Abstract
Fanconi anemia (FA) proteins are thought to play a role in chromosome stability and repair of DNA cross-links; however, these functions may not fully explain the developmental abnormalities and bone marrow failure that are characteristic of FA individuals. Here we associate the FA proteins with the Notch1 developmental pathway through a direct protein-protein interaction between the FA core complex and the hairy enhancer of split 1 (HES1). HES1 interaction with FA core complex members is dependent on a functional FA pathway. Cells depleted of HES1 exhibit an FA-like phenotype that includes cellular hypersensitivity to mitomycin C (MMC) and lack of FANCD2 monoubiquitination and foci formation. HES1 is also required for proper nuclear localization or stability of some members of the core complex. Our results suggest that HES1 is a novel interacting protein of the FA core complex.
Introduction
Fanconi anemia (FA) is a congenital form of aplastic anemia and is transmitted through an autosomal and X-linked recessive mode. FA is manifested by bone marrow failure, congenital abnormalities, and a predisposition to malignancy.1 The primary clinical phenotype and major cause of death in patients is the progressive depletion of hematopoietic stem cells (HSCs) leading to bone marrow (BM) failure. This progressive loss of HSCs has been linked to a defect in HSC self-renewal and maintenance.2,3 Currently, FA is defined by 13 complementation groups and cloned genes (A through N).4 Eight FA proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL/PHF9, and FANCM) bind together in a nuclear complex that is termed the FA core complex. This complex, through the E3 ubiquitin ligase activity of FANCL, mediates monoubiquitination of FANCD2 and FANCI in response to DNA cross-link damage and during normal S phase.5-8 Mutations in any of the core complex components disrupt the structural integrity of the complex and prevent activation of the FA pathway as measured by FANCD2 monoubiquitination. Despite the identification of so many genes, a clear picture of how this pathway relates to the major phenotypic feature of FA such as progressive hematopoietic stem cell loss remains elusive.
Several pathways involved in mesoderm patterning and formation such as Notch, were found to be involved in HSC self-renewal.9-11 Notch1 through constitutive expression of its intracellular domain, NICD, as well as overexpression of its downstream effector hairy and enhancer of split homologue 1 (HES1) has been shown to increase HSC numbers in vivo, increase HSC self-renewal, reduce HSC cycling, and preserve the long-term reconstitution ability of primitive hematopoietic cells.10,12-14 In FA, HSCs were shown to have reduced self-renewal and reconstitution abilities and increased cycling activity,2,3,15 suggesting a link between Notch1/HES1 and FA pathways. In an attempt to elucidate the molecular basis of the HSC defect in FA, we found that HES1 directly interacts with several members of the FA core complex, notably FANCA, FANCF, FANCG, and FANCL.
HES1 is a member of the highly conserved family of Hairy-related basic helix-loop-helix (bHLH) proteins. There are 7 described members in the mammalian HES family. Among these, HES1 and HES5 are the only members known to be involved specifically in Notch1 signaling in neural cells and in bone marrow.16,17 HES1 is a repressor-type bHLH that represses expression of its own gene18,19 (autoregulatory mechanism) and antagonizes bHLH activators.20
In this study, we provide evidence, using several approaches, that HES1 is a novel interactor of the FA core complex. We also show that HES1 is required for cellular resistance to mitomycin C (MMC), FANCD2 monoubiquitination, proper cellular localization of FANCA and FANCL, and protein stability of several FA core complex members.
Methods
Cell lines and culture conditions
293T, HeLa, PD430 (FA-A), and FancC+/+ and FancC−/− fibroblast cell lines were grown at 37°C, in 5% CO2 in DMEM media supplemented with 10% FCS. Cells were maintained in log phase growth before setting up the experiments. FA group A fibroblasts (PD430; gift from Dr Markus Grompe, Oregon Health & Science University, Portland, OR) and HES1 wild-type and knockout murine cells (gift from Ryoichiro Kageyama, Kyoto University, Kyoto, Japan) were transformed with a plasmid coding for the simian virus 40 (SV40) large T antigen. Transformed cell lines were maintained in DMEM supplemented with 10% FBS. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Burlington, ON) or the calcium-phosphate transfection method.
Plasmids
All FA genes were subcloned into Y2H vectors pGADT7 or pGBKT7 by polymerase chain reaction (PCR). FANCE, FANCF, and FANCG genes obtained from the pREP4 expression plasmids in which they were originally cloned21-23 were subcloned by PCR into the mammalian expression vector pCDNA3 in fusion with a Myc expression protein. FANCC obtained from pFAC324 was subcloned into pCMVzeo in fusion with a 5′ HA-tag sequence. The pIRESneoFANCL plasmid, expressing flag-tagged FANCL, was a gift from Dr Hans Joenje and the pIRESneoFANCD2 plasmid was a gift from Dr Markus Grompe. Y2H HES1 plasmid constructs were generated by reverse-transcription (RT)–PCR from 293T mRNA. Truncated HES1 constructs (HES11-90, HES11-156, HES1104-281, HES1190-281) were generated either by PCR or by restriction digestion of full-length cDNA. The FANCF349-395 deletion mutant was obtained by RT-PCR of cDNA from FA group F cells EUFA121. FANCF and FANCG truncated constructs as well as FANCG mutant constructs were generously provided by Dr Manuel Buchwald. All of the constructs were verified by DNA sequencing.
