Abstract
Hereditary folate malabsorption (HFM) patients harbor inactivating mutations including R113S in the proton-coupled folate transporter (PCFT), an intestinal folate transporter with optimal activity at acidic pH. Here we identified and characterized a novel R113C mutation residing in the highly conserved first intracellular loop of PCFT. Stable transfectants overexpressing a Myc-tagged wild-type (WT) and mutant R113C PCFT displayed similar transporter targeting to the plasma membrane. However, whereas WT PCFT transfectants showed a 22-fold increase in [3H]folic acid influx at pH 5.5, R113C or mock transfectants showed no increase. Moreover, WT PCFT transfectants displayed a 50% folic acid growth requirement concentration of 7 nM, whereas mock and R113C transfectants revealed 24- to 27-fold higher values. Consistently, upon fluorescein-methotrexate labeling, WT PCFT transfectants displayed a 50% methotrexate displacement concentration of 50 nM, whereas mock and R113C transfectants exhibited 12- to 14-fold higher values. Based on the crystal structure of the homologous Escherichia coli glycerol-3-phosphate transporter, we propose that the cationic R113 residue of PCFT is embedded in a hydrophobic pocket formed by several transmembrane helices that may be part of a folate translocation pore. These findings establish a novel loss of function mutation in HFM residing in an intracellular loop of PCFT crucial for folate transport.
Introduction
Folates are essential micronutrients that serve as one-carbon donors and acceptors in a multitude of biosynthetic reactions including de novo biosynthesis of purines, thymidylate, methionine, glycine and methylation reactions.1,2 Hence, folate vitamins are essential for DNA replication and cellular proliferation. However, mammalian organisms are devoid of folate biosynthesis and thus dietary sources must meet their metabolic requirement for folate cofactors. As such, mammalians rely on an efficient intestinal transport of folates that proceeds via a carrier-mediated system displaying optimal transport activity at acidic pH occurring primarily in the acidic microenvironment of the upper intestinal mucosal epithelium including that of the duodenum and the upper jejunum.3-5 Recently, the human proton-coupled folate transporter (PCFT) gene, PCFT/SLC46A1, was identified that encodes for an intestinal folate transporter displaying optimal transport activity at acidic pH.6 PCFT exhibits high affinities (Kt = 0.5-1 μM) for oxidized folates (folic acid) and reduced folates [eg, (6S)5-methylTHF and 5-formylTHF], as opposed to the reduced folate carrier (RFC/SLC19A1) that has high affinity for reduced folates (Kt = 2-4 μM) but low affinity for folic acid (Ki = 200-400 μM).6-8
Recently, several loss-of-function mutations were identified in the PCFT gene from patients with hereditary folate malabsorption (HFM), a congenital disease (OMIM 229050).6,9 HFM is a rare autosomal recessive disorder caused by impaired intestinal folate absorption.6,7,9-11 Patients with HFM present with low folate levels in the blood and cerebrospinal fluid (CSF). HFM manifests within the first few months after birth with anemia, recurrent or chronic diarrhea, hypogammaglobulinemia, severe infections, and failure to thrive. Because of the poor folate levels in the CSF and hence impaired folate uptake to the central nervous system (CNS), neurologic abnormalities and deficits occur, including seizures and mental retardation.
Here we studied the molecular basis of HFM in an Arab Israeli child born to consanguineous parents. We identified a novel loss-of-function R113C mutation that maps to the same R113 residue of PCFT previously shown to be substituted in another HFM patient with an R113S inactivating mutation.9 Although stable transfectants target this mutant R113C protein to the plasma membrane, they fail to transport folic acid. Moreover, this R113 residue localizes to a putative β-turn loop region that has been extremely conserved from bacteria to man and is absolutely crucial for folate transport function. Based on a bioinformatics analysis of homology modeling, we propose that the positively charged R113 residue that resides in a hydrophobic pocket formed by several transmembrane (TM) helices may be part of a folate binding and/or folate substrate translocation core.
Methods
Members of the family of the HFM child were studied according to a protocol approved by the Schneider Children's Medical Center of Israel (no. 4178) in accordance with the Declaration of Helsinki.
Drugs, biochemicals, radiochemicals, and antibodies
Folic acid and methotrexate (MTX) were purchased from Sigma-Aldrich Chemie BV (Zwijndrecht, The Netherlands), fluorescein-MTX (F-MTX) from Molecular Probes (Eugene, OR) and 3′,5′7,9-[3H]folic acid (MT783, 20 Ci/mmol) from Moravek (Brea, CA). Anti-Myc monoclonal antibodies were a generous gift from Prof Ami Aronheim (Rappaport Faculty of Medicine, Technion, Haifa, Israel).
