | 2 (49) | - | - | Unknown region | |||
| f-16 | - | 8 (49) | - | - | Dynein cytoplasmic 1 intermediate chain 1 | Dync1i1 | 13426 | 193 |
| f-17 | - | - | 1 (16) | - | Zinc finger, a20 domain containing 3 | Za20d3 | 65098 | −5 |
| f-18 | - | - | 2 (16) | 1 (13) | Carbonic anhydrase 2 | Car2 | 12349 | 4 |
| f-19 | - | - | 2 (16) | - | Eukaryotic translation initiation factor 4e member 3 | Eif4e3 (RIS) | 66892 | −237 |
| . | F1 . | F2 . | F4.1 . | F4.2 . | . | . | . | . |
|---|---|---|---|---|---|---|---|---|
| f-01 | 6 (59) | - | - | - | Riken cdna c730024g19 gene | C730024G19Rik | 232566 | −98 |
| f-02 | 1 (59) | - | 5 (16) | 3 (13) | Cerebellum postnatal development associated protein 2 | Cadps2 | 320405 | 488 |
| f-03 | 1 (59) | 39 (49) | 6 (16) | 9 (13) | Repeat seq. LINE | |||
| f-04 | 1 (59) | - | - | - | Repeat seq. SINE | |||
| f-05 | 1 (59) | - | - | - | Repeat seq. RNA | |||
| f-06 | 1 (59) | - | - | - | Yth domain family 3 | Ythdf3 | 229096 | 12 |
| f-07 | 1 (59) | - | - | - | Riken cdna 3830405g04 gene | 3830405G04Rik | 70681 | 2 |
| f-08 | 18 (59) | - | - | - | Riken cdna 1300010f03 gene | 1300010F03Rik | 219189 | 178 |
| f-09 | 1 (59) | - | - | - | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 | Slc4a7 (RIS) | 218756 | 79 |
| f-10 | 3 (59) | - | - | - | Riken cdna 2610316d01 gene | 2610316D01Rik | 72511 | 14 |
| f-11 | 1 (59) | - | - | - | N-deacetylase/n-sulfotransferase 4-like | LOC545976 | 545976 | 37 |
| f-12 | 10 (59) | - | - | - | Myo3a myosin IIIA | Myo3a (RIS) | 667663 | −285 |
| f-13 | 13 (59) | - | - | - | Sorting nexin 13 | Snx13 | 217463 | 90 |
| f-14 | 1 (59) | - | - | - | Nuclear receptor coactivator 3 | Ncoa3 (RIS) | 17979 | 17 |
| f-15 | - | 2 (49) | - | - | Unknown region | |||
| f-16 | - | 8 (49) | - | - | Dynein cytoplasmic 1 intermediate chain 1 | Dync1i1 | 13426 | 193 |
| f-17 | - | - | 1 (16) | - | Zinc finger, a20 domain containing 3 | Za20d3 | 65098 | −5 |
| f-18 | - | - | 2 (16) | 1 (13) | Carbonic anhydrase 2 | Car2 | 12349 | 4 |
| f-19 | - | - | 2 (16) | - | Eukaryotic translation initiation factor 4e member 3 | Eif4e3 (RIS) | 66892 | −237 |
Distance to TSS indicates distance to transcription start site.
Numbers in this column indicate the number that each integration appears in the TopoTA analysis and numbers in parentheses correspond to all valid sequences in each analysis. One P96 multiwell was analyzed in all cases, except for primary recipients, in which 2 P96 were analyzed.
LAM-PCR analyses detected a total of 25 integrations in recipients of group B (B1, B2, B4.1, and B4.2), and 19 in recipients of group F (F1, F2, F4.1, and F4.2; Table 1). As expected, the number of integrations (repopulating clones, if only one integration per clone took place) found in primary recipients (25 integrations in recipient B1 and 14 integrations in recipient F1) was much higher than numbers found in quaternary recipients (3 integrations in recipients B4.1 and B4.2, and 5 integrations in recipients F4.1 and F4.2; Table 1). One of the 3 integrations detected in quaternary recipients of group B (integration b-25) was not detected in the primary recipient B1. Similarly, 3 of the 5 integrations detected in quaternary recipients of group F (f-17, f-18, and f-19) were not detected in the primary recipient F1, suggesting that clones associated with these integrations were quiescent in the respective primary recipients (Table 1). Two integrations (f-15 and f-16) appeared in secondary recipient F2 that were not identified in the other recipients (Table 1).