Antibodies
Antibodies used include the following: goat anti-FANCA (Santa Cruz Biotechnology, Santa Cruz, CA); monoclonal anti-FANCA clone 5G9 (gift from Dr Maureen Hoatlin, OSHU); polyclonal anti-FANCC25 ; monoclonal anti-FANCC clone 8F3 (gift from Dr Maureen Hoatlin, OSHU); monoclonal and polyclonal anti-FANCD2 (FI-17; Santa Cruz Biotechnology and Novus Biotechnology [Littleton, CO], respectively); rabbit polyclonal anti-FANCE (Fanconi Anemia Research Fund, Eugene, OR); rabbit polyclonal anti-FANCF (6307; a gift from Dr Maureen Hoatlin, OSHU); rabbit polyclonal anti-FANCG (Fanconi Anemia Research Fund); rabbit polyclonal anti-HES1 (Chemicon [Temecula, CA] and clone H-140; Santa Cruz Biotechnology); monoclonal antitubulin (E7; Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA); rabbit polyclonal anti-BACE1 (AB-1; Calbiochem, San Diego, CA); monoclonal anti-FLAG (M2; Sigma-Aldrich, St Louis, MO); monoclonal anti-cMyc (9E10; Santa Cruz Biotechnology), and monoclonal anti-HA (12CA5; Roche Diagnostics, Indianapolis, IN).
Yeast-2-hybrid
Yeast-2-hybrid analyses were performed according to the manufacturer's instructions for MATCHMAKER 2 Hybrid System 3 (Clontech, Palo Alto, CA). Positive controls included pGBKT7-FANCF with pGADT7-FANCG or pGBKT7-p53 with pGADT7-T antigen. Negative controls included empty pGBKT7 or pGADT7 vectors in combination with the corresponding FA gene coding plasmids. Each construct was sequenced and tested for autonomous Gal-4 activation; as reported, FANCG and FANCL as well as NICD showed self-activation when cloned into pGBKT7.26,27 Each experiment was performed at least 3 times. Each experiment was also performed with each gene cloned into either the pGBKT7 or pGADT7 vector.
Immunoprecipitation and Western blotting procedures
Whole-cell lysates were subjected to either immunoblot analysis (1% to 2% of lysates) or immunoprecipitation (90% of lysates). For immunoprecipitation (IP), cell lysates were incubated with antibodies, as indicated in each figure. Immunoprecipitates were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) on 10%, 12%, or 4% to 20% polyacrylamide gels and subjected to Western blotting with specific antibodies as indicated in each figure. Negative IP controls were performed using either mouse or rabbit serum.
Immunofluorescence
For localization of FA and HES1 proteins, immunofluorescent staining was performed as follows. Cells were fixed in methanol-acetone (3:7 vol/vol), permeabilized with 0.1% saponin and 2% BSA, and stained with specific primary antibodies followed with secondary antibodies (donkey anti–goat Alexafluor-488; goat anti–mouse Alexafluor-488; donkey anti–rabbit Alexafluor-555; and goat anti–rabbit Alexafluor-546). Cell nuclei were labeled with TOPRO-3 (Molecular Probes, Eugene, OR). Cells were visualized using the Nikon E800 fluorescent microscope equipped with a C1 confocal system (Nikon Canada, Mississauga, ON). Fluorescence intensity settings were performed with untreated cells. For HES1 and FANCD2 foci formation, cells were treated with either 50 ng/mL or 120 ng/mL for 4 or 16 hours. Cells were fixed in paraformaldehyde (2%) and permeabilized with Triton 0.3% before immunofluorescent staining. Cells with at least 5 FANCD2 foci were considered to be foci-positive cells. Estimation of the significance of the difference between means was done using the Student paired t test.
Immunofluorescent images were acquired with a CFI Plan apochromat 60×/1.40 working distance 0.22 mm objective on an E800 fluorescent microscope and captured with a C1 scanning system (Nikon Canada, Mississauga, ON).
siRNA analysis
HeLa cells were transfected with Stealth Select RNAi against HES1 (base pairs 318, 469, and 916; Invitrogen) using Lipofectamine 2000 (Invitrogen) at a final concentration of 250 nM according to the manufacturer's protocol. Cells were analyzed for MMC hypersensitivity, survival, and FANCD2 monoubiquitination. For survival, various concentrations of MMC were added 48 hours after RNAi transfection. After 5 days, the colonies were fixed and stained or counted. For FANCD2 monoubiquitination and foci formation analysis, HeLa cells were transfected with Stealth Select RNAi against HES1 for 48 hours and treated with the γ-secretase inhibitor L685458 (20 μM) or DMSO for 24 hours. MMC (120 ng/mL) was added 16 hours before the cells were harvested. Either the cells were stained for immunofluorescence experiments or protein extracts were prepared for immunoblotting.