Identification of a novel PCFT mutation in an HFM patient
The clinical features of the HFM child studied here were recently described.11 The healthy consanguineous parents and their HFM child were studied according to a protocol approved by the Schneider Children's Medical Center of Israel (no. 4178). Genomic DNA and total RNA were isolated from peripheral blood lymphocytes; RNA was reverse-transcribed and cDNA and genomic DNA were amplified using Expand polymerase (Roche Applied Science, Indianapolis, IN) as previously described.12,13 The primers for genomic polymerase chain reaction (PCR) and RT-PCR are listed in Tables S1 and S2 (available on the Blood website; see the Suppplemental Materials link at the top of the online article). PCR was performed as detailed recently.12,13 After fractionation on 1% agarose gels, PCR products were purified using a gel purification kit (Promega, Madison, WI) and DNA sequencing was carried out by the fluorescent dideoxy chain termination method using an ABI 377 sequencer (Hy-Labs, Rehovot, Israel).
Tissue cultures
Cells were grown under monolayer conditions in RPMI-1640 medium (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (FCS), 2 mM glutamine, 100 μg/mL penicillin, and 100 units/mL streptomycin (Biological Industries, Beth-Haemek, Israel) in a humidified atmosphere of 5% CO2. The growth medium of transfectant cells also contained 500 μg/mL G-418 (Calbiochem, San Diego, CA).
Site-directed mutagenesis and construction of a C-terminally Myc-tagged PCFT
The single nucleotide mutation 337C>T encoding for R113C was introduced into the WT PCFT gene and cloned into pCDNA 3.1(+) using Pfu Turbo DNA polymerase (QuickChange kit, Stratagene, La Jolla, CA). The coding region of PCFT was amplified using pCDNA 3.1(+) containing the WT PCFT gene as a template and primers containing EcoRI (forward primer) and BamHI (reverse primer) restriction sites. These primers were as follows:
Forward primer: 5′-CTGAATTCATGGAGGGGAGCGCGAGC-3′
Reverse primer: 5′-CTGGATCCGGGGCTCTGGGGAAACTG-3′
The resulting PCFT gene was then directionally cloned into pCDNA 3.1(−) (Invitrogen) containing a His Myc tag, and the 337C>T mutation was introduced into it.
Establishment of stable and Myc-tagged PCFT transfectants
RFC transport–null Chinese hamster ovary (CHO) C5 MTXR0.15 cells12 (2 × 107) were trypsinized and then transfected by electroporation (1000 μF, 234 V) with 10 μg of the following constructs: pCDNA 3.1(+) harboring the WT PCFT, the mutant R113C PCFT, and the empty vector in a final volume of 0.4 mL of serum-free RPMI-1640 medium containing 10 mM glucose and 0.1 mM DTT. Cells were then diluted in 10 mL of complete preheat RPMI-1640 medium, allowed to recover for 48 hours, and grown in medium containing 500 μg/mL G-418.
Folate growth requirement
Folic acid growth requirement was determined as previously described14 with some modifications as detailed in “WT but not mutant R113C PCFT markedly decreases the folic acid growth requirement in transfectant cells.” EC50 is defined as the folic acid concentration necessary to produce 50% of maximal cell growth.
Assay of F-MTX staining and competition with MTX
PCFT transfectants were seeded in 60-mm Petri dishes and incubated in growth medium containing 2 μM F-MTX for 8 hours at 37°C to saturate intracellular DHFR, after which MTX displacement analysis was performed as previously described.15 DC50 is defined as the MTX concentration necessary to displace 50% of maximal fluorescence.
Immunohistochemistry
Subcellular localization of Myc tagged PCFT in stable transfectants was determined by immunohistochemistry as recently described14 using a mouse anti-Myc monoclonal antibody (1:100 dilution) and a horseradish peroxidase-conjugated goat anti–mouse IgG as a secondary antibody (1:100 dilution, Jackson Immunoresearch Labs, West Grove, PA). Cells were then examined with a Leica DMIRE 2 fluorescence microscope (Wetzlar, Germany).