The 34 genes targeted by LVs in repopulating clones corresponding to primary, secondary, and quaternary recipients are shown in Table 1. As deduced from the analysis of the Retroviral Tagged Cancer Database (http://rtcgd.abcc.ncifcrf.gov), we noted that none of the 34 targeted genes corresponded to common integration sites (CISs), although 9 of them were represented in the database as retroviral integration sites (RISs), such as the Activated leukocyte cell adhesion molecule (Alcam, integration b-03), the atpase class vi-type 11b (Atp11b, integration b-14), the Thymoma viral proto-oncogene 3 (Akt3; targeted by integration b-15), the Zinc finger protein 804A (Zfp804a, integration b-20), the Similar to svh protein gene (LOC382626, integration b-25), the Sodium bicarbonate cotransporter, member 7 (Slc4a7, integration f-09), the Myo3a myosin IIIA (Myo3a, integration f-12), the Nuclear receptor coactivator 3 (Ncoa3, integration f-14), and the Eukaryotic translation initiation factor 4e member 3 (Eif4e3, integration f-19). Some of them would play a role in cell repopulation and tumorigenesis, such as Ncoa3 (also known as SRC-3).47,48
To estimate the contribution of a given clone to the hematopoiesis of transplant recipients, frequencies of a given sequence after TopoTA cloning were determined. As shown in Table 1, most of the integrations appeared at frequencies lower than 3 positives in a P96 multiwell. Integration b-01 appeared, however, at a relatively high frequency in the B1 primary recipient (15 of 112 correct sequences analyzed from 2 P96 multiwell) and in the B4.2 quaternary recipient (14 of 15 correct sequences), suggesting a high prevalence of this targeted clone in the BM of these recipients. Other integrations (b-07, b-19, b-20, b-21, b-22, f-01, f-03, f-08, f-12, f-13, and f-16) also appeared at high-intermediate frequencies in the BM of recipient mice. Significantly, 7 of the 9 integrations that targeted RIS genes were detected only in 2 primary recipients (B1 and F1) and 1 in secondary recipient (B2) also, but at relatively low frequencies, and were never detected in quaternary recipients. Two of the 9 RISs, LOC382626 (b-25) and Eif4e3 (f-19), have been detected only in quaternary recipients.
Finally, genes that were targeted in clones characterized by a very extensive repopulation potential—capable of repopulating irradiated recipients after 4 rounds of BM transplantation—have no reported activity on cell proliferation or survival. This final observation demonstrates that the extensive repopulation potential of these clones was not produced as a consequence of insertional mutagenesis events produced by LV integration.
Discussion
Previous clinical trials have demonstrated the efficacy of gRVs for the treatment of several monogenic diseases.6-9 Nevertheless, risks of insertional oncogenesis are currently limiting the use of these vectors in the clinics.21,22,24 The strong enhancer activity of the LTRs present in the first generation of gRVs accounted for risks of insertional oncogenesis associated with these vectors, suggesting the convenience of using weaker promoters to minimize risks of oncogene trans-activation.33,38,39,49,50
Also due to the strong trans-activation capacity of standard gRVs, previous conclusions on HSC dynamics based on retroviral gene marking approaches have been recently questioned. In this respect, early studies described that long term after BM transplantation, hematopoiesis is oligoclonal or even monoclonal, indicating the enormous proliferation potential of individual HSCs.1-4 The recent studies of Kustikova et al showed that dominant HSC clones capable of extensive repopulation in irradiated recipients harbored provirus inserts in the vicinity of genes with a role in self-renewal or cell survival. This observation allowed the authors to propose that retroviral integrations themselves may trigger nonmalignant clonal expansion in mice that underwent transplantation.39,40 Because similar insertional effects may have also occurred in earlier marking studies, the HSC makeup described upon the use of gRV inserts may not necessarily reflect a physiological property of these cells.