Results
HES1 is a novel FA interacting protein
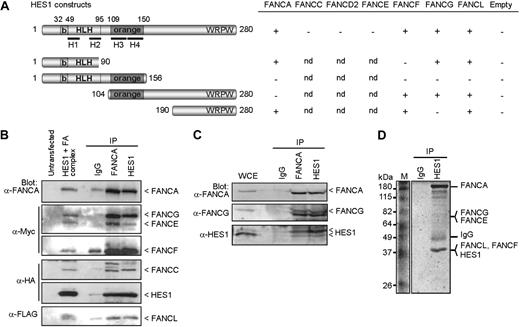
To determine the basis of the hematopoietic and developmental defects found in FA, we used the yeast-2-hybrid system to test for possible interaction between FA core complex proteins and both the Notch1 intracellular domain (NICD) and its downstream effector HES1. Although no direct interactions were detected between NICD and specific FA proteins (FANCA, FANCC, FANCE, and FANCF), we found that HES1 interacts directly with FA core complex components including FANCA, FANCF, FANCG, and FANCL but not FANCC, FANCD2, or FANCE (Figure 1A). Mapping of the interacting sites indicates that FANCA binds HES1 through its orange domain, whereas both FANCF and FANCG require the HES1 C-terminus region. We found that full-length FANCF is required for HES1 binding (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article) as for its interaction with FANCG.26 The FANCFΔ349-395 patient-derived pathogenic mutation21 disrupts both HES1 and FANCG binding (Figure S1A). In addition, the interaction of FANCG with HES1 occurs via its central region, which corresponds to the TPR4 and TPR5 domains (Figure S1B). The FANCG patient-derived pathogenic mutation, FANCG-L71P,22,28 disrupted FANCG interaction with HES1 but not with FANCA as previously reported,26 whereas mutation in the TPR6 domain (FANCG-G546R22,28 ) did not affect either FANCA or HES1 binding (Figure S1B).
FA proteins interact with HES1. (A) Yeast-2-hybrid assay with HES1 and FA proteins. Yeast strain AH109 was cotransformed with HES1 constructs expressing full-length or truncated HES1 protein with FA proteins as indicated and assayed for interaction as described in “Yeast-2-hybrid.” A positive interaction is indicated as +. Negative controls included pGBK-HES1 cotransformed with pGADT7 empty vector. (B) FA core complex components coimmunoprecipitate with HES1. 293T cells were cotransfected with HA-tagged HES1 and FA coding vectors and were subjected to immunoprecipitation (IP) with either anti-FANCA antibodies, anti-HES1 antibodies, or control IgG. IP was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. (C) Co-IP of endogenous proteins from 293T cell extracts using anti-HES1 or anti-FANCA antibodies. (D) Coomassie gel staining of endogenous protein extracts of 293T cells subjected to IP using anti-HES1 antibodies. Major bands were extracted from gel slices and the indicated proteins were identified by mass spectrometry.
FA proteins interact with HES1. (A) Yeast-2-hybrid assay with HES1 and FA proteins. Yeast strain AH109 was cotransformed with HES1 constructs expressing full-length or truncated HES1 protein with FA proteins as indicated and assayed for interaction as described in “Yeast-2-hybrid.” A positive interaction is indicated as +. Negative controls included pGBK-HES1 cotransformed with pGADT7 empty vector. (B) FA core complex components coimmunoprecipitate with HES1. 293T cells were cotransfected with HA-tagged HES1 and FA coding vectors and were subjected to immunoprecipitation (IP) with either anti-FANCA antibodies, anti-HES1 antibodies, or control IgG. IP was analyzed by SDS-PAGE and Western blotting using the indicated antibodies. (C) Co-IP of endogenous proteins from 293T cell extracts using anti-HES1 or anti-FANCA antibodies. (D) Coomassie gel staining of endogenous protein extracts of 293T cells subjected to IP using anti-HES1 antibodies. Major bands were extracted from gel slices and the indicated proteins were identified by mass spectrometry.
To confirm the interaction between FA core complex proteins and HES1, coimmunoprecipitation studies were performed in human cells. FA proteins were expressed, together or separately, with HA-tagged HES1 and subjected to immunoprecipitation. HES1 coimmunoprecipitated with each FANCA, FANCF, FANCG, and FANCL, but not with FANCC, FANCE, or the control protein EGFP (Figure S2). These results confirmed the yeast-2-hybrid findings. When FA core complex members were expressed together, immunoprecipitates obtained with either anti-FANCA or anti-FANCC antibodies, but not control IgG, were found to contain HES1 and all complex members that were tested, FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL (Figure 1B; Figure S2G). In addition, immunoprecipitates obtained with anti-HES1 antibodies contained FA core complex components including FANCA, FANCC, FANCE, FANCF, and FANCG.