Results
Identification of a homozygous PCFT mutation in a child with HFM
Nucleotide sequencing was performed on both genomic PCR and overlapping RT-PCR products (Tables S1,S2) spanning the entire coding region of the PCFT gene. A single homozygous nucleotide 337C>T substitution was identified in exon 2 from both genomic DNA and cDNA of the HFM patient, whereas his parents were heterozygous. This mutation resulted in a homozygous R113C substitution residing in a region highly conserved from bacteria to man (see below); R113 localizes to the first intracellular loop connecting transmembrane helix 2 (TM2) and TM3 (Figure S1).
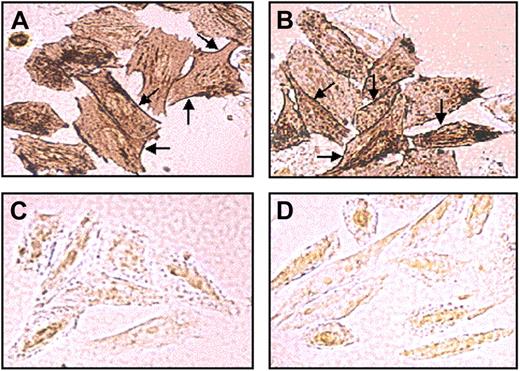
Mutant R113C PCFT is targeted to the plasma membrane in stable transfectants
To explore cellular expression and subcellular trafficking, the WT and mutant R113C PCFT and their C-terminally Myc-tagged cDNAs were stably transfected into RFC-null CHO cells.12 Semiquantitative RT-PCR analysis of the WT and mutant R113C PCFT transfectants showed comparable transcript levels relative to the hamster β-actin control (Figure S2A). Consistently, immunofluorescence and flow cytometric analysis revealed that both the WT and mutant R113C Myc-tagged PCFT proteins were overexpressed at similar levels in transfectant cells (Figure S2B). Furthermore, immunohistochemical analysis of the sub-cellular localization of the WT (Figure 1A) and mutant R113C PCFTs (Figure 1B) revealed comparable targeting to the plasma membrane (see arrows) along with some cytoplasmic localization. In contrast, mock transfected cells (Figure 1C) and untransfectants (Figure 1D) failed to show any significant immunostaining with the Myc-specific monoclonal antibody. Consistent with a previous report,9 the mutant Myc-tagged R113S PCFT was found here to be retained in the cytoplasm (data not shown).
Subcellular localization of the WT and mutant PCFT in stable transfectants. Immunohistochemical analysis of Myc-tagged WT (A) and mutant R113C PCFT (B) was performed with stably transfected RFC-null CHO cells using a Myc-tag–specific monoclonal antibody and 3,3′-diaminobenzidine as a chromogen. No specific immunostaining was observed in mock (C) and untransfected cells (D). Note the similar expression and the intense plasma localization (arrows) of the WT and mutant R113C PCFT.
Subcellular localization of the WT and mutant PCFT in stable transfectants. Immunohistochemical analysis of Myc-tagged WT (A) and mutant R113C PCFT (B) was performed with stably transfected RFC-null CHO cells using a Myc-tag–specific monoclonal antibody and 3,3′-diaminobenzidine as a chromogen. No specific immunostaining was observed in mock (C) and untransfected cells (D). Note the similar expression and the intense plasma localization (arrows) of the WT and mutant R113C PCFT.
Loss of folic acid transport in mutant R113C PCFT transfectants
To explore the folate transport function, initial rates of [3H]folic acid uptake were determined at pH 5.5 in stable RFC-null transfectants overexpressing the WT or mutant R113C PCFT (Figure 2). Untransfected cells as well as mock and mutant R113C PCFT transfectants all displayed the same basal [3H]folic acid influx (0.85 pmol/107 cells/min; P = .87), whereas stable transfection of the WT PCFT resulted in a 22-fold increase in the [3H]folic acid uptake rate (P < .001).
[3H]Folic acid uptake in cells stably transfected with the WT and mutant PCFT. To assess the functionality of the mutant R113C PCFT, [3H]folic acid influx into stable transfectants overexpressing the WT and mutant R113C PCFTs was determined as detailed in Document S1. Note the statistically significant, marked increase in folic acid uptake in WT PCFT transfectants but not in mutant R113C or mock transfectants. Data depicted are means plus or minus SD from 3 independent experiments performed on separate days.
[3H]Folic acid uptake in cells stably transfected with the WT and mutant PCFT. To assess the functionality of the mutant R113C PCFT, [3H]folic acid influx into stable transfectants overexpressing the WT and mutant R113C PCFTs was determined as detailed in Document S1. Note the statistically significant, marked increase in folic acid uptake in WT PCFT transfectants but not in mutant R113C or mock transfectants. Data depicted are means plus or minus SD from 3 independent experiments performed on separate days.