Studies conducted with other vectors with a much weaker trans-activation activity would shed new light on whether monoclonality or oligoclonality frequently associated with hematopoiesis of transplant recipients (particularly after several rounds of BM transplantation) is innate to normal HSC dynamics and not due to side effects due to insertional mutagenesis. Compared with gRVs, LVs have a lower preference for integration close to TSSs of genes.35,36,51 This characteristic, together with the possibility of using weak promoters in sinLVs, accounts for the lower genotoxicity of sinLVs, compared gRVs,35-38 suggesting that these vectors would be good candidates for clarifying the relevance of insertional mutagenesis in HSC clonal dynamics.
To investigate whether repopulating advantages conferred by gRVs in HSCs were reproduced with sinLVs, we conducted in vivo competition experiments involving the coinfusion of BM cells transduced with the LVs (Ly5.1+/EGFP+ cells), cells exposed—but not transduced—to the LVs (Ly5.1+/EGFP− cells), and cells subjected to identical in vitro manipulation conditions, but not exposed to the LV particles. Although one could consider that the fraction of Ly5.1+/EGFP− cells could be in fact Ly5.1+/EGFP+ cells that lost the expression of the transgene, data in Figure S2 show the good correlation that was observed between the proportion of EGFP+ cells and the mean proviral copy numbers per cell determined in the BM of mice that underwent serial transplantation. These data show the stability of EGFP expression in vivo, consistent with previous data from our laboratory,32,52 and indicate that most Ly5.1+/EGFP− cells can be considered LV-exposed populations that are not stably transduced by the vector.
Although the individual analysis of primary and secondary recipients showed a somewhat heterogeneous pattern of repopulation of the animals, no consistent differences in the dynamics of engraftment were observed in LV-transduced HSCs, compared with HSCs not exposed to the LVs, or to HSCs exposed but not transduced with the LVs (Figures 2,3). Consistent with these observations, no evidence of myelodysplasias or leukemias was observed in any of our animals, despite the serial transplantation rounds subjected to the transduced grafts.
Although the competitive repopulation ability (CRA) of LV-transduced grafts was not modified at a population level, we aimed to investigate in individual recipients whether repopulating clones with very extensive repopulating properties harbored LV insertions in the vicinity of genes with a role in self-renewal or cell survival. For this purpose, 4 rounds of BM transplantation were conducted and proviral inserts analyzed both in primary and quaternary recipients. Although our studies were conducted with the specific purpose of determining whether clones with extensive repopulation potential harbored integrations in the vicinity of genes playing a role in self-renewal or cell proliferation, the distribution of the proviral inserts in all our recipients was consistent with previous studies,34-36 since one half of the LV integrations took place within genes. Concerning the distributions of integrations close to the TSSs, 16.6% of the targeted genes harbored the LV provirus plus or minus 10 kb around the TSS. This value is also consistent with previous observations of LV integrations in murine and human CD34+ cells,38,51,53 and is significantly lower compared with values obtained in cells transduced with gRVs.35,38,52,54
Significantly, no integration sites were localized within or near to common insertion sites represented in the Retroviral Tagged Cancer Database. Neither did we find integrations in genes included in the Insertional Dominance Database (IDDb).40 Although 9 clones corresponded to previously defined RISs, it is of significance that none was characterized as CIS, and none was present in quaternary recipients. Moreover, genes targeted in clones detected in quaternary recipients have no reported activity in tumorigenesis or in other properties conferring cell repopulation advantage (ie, self-renewal, cell survival, or cell proliferation).
Although recent studies have shown that γ-globin lentiviral vectors containing enhancer elements from the β-globin locus control region can cause insertional dysregulation of cellular genes, insertions that mediated alterations in cellular gene expression did not affect the proliferation or survival of repopulating clones, and were not dominant in those animals.55 Data obtained by these authors in primary recipients are consistent with our observations after 4 rounds of BM transplantation showing that the clonal simplification observed in quaternary recipients can hardly be accounted for by a repopulation advantage conferred by the LV integration in genes with a role in self-renewal or cell survival, as already shown in HSCs targeted with gRVs.39,40 Although our gene marking studies with LVs do not contradict data obtained by Kustikova et al39,40 with gRVs, we propose that the clonal dominance observed after extensive repopulation stress can be accounted for by the progressive physiological expansion of a reduced population of very primitive HSCs with remarkable repopulating properties, as suggested in pioneering studies of HSC gene marking.1-4
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The technical assistance of Elena Lopez, Aurora de la Cal, Sergio Garcia, Israel Orman, Jesus Martinez, Edilia de Almeida and the support of the animal facility team of the CIEMAT are gratefully acknowledged. The authors also thank the Fundación Marcelino Botín for promoting translational research at the Hematopoiesis and Gene Therapy Division at CIEMAT.