To confirm that the FA core complex-HES1 interaction occurs endogenously, immunoprecipitation of endogenous proteins was performed. Western blot analysis of immunoprecipitates of endogenous proteins obtained with either anti-FANCA or anti-HES1 antibodies revealed that FANCA and FANCG coimmunoprecipitated with HES1 (Figure 1C). These results confirm that HES1 interacts with the endogenous FA core complex. In addition, HES1 immunocomplexes were purified and evaluated by mass spectrometry. Mass spectrometric analysis of major bands revealed the presence of several FA core complex members, notably FANCA, FANCE, FANCF, FANCG, and FANCL as well as known FA interacting proteins such as HSP70, HSP90, and α-spectrin (Figure 1D; Table S1).
To assess the cellular localization of HES1 and FA core complex proteins, immunofluorescence experiments were performed in HeLa cells. As expected, confocal analysis of endogenous proteins showed that HES1 localizes to the nucleus with FANCA, FANCG, FANCL, and with nuclear FANCC, which supports the immunoprecipitation findings (Figure S3). Together, our data suggest that HES1 is a novel FA interacting protein.
HES1 interaction is FA pathway dependent
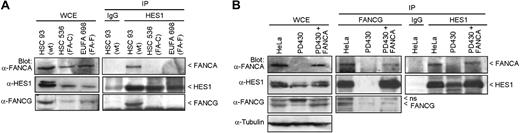
To determine whether the interaction of HES1 with FA core complex members requires a functional FA complex/pathway, we performed coimmunoprecipitation studies in FA mutant cell lines. Cellular extracts from FA groups C (HSC536) and F (EUFA698) mutant cell lines were subjected to immunoprecipitation using anti-HES1 antibodies. Endogenous FANCA and FANCG failed to coimmunoprecipitate with HES1 in both FA groups C and F mutant cells compared with normal cells (HSC93; Figure 2A). Similarly, endogenous HES1 failed to coimmunoprecipitate with FANCG in FA-A (PD430) mutant cells, whereas correction of FA-A cells with FANCA restored HES1 interaction with FANCG (Figure 2B).
HES1 dependence on the Fanconi anemia pathway. (A) FANCA and FANCG failed to immunoprecipitate with HES1 in FA mutant cells. Normal HSC93 and FA mutant lymphoblasts (FA-C: HSC536; FA-F: EUFA698) were subjected to immunoprecipitation with anti-HES1 antibodies. The immunoprecipitates were analyzed by Western blotting with the indicated antibodies. (B) Cell extracts from HeLa, PD430 (FA-A), and PD430 cells that were complemented with the FANCA gene (FA-A + FANCA) were subjected to immunoprecipitation using anti-FANCG or anti-HES1 antibodies and were immunoblotted with the indicated antibodies.
HES1 dependence on the Fanconi anemia pathway. (A) FANCA and FANCG failed to immunoprecipitate with HES1 in FA mutant cells. Normal HSC93 and FA mutant lymphoblasts (FA-C: HSC536; FA-F: EUFA698) were subjected to immunoprecipitation with anti-HES1 antibodies. The immunoprecipitates were analyzed by Western blotting with the indicated antibodies. (B) Cell extracts from HeLa, PD430 (FA-A), and PD430 cells that were complemented with the FANCA gene (FA-A + FANCA) were subjected to immunoprecipitation using anti-FANCG or anti-HES1 antibodies and were immunoblotted with the indicated antibodies.
HES1 is essential for FA core complex integrity
Each of the FA core complex proteins has been shown to be essential for the complex integrity and nuclear localization of core complex proteins, which reflects an interdependency between FA core proteins.8,29-34 To determine whether HES1, as a core complex interacting protein, is essential for the complex integrity, immunoprecipitation and immunofluorescence studies were performed in HES1-depleted cells. HeLa cells treated with the γ-secretase inhibitor L685,458, which blocks cleavage and activation of Notch1 and prevents HES1 expression,35 were subjected to immunoprecipitation using anti-FANCA and anti-FANCC antibodies. Immunoprecipitation of FANCA in HES1-depleted cells failed to coimmunoprecipitate FANCG (Figure 3A). Similarly, FANCA and FANCG failed to coimmunoprecipitate with FANCC in HES1-depleted cells, suggesting that HES1 is required for FA complex formation.
HES1 affects FANCA and FANCL nuclear localization. (A) Immunoprecipitation of FANCA and FANCC in HeLa cells treated with the γ-secretase inhibitor L685,458 (10 μM, 5 hours) or DMSO (control) and Western blotted for FANCA, FANCC, FANCG, and HES1. (B) Immunofluorescence staining of endogenous FANCA in Hes1−/− and wild-type fibroblasts cells with and without treatment with L685,458 (10 μM, 5 hours) or DMSO control. (C) Immunofluorescence staining of endogenous FA proteins, as indicated, or HES1 in HeLa cells treated with the γ-secretase inhibitor L685,458 (10 μM, 5 hours) or DMSO (control). The staining was visualized by confocal microscopy. Confocal fluorescence settings were established with untreated cells. (D) FA protein levels from HeLa cell extracts treated with L685,458 (10 μM, 5 hours) and analyzed by Western blotting.