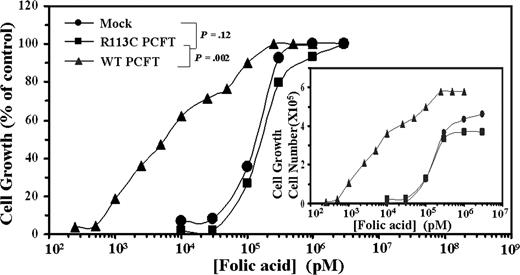
WT but not mutant R113C PCFT markedly decreases the folic acid growth requirement in transfectant cells
To further corroborate the loss of the folate transport function in the mutant R113C PCFT, transfectants were examined for their folic acid growth requirement (Figure 3). WT PCFT transfectants displayed a 50% folic acid growth requirement concentration (EC50) of 7.1 plus or minus 2.5 nM, whereas mock and mutant PCFT transfectants exhibited 24- and 27-fold higher EC50 values, respectively, thereby further establishing the loss of folic acid uptake in the mutant R113C PCFT.
Folic acid growth requirement of WT and mutant R113C PCFT transfectants. Exponentially growing PCFT transfectants were washed 3 times with PBS, transferred to folic acid-free RPMI-1640 medium supplemented with 10% dialyzed FCS (Invitrogen). After 10 days of deprivation in folic acid–free medium, cellular growth was arrested. Cells were then seeded in 24-well microplates (2 × 104 cells/well) containing increasing concentrations of folic acid ranging from 0.25 nM to 1 μM for WT PCFT transfectants and 10 nM to 3 μM for R113C PCFT as well as mock transfectants. After 7 days of incubation at 37°C, viable cell numbers were determined by hemocytometer count using trypan blue exclusion. A representative curve is shown from a total of 3 independent experiments. Actual cell numbers obtained for each folic acid concentration in the various transfectants are shown in the inset. The 100% control group used was the cell numbers obtained at the highest concentration of folic acid, ie, 3 μM for mock and R113C PCFT transfectants as well as 1 μM for folic acid for WT PCFT transfectants. Cell numbers obtained in these 100% control groups of mock, R113C PCFT, and WT PCFT transfectants were 3.7 × 105, 4.6 × 105, and 5.8 × 105, respectively.
Folic acid growth requirement of WT and mutant R113C PCFT transfectants. Exponentially growing PCFT transfectants were washed 3 times with PBS, transferred to folic acid-free RPMI-1640 medium supplemented with 10% dialyzed FCS (Invitrogen). After 10 days of deprivation in folic acid–free medium, cellular growth was arrested. Cells were then seeded in 24-well microplates (2 × 104 cells/well) containing increasing concentrations of folic acid ranging from 0.25 nM to 1 μM for WT PCFT transfectants and 10 nM to 3 μM for R113C PCFT as well as mock transfectants. After 7 days of incubation at 37°C, viable cell numbers were determined by hemocytometer count using trypan blue exclusion. A representative curve is shown from a total of 3 independent experiments. Actual cell numbers obtained for each folic acid concentration in the various transfectants are shown in the inset. The 100% control group used was the cell numbers obtained at the highest concentration of folic acid, ie, 3 μM for mock and R113C PCFT transfectants as well as 1 μM for folic acid for WT PCFT transfectants. Cell numbers obtained in these 100% control groups of mock, R113C PCFT, and WT PCFT transfectants were 3.7 × 105, 4.6 × 105, and 5.8 × 105, respectively.
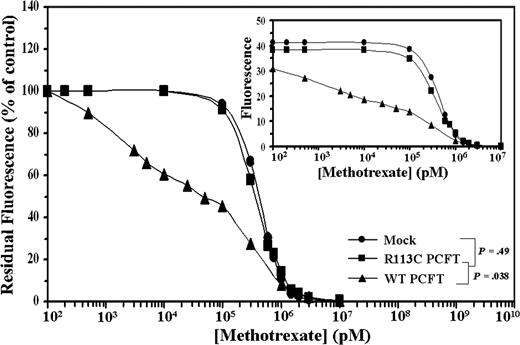
WT but not mutant R113C PCFT markedly decreases the 50% MTX displacement concentration in transfectants labeled with F-MTX
Further analysis was undertaken to explore the transport function of the WT and mutant PCFT. After saturation of intracellular DHFR with F-MTX, cells were exposed to competition with MTX and residual fluorescence was determined by flow cytometry. WT PCFT transfectants showed a low 50% MTX displacement concentration (DC50) of 49.8 plus or minus 0.3 nM, whereas mutant R113C PCFT and mock transfectants exhibited 12.2- and 14.2-fold higher DC50 values, respectively (Figure 4; P = .038). These results further establish that mutant R113C PCFT has lost both folic acid and MTX transport activities.