This work was supported by grants from the European Program Life Sciences, Genomics and Biotechnology for Health (CONSERT; Ref 005242), Comisión Interministerial de Ciencia y Tecnología (SAF2005-00058 and SAF2005-02381) and Center for Biomedical Research on Rare Diseases (CIBERER-ISCIII).
Authorship
Contribution: A.G.-M. performed research, analyzed data, and wrote the paper; M.L.L. performed research; E.M. performed research and analyzed data; J.A.B. designed research and wrote the paper; and G.G. designed research, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guillermo Guenechea, Hematopoiesis and Gene Therapy Division, CIEMAT and CIBERER, Edificio 70A, Av. Complutense 22, 28040 Madrid, Spain; e-mail: g.guenetxea@ciemat.es.

![Figure 2. Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 1). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they underwent transplantation on an individual basis (n = 10). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells []; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells []; non–LV-exposed cells: Ly5.2+ cells [▭]). Because of the efficacy of the LV used in these experiments to express the EGFP reporter gene, LV-transduced cells are defined as EGFP+ cells (“Results”).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/8/10.1182_blood-2008-03-142661/4/m_zh80200825800002.jpeg?Expires=1763502845&Signature=I-FiDq47DI1aqRILKy7JyFNHi5tsxuXlC8~G3To9u5t95FpK2~Vc03pI1HvLViEE32BgY63An5~32GNaIKleuTuyPxgfF3fwwGvsP2fMJ-Pts9KzaDN2vw2S6DykTovVL1z6zoBml8FDlyrvvt0cZyfWdchcpWFeSQrXx~g8uWEuOvhcLjsYTGBDSoL5j7bGu2D8BdXJAyfjtRjHRpKdu31-i16lVahyQnV1QhSp54S-W8riUt9PT6uIdHUkVl~Xte6uBCmHoeT0pZqhEpD5fYIpIm0LSQY4lrJM1Qksm5XysRPWkFIqEYMhYxCH4cLViqnOJ-vPShkktOJSjR1usw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Analysis of the competitive repopulating ability (CRA) of HSCs transduced with LVs as deduced from the analysis of primary and secondary recipients (experiment no. 2). (A) CRA of HSCs exposed to LVs relative to non–LV-exposed HSCs (percentage of Ly5.1-positive cells with respect to the whole population), as deduced from the analyses of PB from primary and secondary recipients. (B) CRA of HSCs transduced with LVs relative to HSCs exposed to, but not transduced with, the LVs (percentage of EGFP-positive cells inside Ly5.1+ population) in primary and secondary recipients. In panels A and B, each symbol corresponds to individual recipients. The same symbol is maintained for primary and secondary recipients as they were underwent transplantation on an individual basis (n = 9). (C) The mean contribution of each of the populations that reconstituted primary and secondary recipients with donor cells (LV-transduced cells: double-positive Ly5.1+ and EGFP+ cells []; LV-exposed but not LV-transduced cells: Ly5.1+ EGFP− cells []; non–LV-exposed cells: Ly5.2+ cells [▭]). In this experiment, 2 secondary recipients died on days 9 and 12 after transplantation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/8/10.1182_blood-2008-03-142661/4/m_zh80200825800003.jpeg?Expires=1763502845&Signature=Ta5POYJ1m~ChiNLsAs1hC2nHHQnUUlOqZNbZhwaoUtVWalokRy-RXtc03kCWmH3r1k6O1-ZoXWKpVMngC8e~IMxWLgZ-HphUu8TFiB83mNrc1TBL83KqWGK7cx9chSygWaTMDvgoC-WKPlV7Huj1FJxvaESKQoIw8YRaZYGsWR3pB~UwtGB-VQs~LJaQid45-65WuCWN8RzLNfPRWqAEmaUibF8~d0qIz5an4cecMA3vog8ZWmgVctke8blBiaEdCGtKYQKRLTct~j~iHR5O5O7yA-jNK0E2dDsfwzUz1dpYNDo4Se5egNEm5MW0i4MWRgsfXapK~uky7~vIYanT2Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