HES1 affects FANCA and FANCL nuclear localization. (A) Immunoprecipitation of FANCA and FANCC in HeLa cells treated with the γ-secretase inhibitor L685,458 (10 μM, 5 hours) or DMSO (control) and Western blotted for FANCA, FANCC, FANCG, and HES1. (B) Immunofluorescence staining of endogenous FANCA in Hes1−/− and wild-type fibroblasts cells with and without treatment with L685,458 (10 μM, 5 hours) or DMSO control. (C) Immunofluorescence staining of endogenous FA proteins, as indicated, or HES1 in HeLa cells treated with the γ-secretase inhibitor L685,458 (10 μM, 5 hours) or DMSO (control). The staining was visualized by confocal microscopy. Confocal fluorescence settings were established with untreated cells. (D) FA protein levels from HeLa cell extracts treated with L685,458 (10 μM, 5 hours) and analyzed by Western blotting.
Immunofluorescence studies performed with Hes1−/− cells using a polyclonal anti-FANCA antibody that recognizes the murine FancA protein showed that the absence of Hes1 affected FancA nuclear localization (Figure 3B top right panel). Similarly, treatment of murine wild-type cells with L685,458 showed that FancA was predominantly located in the cytoplasm (Figure 3B bottom left panel), as it is in Hes1−/− cells, confirming that treatment of wild-type cells with L685,458 mimics Hes1−/− cells. This effect is reminiscent of the dependence of FANCA nuclear localization on other core complex components.8,34
HES1 depletion in human cells using either siRNA or L685,458 induced endogenous FANCA cellular localization to both the cytoplasmic and nuclear compartments (Figure 3C; Figure S3). In addition, the majority of FANCL is found in the cytoplasm following HES1 depletion (Figure 3C; Figure S3). Moreover, HES1 depletion affected the cellular localization of other core complex proteins with increased nuclear FANCC and cytoplasmic FANCF. HES1-depleted cells displayed reduced FANCG levels concomitant with HES1 depletion, whereas no change in FANCD2 localization was observed following HES1 depletion (Figure 3C). Western blot analysis of protein extracts from L685,458-treated HeLa cells showed a slight reduction in endogenous FANCA but no change in the FANCL protein level, suggesting that the absence of HES1 affects mainly their cellular localization. Increased endogenous FANCF protein levels and decreased FANCG levels concomitant with HES1 depletion were also observed by Western blotting (Figure 3D) correlating with immunofluorescence data (Figure 3C; Figure S3). No change in FANCD2 protein levels was observed following treatment with L685,458, suggesting that HES1 specifically affects core complex members.
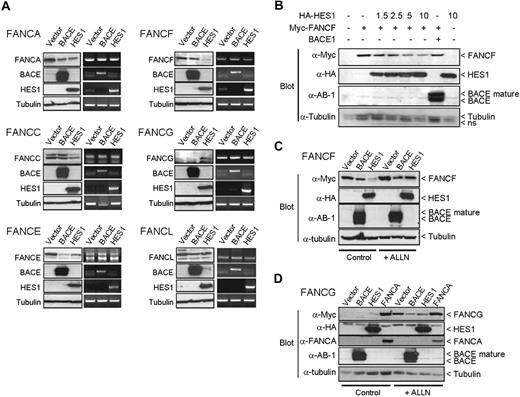
Reduced FANCF and increased FANCG protein levels were also observed when coexpressed with HES1 (Figure 4A). This effect seemed specific to HES1 and was not the result of other proteins such as BACE1. HES1 expression did not influence FANCF or FANCG transcription or other FA genes tested since mRNA levels were similar in cells transfected with either the HES1 expression plasmid, empty vectors, or the BACE1 expression plasmid. No change in FANCA, FANCE, and FANCL protein levels while only a slight decrease in FANCC protein levels were observed when cotransfected with HES1 (Figure 4A). A HES1 dose-dependent negative effect on FANCF protein levels (Figure 4B) resulted from increased protein degradation (Figure 4C). We also tested whether the HES1-mediated increase in FANCG protein levels resulted from reduced protein degradation (Figure 4D). FANCA has been shown to stabilize FANCG protein levels31 and was used as a positive control. We show that the HES1-mediated increase in FANCG protein levels correlated with reduced protein degradation, suggesting an increase in protein stability (Figure 4D). Together, these results strongly suggest that HES1 is essential for the stability and nuclear localization of FA core complex proteins.
HES1 affects FANCF and FANCG protein stability. (A) 293T cells were cotransfected with FA gene expression plasmids as indicated with either empty, or BACE1 or HES1 coding vectors. Both protein and RNA were extracted and subjected to either SDS-PAGE (left panel) or RT-PCR (right panel) of each gene as indicated. Endogenous tubulin protein and GAPDH RNA expression were used as internal controls. (B) Dose-dependent effect of HES1 on FANCF protein stability. 293T cells were cotransfected with FANCF expression plasmids along with increasing amounts of HES1 coding vector as indicated (μg). The total amount of transfected plasmid was equalized for all strategies with control empty vectors. (C,D) 293T cells cotransfected with FANCF or FANCG along with either empty, or BACE1 or HES1 expression plasmids were treated with the proteasome inhibitor ALLN (50 μM) for 16 hours following transfection. The FANCA expression plasmid was used as a positive control for FANCG stabilization.