Flow cytometric analysis of cellular F-MTX staining and its displacement by MTX. After intracellular saturation of DHFR with 2 μM F-MTX for 8 hours, cells were incubated for 3 hours with increasing concentrations of MTX to initiate cellular F-MTX displacement. Residual F-MTX labeling was then determined by flow cytometry. A representative graph is shown from a total of 3 independent experiments. Actual cell numbers obtained for each MTX concentration in the various transfectants are shown in the inset.
Flow cytometric analysis of cellular F-MTX staining and its displacement by MTX. After intracellular saturation of DHFR with 2 μM F-MTX for 8 hours, cells were incubated for 3 hours with increasing concentrations of MTX to initiate cellular F-MTX displacement. Residual F-MTX labeling was then determined by flow cytometry. A representative graph is shown from a total of 3 independent experiments. Actual cell numbers obtained for each MTX concentration in the various transfectants are shown in the inset.
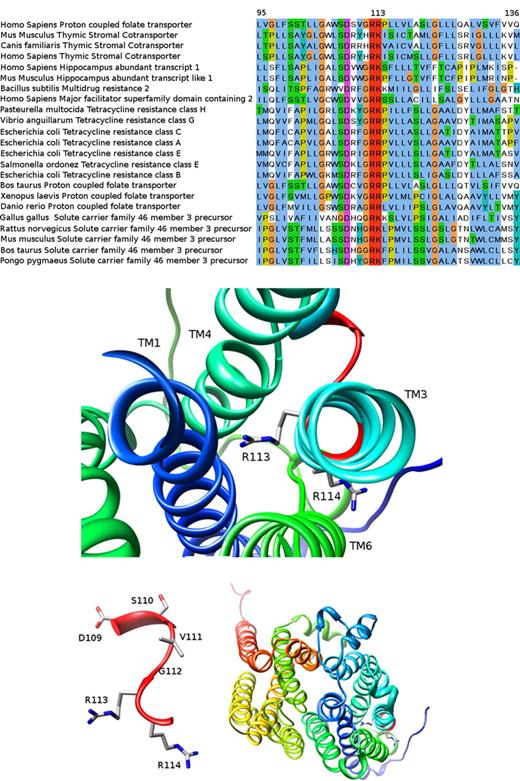
Bioinformatics analysis of the R113 residue
Here we identified a novel loss-of-function R113C mutation residing in the first cytoplasmic loop of PCFT, hence targeting the same R113 residue recently found to be inactivated in another HFM patient harboring an R113S mutation.9 Given the small number of people afflicted with HFM worldwide,10,11 this surprising cluster of mutations prompted us to explore the putative functional role of the R113 residue in greater detail. We identified a conserved G-R-[KR] motif (ie, residues 112-114), which is part of a larger sugar transport protein family signature: [LIVMSTAG]-[LIVMFSAG]-{SH}-{RDE}-[LIVMSA]-[DE]-{TD}-[LIVMFYWA]-G-R-[RK]-x(4,6)-[GSTA]. Evolutionary conservation analysis (see Document S1 for bioinformatics methodology) revealed that this region of PCFT is highly conserved among various transporters from bacteria to man (Figure 5A). Focusing our bioinformatics analysis around this G112R113R114 sub-motif we found that the 13-residue window encompassing amino acids 107-119 (including the G-R-[KR] signature) has a conservation average score of 6.1 ConSurf units,16 a high value compared with the average 5.0 (± 1.0), obtained for all 13 residue windows. This conserved region was further analyzed structurally as follows: we initially used the HHpred server17 (see Document S1 for bioinformatics methodology) to search for PCFT homologues with a known crystal structure available in the PDB.18 HHpred found 65 such homologues, of which the 3 closest homologous proteins were Escherichia coli transporters containing 12 TM helices; the glycerol-3-phosphate transporter (PDB 1pw4),19,20 EmrD, a multidrug transporter (PDB 2gfp)21 and the lactose permease (PDB 2cfq).22 The alignment between these 3 homologues and PCFT covers most of the sequence, yielding a relatively low sequence identity (approximately 14% for all of them), yet a statistically highly significant E-value (4.8 × 10−29, 1.0 × 10−25 and 5.6 × 10−25, respectively). Hence, the glycerol-3-phosphate transporter (GlpT) from E coli was selected to create the putative structural model of the PCFT as it has the lowest E-value and the highest alignment score, thereby resulting in a sequence identity of 13.8% over 424 aligned residues (from a total of 459). We hence built a 3D model for PCFT (Figure 5B-D) using the MPI implementation of MODELLER23 (see Document S1 for bioinformatics methodology). As evident from Figure 5B, 4 TM helices including TM1, TM3, TM4, and TM6 form a putative hydrophobic pocket with an average of 0.7 units in the Kyte and Doolitle scale24 toward which the extremely conserved, positively charged