HES1 affects FANCF and FANCG protein stability. (A) 293T cells were cotransfected with FA gene expression plasmids as indicated with either empty, or BACE1 or HES1 coding vectors. Both protein and RNA were extracted and subjected to either SDS-PAGE (left panel) or RT-PCR (right panel) of each gene as indicated. Endogenous tubulin protein and GAPDH RNA expression were used as internal controls. (B) Dose-dependent effect of HES1 on FANCF protein stability. 293T cells were cotransfected with FANCF expression plasmids along with increasing amounts of HES1 coding vector as indicated (μg). The total amount of transfected plasmid was equalized for all strategies with control empty vectors. (C,D) 293T cells cotransfected with FANCF or FANCG along with either empty, or BACE1 or HES1 expression plasmids were treated with the proteasome inhibitor ALLN (50 μM) for 16 hours following transfection. The FANCA expression plasmid was used as a positive control for FANCG stabilization.
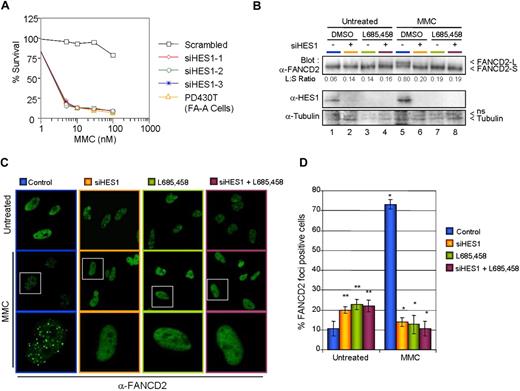
HES1 is required for MMC resistance and FANCD2 monoubiquitination
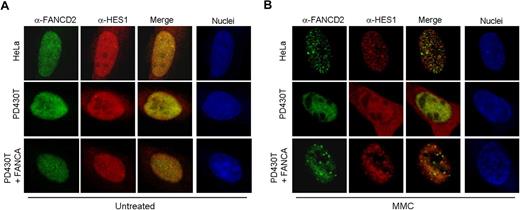
Cellular hypersensitivity to DNA cross-linking agents such as mitomycin C (MMC) is a hallmark of Fanconi anemia cells.36 As a novel interacting protein of the FA core complex, we tested the response of HES1 following MMC treatment. Immunofluorescence experiments were performed using HeLa, PD430T (FA-A), and PD430T cells complemented with FANCA. Confocal microscopy revealed that endogenous HES1 is localized to MMC-induced foci in MMC-treated HeLa cells and seems to be partially localized to FANCD2-containing foci (Figure 5). HES1 foci formation is impaired in FA-A mutant cells, whereas complementation of these cells with FANCA restored HES1 foci formation as well as FANCD2 foci in response to MMC. Surprisingly, HES1 nuclear localization seemed to be affected in FA-A mutant cells with cellular localization in both the nucleus and cytoplasm following MMC treatment (Figure 5B).
HES1 forms MMC-induced foci. Localization of endogenous HES1 in HeLa, PD430T (FA-A), and PD430T complemented with the FANCA gene. Cells were double stained with anti-HES1 and anti-FANCD2 antibodies at 0 (A: untreated) or 4 (B) hours following treatment with MMC (50 ng/mL) and processed for immunofluorescence. Nuclei were labeled with TOPRO-3.
HES1 forms MMC-induced foci. Localization of endogenous HES1 in HeLa, PD430T (FA-A), and PD430T complemented with the FANCA gene. Cells were double stained with anti-HES1 and anti-FANCD2 antibodies at 0 (A: untreated) or 4 (B) hours following treatment with MMC (50 ng/mL) and processed for immunofluorescence. Nuclei were labeled with TOPRO-3.
To determine whether HES1 as a FA core complex interacting protein is required for MMC resistance, HeLa cells were transfected with 3 different nonoverlapping Stealth Select small interfering RNAs against HES1 or the corresponding HES1 scrambled sequence. Following transfection, cells were incubated for 5 days with various concentrations of MMC. After MMC treatment, cells were fixed and either stained (Figure S4A) or counted for cell viability (Figure 6A). HES1-depleted cells showed a dramatic increase in MMC sensitivity compared with scrambled RNAi–transfected cells.
HES1 is required for MMC resistance. (A) HeLa cells transfected with 3 different Stealth RNAi against HES1 or scrambled sequences were treated with various concentration of MMC. The cells were incubated for 5 days before counting. (B) HES1 is required for FANCD2 monoubiquitination. Ubiquitination analysis of FANCD2 in HeLa cells that were depleted of HES1 by either Stealth RNAi against HES1, HES1 scrambled sequences (negative control), or the γ-secretase inhibitor L685,458. (C) FANCD2 foci formation in HeLa cells. HeLa cells depleted of HES1 using either Stealth RNAi against HES1, HES1 scrambled sequences (negative control), or the γ-secretase inhibitor L685,458 were treated with 120 ng/mL MMC and subjected to immunofluorescence analysis 16 hours later. White boxes indicate selected cells (middle panel) shown at higher magnification in the bottom panel. (D) Reduction in the number of DNA damage–induced FANCD2-containing foci cells following HES1 depletion. Cells containing more than 5 bright foci were scored as FANCD2-positive cells. Mean (± SD) of 3 separate experiments each from 100 cells. *P < .01; **P < .05.
HES1 is required for MMC resistance. (A) HeLa cells transfected with 3 different Stealth RNAi against HES1 or scrambled sequences were treated with various concentration of MMC. The cells were incubated for 5 days before counting. (B) HES1 is required for FANCD2 monoubiquitination. Ubiquitination analysis of FANCD2 in HeLa cells that were depleted of HES1 by either Stealth RNAi against HES1, HES1 scrambled sequences (negative control), or the γ-secretase inhibitor L685,458. (C) FANCD2 foci formation in HeLa cells. HeLa cells depleted of HES1 using either Stealth RNAi against HES1, HES1 scrambled sequences (negative control), or the γ-secretase inhibitor L685,458 were treated with 120 ng/mL MMC and subjected to immunofluorescence analysis 16 hours later. White boxes indicate selected cells (middle panel) shown at higher magnification in the bottom panel. (D) Reduction in the number of DNA damage–induced FANCD2-containing foci cells following HES1 depletion. Cells containing more than 5 bright foci were scored as FANCD2-positive cells. Mean (± SD) of 3 separate experiments each from 100 cells. *P < .01; **P < .05.
The FA core complex, but not its downstream components, has been shown to be required for FANCD2 monoubiquitination in response to DNA damage, whereas mutations in any FA core complex protein impair FANCD2 monoubiquitination and, thus, its localization to DNA damage– or S-phase–induced foci.5,37-42 To determine whether HES1 is required for FANCD2 monoubiquitination and foci formation, HeLa cells were transfected with Stealth Select RNAi against HES1 or with the HES1 RNAi scrambled sequence and were subsequently incubated with MMC. Cell extracts were subjected to SDS-PAGE and were immunoblotted for FANCD2 and HES1 (Figure 6B). Depletion of HES1 affected the monoubiquitination of FANCD2 as shown by a reduction of the long form of FANCD2 (FANCD2-L) in RNAi-treated cells (Figure 6B lanes 6 and 8). Similarly, treatment of HeLa cells with the γ-secretase inhibitor L685,458 prior to DNA damage resulted in diminished monoubiquitination of FANCD2 (Figure 6B lanes 7,8). Since FANCD2 monoubiquitination is required for its localization to DNA damage–induced nuclear foci, we looked at FANCD2 foci formation in HES1-depleted cells. As expected, HES1 depletion in both RNAi- and L685,458-treated cells resulted in a dramatic reduction of DNA damage–induced FANCD2 foci expressed as a decrease in the number of cells with 5 or more foci (Figure 6C,D). In the absence of DNA damage, HES1 depletion increased the number of cells with FANCD2 foci.
Among all FA complementation groups, only cells with mutations in the downstream components (FANCD1/BRCA2, FANCJ/BRIP1/BACH1, and FANCN/PALB2) have an impaired Rad51 foci phenotype.38,43 Depletion of HES1 had no effect on Rad51 foci formation (Figure S4B), which further supports a role of HES1 with FA core complex members. Taken together, these results support the requirement of HES1 for a functional FA core complex response to DNA cross-link damage.
Discussion
Although the FA pathway has been linked to DNA-damage responses, the specific biochemical function of each FA protein (separately or in the context of the FA complex/pathway) remains unclear. In particular, functions that would explain the disease phenotype, stem cell defects leading to bone marrow failure, and developmental abnormalities have been elusive. Our data presented here provide the first evidence linking FA proteins to HES1, a protein involved in stem cell function and developmental processes. We found that FA core complex components (FANCA, FANCG, FANCF, and FANCL) interact directly with HES1 and are present in HES1 immunocomplexes, implying that HES1 is a novel FA core complex interacting protein. We also found that HES1 is detected in immunocomplexes using antibodies directed against multiple FA core complex proteins (FANCA, FANCG, and FANCC), whereas reciprocal immunoprecipitation using anti-HES1 antibodies showed the presence of FA core complex members. Mass-spectrometric analysis revealed the presence of FA core complex members along with HES1 and other known FA interacting proteins such as HSP70, HSP90, GRP94, and α-spectrin found by several groups using different methods.44-49 We did not find BRAFT complex components such as BLM, RPA, or Topo IIIα identified by Wang's group (Meetei et al).33 These are associated with DNA repair functions and may be part of different FA subcomplexes49 other than the HES1/FA association. In addition, we believe that Meetei et al33 did not identify HES1 in their BRAFT complexes because only one HES1 signature peptide following tryptic digestion can be detected by mass spectrometry.