R113 residue surprisingly protrudes. Moreover, R113 is completely buried within this pocket; the surface accessible area (SAS) is 0 Å,2 whereas the next R114 residue faces outward and is accessible to water (SAS 45 Å2 ). A closer view of the D109SVGRR114 short loop between TM2 and TM3 reveals that R113 is the only residue in this loop predicted to face the intramembrane hydrophobic pocket lined by TM1, TM3, TM4, and TM6 (Figure 5C). Moreover, a general view of the modeled PCFT structure reveals the location of the R113 and R114 residues (Figure 5C in sticks on the bottom right corner) and the 12 TM helices (Figure 5D). We also carried out a second modeling experiment using the crystal structure of the EmrD transporter (PDB 2gfp)21 as a template. This model revealed a similar arrangement of the TM helices and here, too, the R113 was found to face the hydrophobic pocket region formed by TM1, TM3, TM4, and TM6, albeit, as expected, with a different rotameric conformation (data not shown).
Homology of the human PCFT to various transporters and 3D modeled structure of PCFT based on the crystal structure of the glycerol-3-phosphate transporter. The bioinformatics analysis performed here is given in great detail in Document S1. (A) A section of the multiple sequence alignment created by MUSCLE (residues 95-136 according to the human PCFT sequence; see bioinformatic methodology in Document S1). The human PCFT appears in the first line. Note the remarkable degree of conservation at this segment and especially human PCFT residue R113 that is absolutely conserved in many transporters from bacteria to man. This figure was prepared with JALVIEW31 and edited with GNU Image Manipulation Program (GIMP; http://www.gimp.org). (B-D) The 3D modeled structure of human PCFT viewed from the extracellular face. TM helices are presented in ribbon and residue side chains are shown in sticks and colored by element (carbon in gray, oxygen in red, and nitrogen in blue). (B) TM1, TM3, TM4, and TM6 in the vicinity of R113 and R114. (C) The D109-R114 short loop between TM2 (not shown) and TM3 (see panel A). (D) Overall view of the model. The 4 TM regions surrounding the conserved loop appear on the right bottom corner of the figure. Molecular graphics images were produced using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR-01 081).
Homology of the human PCFT to various transporters and 3D modeled structure of PCFT based on the crystal structure of the glycerol-3-phosphate transporter. The bioinformatics analysis performed here is given in great detail in Document S1. (A) A section of the multiple sequence alignment created by MUSCLE (residues 95-136 according to the human PCFT sequence; see bioinformatic methodology in Document S1). The human PCFT appears in the first line. Note the remarkable degree of conservation at this segment and especially human PCFT residue R113 that is absolutely conserved in many transporters from bacteria to man. This figure was prepared with JALVIEW31 and edited with GNU Image Manipulation Program (GIMP; http://www.gimp.org). (B-D) The 3D modeled structure of human PCFT viewed from the extracellular face. TM helices are presented in ribbon and residue side chains are shown in sticks and colored by element (carbon in gray, oxygen in red, and nitrogen in blue). (B) TM1, TM3, TM4, and TM6 in the vicinity of R113 and R114. (C) The D109-R114 short loop between TM2 (not shown) and TM3 (see panel A). (D) Overall view of the model. The 4 TM regions surrounding the conserved loop appear on the right bottom corner of the figure. Molecular graphics images were produced using the Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR-01 081).
Discussion
Several lines of evidence support the conclusion that R113C is a loss-of-function mutation. (1) Despite the similar targeting of the WT and mutant R113C PCFTs to the plasma membrane, R113C transfectants failed to display an increase in the [3H]folic acid influx beyond the basal transport activity observed at pH 5.5 in untransfected or mock-transfected cells. In contrast, WT PCFT transfectants exhibited a 22-fold increase in folic acid influx. (2) Stable transfection of the mutant R113C PCFT did not decrease the high folic acid growth requirement of the untransfected RFC-null CHO cells at pH 7.4, whereas WT PCFT transfection resulted in a 27-fold decrease in the folic acid growth requirement, hence being in concordance with the 22-fold increase in the folic acid influx in the WT PCFT transfectants. These results establish that the WT PCFT but not the mutant R113C PCFT efficiently takes up folic acid at acidic pH and to some extent at physiologic pH. (3) Transfection of the mutant R113C PCFT did not alter the DC50 value for MTX upon cellular F-MTX staining and flow cytometric analysis. In contrast, WT PCFT transfectants displayed a marked decrease (14-fold) in the DC50 value of MTX compared with untransfected cells. These findings demonstrate that the WT PCFT but not the mutant PCFT was active in MTX uptake at physiologic pH.
The following findings and considerations suggest that the R113 residue of PCFT and its close vicinity including cytoplasmic loop 1 are crucial for the folate transport function. (1) The current R113C mutation colocalizes to the same R113 residue recently found to be inactivated in an HFM patient with an R113S mutation that also nullified the positive charge of this arginine 113 residue.9 Hence, in the current study, R113C transfectants did not show any increase in [3H]folic acid influx, compared with the dramatic increase in the rate of folic acid uptake obtained in the WT PCFT transfectant. Consistently, stable transfectants expressing the R113S mutation failed to transport the physiologic blood folate, (6S)5-methylTHF.9 (2) A structure-based bioinformatics analysis revealed that the highly conserved cytoplasmic loop 1 motif D109XXGRR114 connecting TM2 and TM3 in PCFT (Figure 5) presumably forms a β-turn25 and is present in a spectrum of transporters of the major facilitator superfamily (MFS).26 Progressive alanine-substitution of amino acids within this consensus motif revealed that ablation of the putative β-turn structure abolished apical folic acid transport in polarized MDCK cell transfectants.25 This was associated with retention of the presumably misfolded mutant proteins in the endoplasmic reticulum (ER) and consequent lack of apical membrane targeting. Furthermore, in the metal-tetracycline/H + antiporter of E coli, site-directed mutagenesis of the Gly residues at positions 62 and 69 in this highly conserved motif abolished transport activity due to disruption of the β-turn structure of the peptide backbone.27 (3) Although comparative modeling of proteins with less than 30% sequence identity may appear challenging, recent studies have demonstrated that structural conservation between related proteins is particularly high within conserved functional sites and information regarding these regions can be often extracted from homology models, even in cases where sequence identity is low.28,29 Using modern software for sensitive protein homology detection including HHpred,17 we here identified a homology between PCFT and the E coli glycerol-3-phosphate transporter (GlpT), for which the crystal structure has been recently reported (PDB 1pw4).18 Although a relatively low degree of global sequence identity (∼14%) exists between PCFT and GlpT, their alignment yielded a highly significant E-value 4.8 × 10−29 (ie, an E-value much lower than 1 indicates a statistically significant match), thus suggesting that they may share some common structure-function features. Most importantly, sequence identity is markedly higher at the vicinity of the conserved intracellular loop 1 connecting TM2 and TM3 (∼24%), indicating that this region is more essential for the maintenance of the structure of this type of fold and for the transport function. As PCFT and GlpT share several structural and functional characteristics including 12 TM helices, similar size (452 aa and 459 aa, respectively) and anion substrate translocation, one can suggest that the general fold of PCFT is similar to that of GlpT. Hence, we constructed a homology-based model using the GlpT crystal structure as the template and subsequently analyzed the structure of the DXXGRR motif region. This analysis predicts that the cationic R113 residue of PCFT is completely buried in a hydrophobic cavity, the walls of which comprise TM1, TM3, TM4, and TM6 (Figure 5B). Interestingly, in GlpT, 2 crucial arginine residues which reside at the beginning of the translocation pore comprise the positively charged substrate-binding site for the anionic substrate, glycerol-3-phosphate.19,20 Hence, because this hydrophobic cleft is part of the glycerol-3-phospate substrate translocation core in the bacterial GlpT, the R113 of PCFT may possibly participate in the binding and/or translocation of the negatively charged folate substrate. In contrast, the following R114 residue in the PCFT model points out of this hydrophobic cleft and is significantly more exposed to the hydrophilic environment (Figure 5B,D). Moreover, the site-directed R114A mutant PCFT neither impaired its apical targeting nor interfered with folic acid transport activity.25 Hence, R113 (but not R114) is crucial for folate transport activity.
It was recently reported that PCFT displays a very low level of folic acid and MTX transport activity at physiologic pH (pH 7.4), compared with the excellent transport activity at pH 5.5.6,7 Specifically, the transport Kt for folic acid (Kt = 0.5 μM at pH5.5) increased 40-fold and the transport Vmax decreased by a factor of 2.5 as the pH was increased from pH 5.5 to pH 7.4. Similarly, the transport Kt for MTX (Kt =3.4 μM at pH 5.5) increased by a factor of 38 as the pH was increased from pH 5.5 to pH 7.4. Hence, these findings strongly suggested that PCFT plays a marginal role in folic acid and MTX uptake at physiologic pH. This presumption was functionally tested for the first time in the present study using 2 different assays: folic acid growth requirement as well as displacement of intracellular F-MTX labeling by exogenous MTX.15 Surprisingly, stable introduction of the WT PCFT but not the mutant R113C PCFT into RFC-null CHO cells resulted in a dramatic reduction in the folic acid growth requirement at an initial extracellular medium pH of 7.4. Specifically, whereas R113C PCFT and mock transfectants exhibited a folic acid EC50 as high as 200-220 nM, WT PCFT transfectants displayed a very low folic acid EC50 of only about 7 nM. Consistently, whereas the MTX DC50 value was as high as about 700 nM in R113C PCFT and mock transfectants, WT PCFT transfectants showed an efficient displacement of intracellular F-MTX with MTX DC50 values of 50 nM. Hence, the current study provides the first functional evidence in viable mammalian cells that WT PCFT has a substantial folic acid and MTX transport activity at neutral (physiologic) pH. Clearly, these findings may have important clinical implications for PCFT-dependent folate and antifolate transport at physiologic pH. This is true not only for the oxidized folate, folic acid, but particularly for the more physiologic reduced folate substrate (6S)5-methylTHF which retains a substantial transport activity at pH 7.4. Nevertheless, we find that the 7 days of folic acid growth requirement assay used here without medium refreshment can result in some acidification of the growth medium (∼pH 7.0) as previously shown,30 thereby further contributing to the retention of a significant PCFT-dependent folic acid transport activity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Idit Kosti for her assistance with the structural bioinformatics analysis.
This work was supported by a grant from The Fred Wyszkowski Cancer Research Fund, Haifa, Israel (to Y.G.A.).
Authorship
Contribution: Y.G.A. designed and wrote the paper; I.L. performed the majority of the experiments to which B.B., S.D., H.B., and G.J. contributed; R.S., Y.S., and M.S. provided patient specimens and clinical data; and F.G. performed most of the functional bioinformatics analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Yehuda G. Assaraf, The Fred Wyszkowski Cancer Research Laboratory, Department of Biology, Technion-Israel Institute of Technology, Haifa 32000, Israel; e-mail: assaraf@tx.technion.ac.il.

![Figure 2. [3H]Folic acid uptake in cells stably transfected with the WT and mutant PCFT. To assess the functionality of the mutant R113C PCFT, [3H]folic acid influx into stable transfectants overexpressing the WT and mutant R113C PCFTs was determined as detailed in Document S1. Note the statistically significant, marked increase in folic acid uptake in WT PCFT transfectants but not in mutant R113C or mock transfectants. Data depicted are means plus or minus SD from 3 independent experiments performed on separate days.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/5/10.1182_blood-2008-04-150276/5/m_zh80170823430002.jpeg?Expires=1769099186&Signature=agW2QnUyVuX3DtcDi59dVNUK-TmToItZ4rnveWxEkMEvYrN~XAoRPzegKpdYvaWUBaQXu4rJXycmRJCkaRXYcc38b-rW8sq8y8j-9EH9aBa-GLOYwvTnPyCdDPKbXA1Ku~syPXGBklQr6d-8mVObG41~6i2P8S~m-6Fe52yKW4zw~NBECMFaS8fT66b-49avwEPLNi5bn2~WQ3rlt7pxEG-z3oL6yGt13JkL17o7eGiHUDetEeROUUjn3Xsdnp8OYK4PJUhYT5EBGhV-2wwoxFWuvrn41XjIuUafk4~PyY~djDpnUz1ouQcpm2yQhb0QfkbjmbvWyExt~KVBwfULaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)