We found that HES1 interaction with the FA core complex requires a functional FA pathway as shown by absence of FA proteins in HES1 immunoprecipitates from FA mutant cells. As a core complex interacting protein, HES1 seems to share the interdependency attributed to FA core complex proteins where lack of one core protein affects the stability and/or cellular localization of the others, and thus the integrity of the core complex.8,29-34 This phenomenon is observed in the absence of HES1, achieved by using Hes1 knockout cells, siRNA against HES1, or a pharmacological inhibitor of HES1. In these cells, the core complex formation, the cellular distribution and stability of some core proteins are affected. Specifically, FANCA and FANCL become predominantly cytoplasmic while minor redistribution of FANCC, FANCF, and FANCG is observed. Moreover, depletion of HES1 affects the stability of some core complex proteins, more dramatically by increasing FANCF protein levels and decreasing FANCG, whereas overexpression of HES1 produces the opposite effect, decreased FANCF and increased FANCG levels. All these effects correlate with the absence of core complex formation. Together, these observations strongly suggest that HES1 is a crucial FA core complex interacting protein that seems required for the integrity of the core complex.
The phenotype of cells from FA individuals with mutations in core complex genes includes hypersensitivity to DNA cross-linking agents such as mitomycin C (MMC), impaired FANCD2 and FANCI activation through monoubiquitination, and consequently impaired FANCD2 and FANCI foci formation.36,50 An interesting finding is that depletion of HES1 leads to MMC hypersensitivity where HES1-depleted cells show an EC50 of 2.3 nM comparable to those reported for FA cells.51 Depletion of HES1 by a pharmacological approach or with siRNA impairs activation of FANCD2 through monoubiquitination and consequently prevents FANCD2 foci formation in response to DNA cross-link damage. HES1 depletion does not affect Rad51 foci formation in response to cross-link damage, which is consistent with a function within the core complex.38,43 These observations are reminiscent of a nonfunctional FA core complex and underscore the importance of HES1 on FA core proteins cellular localization and stability.
One characteristic of FA proteins is the formation of DNA cross-link–induced foci.5 HES1 forms cross-link–induced foci, which are dependent on a functional FA pathway. Since HES1 is a known transcriptional repressor, HES1-containing foci may be sites of transcriptional repression in response to cross-link damage. This idea is also supported by the fact that HES1-containing foci do not colocalize with FANCD2-containing foci, which are thought to be sites of DNA repair, thus HES1 foci may be distinct from sites of DNA repair.
Our results suggest a cross talk between the FA pathway and HES1 signaling. This cross talk is reflected by the various similarities between HES1 and FA function. For instance, HES1 has been linked to both neuronal and hematopoietic stem cell self-renewal abilities,10,12,13,16,52 a function that is defective in both neuronal and hematopoietic FA stem cells.2,3,53 Forced expression of HES1 has been shown to reduce cycling of short-term repopulating cells,14,54 whereas FancC−/− BM progenitor cells were shown to have increased cell cycle activity.15 Hes1−/− knockout mice are embryonic lethal due to severe neural tube defects,55 and embryonic lethality has been reported in FA knockout mice of both core complex genes and downstream components.56-58 In both Hes1 and FA mutant mice, eye development is abnormal.56,57,59-61 Together, these observations suggest that FA proteins may act in concert with the Notch1/HES1 signaling pathway to regulate stem cell function, whereas disruption of a functional FA/HES1 cross talk may lead to the HSC defect causing the Fanconi-type anemia. These observations also raise the possibility that in humans HES1 mutations that affect specifically their interaction with core complex members could cause FA, although such mutations have yet to be identified. Nevertheless, the fact that HES1 is a molecular partner of the FA core complex supports a function of the FA pathway in stem cell maintenance and developmental processes. These findings also support the idea that phenotypic features of FA, specifically those related to developmental defects, might stem from other functional impairments than DNA repair defects, as is the case for the syndrome trichothiodystrophy (TTD) where a dysfunction in basal transcription has been shown to underlie part of the TTD phenotype.62,63 With the identification of HES1 as a novel FA core complex interacting protein, functions of the FA pathway in developmental processes can now be assessed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr T. Taniguchi for FANCD2 foci immunostaining protocols.
This work was supported by a grant from the Sickkids Foundation/Institute for Human Development, Child and Youth Health of the Canadian Institutes of Health Research (CIHR, Ottawa, ON; XG 05-014R: Role of Fanconi anemia proteins in development; M.C. and G.L.), a CIHR junior investigator award (M.C.), FRSQ (Montreal, QC) senior and junior II investigator awards (G.L. and M.C.), and training awards from La Fondation de la Recherche sur les Maladies Infantiles (C.S.T., F.F.H., and O.H.), NSERC, (Ottawa, ON; C.C.H.), and FRSQ (C.S.T.).
Authorship
Contribution: C.S.T. designed and performed research, analyzed data, made the figures, and contributed to writing the paper; F.F.H., O.H., C.C.H., and C.G. performed research; G.L. designed research and contributed to writing the paper; and M.C. designed research, supervised the study, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Madeleine Carreau, Unité de recherche en Pédiatrie, Centre de recherche du CHUL, 2705 Boulevard Laurier, RC-9800, Québec, QC, Canada, G1V 4G2; e-mail: madeleine.carreau@crchul.ulaval.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal