Abstract
Diamond-Blackfan anemia (DBA) is an inherited bone marrow failure syndrome characterized by anemia, congenital abnormalities, and cancer predisposition. Small ribosomal subunit genes RPS19, RPS24, and RPS17 are mutated in approximately one-third of patients. We used a candidate gene strategy combining high-resolution genomic mapping and gene expression microarray in the analysis of 2 DBA patients with chromosome 3q deletions to identify RPL35A as a potential DBA gene. Sequence analysis of a cohort of DBA probands confirmed involvement RPL35A in DBA. shRNA inhibition shows that Rpl35a is essential for maturation of 28S and 5.8S rRNAs, 60S subunit biogenesis, normal proliferation, and cell survival. Analysis of pre-rRNA processing in primary DBA lymphoblastoid cell lines demonstrated similar alterations of large ribosomal subunit rRNA in both RPL35A-mutated and some RPL35A wild-type patients, suggesting additional large ribosomal subunit gene defects are likely present in some cases of DBA. These data demonstrate that alterations of large ribosomal subunit proteins cause DBA and support the hypothesis that DBA is primarily the result of altered ribosomal function. The results also establish that haploinsufficiency of large ribosomal subunit proteins contributes to bone marrow failure and potentially cancer predisposition.
Introduction
Diamond-Blackfan anemia (DBA, MIM #105650)1 is an autosomal dominant bone marrow failure syndrome characterized by anemia typically presenting in infancy or early childhood. Similar to other inherited bone marrow failure syndromes, the central hematopoietic defect is of enhanced sensitivity of hematopoietic progenitors to apoptosis2 along with evidence of stress erythropoiesis, including elevations in fetal hemoglobin and mean red cell volume (MCV).3 In addition, the majority of patients exhibit an increase in erythrocyte adenosine deaminase activity.4 Occasionally, neutropenia and thrombocytopenia are also observed. Congenital abnormalities, including craniofacial, cardiac, genitourinary, and upper limb/hand malformations, are found in 40% to 50% of patients.5,6 In addition to anemia and abnormalities of embryogenesis, DBA is associated with an increased risk of cancer, most commonly hematologic malignancies and osteogenic sarcoma.5,7
Approximately 25% of affected patients have heterozygous alterations of RPS19, which encodes a protein component of the small ribosomal subunit.8 Haploinsufficiency of the RPS19 gene product has been demonstrated in a subset of cases9 and appears to be sufficient to cause DBA. There is as yet no definitive explanation of the restricted clinical phenotype of DBA given that the RPS19 gene product is expressed in most somatic tissues. Potential extra-ribosomal functions for Rps19 have been demonstrated,10 although how these might preferentially affect erythropoiesis and/or embryogenesis in DBA remains unclear.
A role for Rps19 in 18S rRNA maturation and small ribosomal subunit assembly has been established in yeast11 and in mammalian hematopoietic cell lines.12,13 Abnormalities of 18S rRNA maturation have also been observed in primary DBA fibroblasts, Epstein-Barr virus (EBV) lines, and bone marrow progenitors from patients with RPS19 mutations, suggesting that an abnormality of ribosomal function might be the predominant pathophysiologic abnormality underlying DBA.12-14 Additional evidence in support of a ribosome-mediated abnormality in DBA comes from the finding of mutations in genes encoding 2 additional small ribosomal subunit proteins, Rps24 and Rps17, which have also been identified as mutated in a small percentage of DBA patients.15,16 Haploinsufficiency resulting from somatic chromosomal deletions of another small subunit ribosomal protein gene, RPS14, has been implicated in the 5q-myelodysplastic syndrome, suggesting a critical role for abnormalities of the small ribosomal subunit in both inherited and acquired disorders of erythropoiesis and providing further evidence of a link between ribosomal abnormalities and cancer.17 Furthermore, abnormalities of ribosomal metabolism have been implicated in other inherited bone marrow failure syndromes, including Shwachman-Diamond syndrome, cartilage hair hypoplasia, and X-linked dyskeratosis congenita.18 Here we report the novel finding of a large ribosomal protein gene abnormality in DBA that affects rRNA processing, ribosome biogenesis, and selective cell proliferation and apoptosis, thus demonstrating a central role of the ribosome in this bone marrow failure syndrome.
Methods
Patients
A total of 150 DBA families participated in the study. Twenty-four were from multiplex families, whereas 126 included only one affected patient. Eleven probands had known mutations in RPS19 and 3 had RPS24 mutations. Informed consent was obtained from all patients and their family members under participating institutional protocols and in accordance with the Declaration of Helsinki. The diagnosis of DBA in all probands was based on the presence of sufficient classical criteria, including anemia presenting before the first year of life, reticulocytopenia, normal platelet and neutrophil counts, normocellular marrow with a paucity of erythroid precursors, and supportive criteria, including family history of DBA, red blood cell macrocytosis, elevated fetal hemoglobin, or elevated erythrocyte adenosine deaminase activity (eADA). A diagnosis of other inherited or acquired bone marrow failure syndromes, including Fanconi anemia, dyskeratosis congenital, and Shwachman Diamond syndrome, was excluded.
Institutional Review Board approval for the procurement and testing of clinical DBA blood samples for mutations in potential disease-related genes, establishment of cell lines, and banking was obtained from institutional review boards of Johns Hopkins University School of Medicine, Schneider Children's Hospital, and Children's Hospital of Boston.
RPL35A sequencing
Genomic DNA samples from 150 unrelated DBA probands (including 2 with 3q deletions) enrolled in the study and 180 control patients were amplified by polymerase chain reaction (PCR) and sequenced for mutations in the RPL35A gene. Primers were designed to amplify the coding exons, intron/exon boundaries of the RPL35A gene, and 250 base pairs (bp) upstream of the transcriptional start site. PCR products were directly sequenced from the forward and reverse primers. Mutations were confirmed by sequencing from 2 or more independent PCR reactions. DNA from 180 control patients was sequenced to determine whether the observed sequence variations were pathogenic mutations or polymorphisms. DNA samples from available family members were sequenced to determine whether the mutation cosegregated with the DBA phenotype within the pedigree. Primer sequences are included in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell culture
UT-7/Epo cells were maintained in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 U/mL erythropoietin. TF-1 cells were maintained in modified RPMI 1640 supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 4 μg/mL granulocyte-macrophage colony-stimulating factor. EBV-transformed lymphoblastoid cells lines from normal control subjects and DBA patients were derived from peripheral blood mononuclear cells using standard techniques.
shRNA design, lentivirus production, and target cell infection
Three short-hairpin interfering RNA sequences targeting the coding sequence and one targeting the 3′ untranslated region of RPL35A were investigated (Figure S1). An shRNA targeting firefly luciferase was used as a control.19 The 4 RPL35A shRNA constructs and the luciferase control sequence were directionally cloned into a lentilox vector (pLL3.7) downstream of the U6 promoter.20 Lentivirus-transduced cells were purified by fluorescence-activated cell sorting (FACSVantage SF; BD Biosciences, San Jose, CA) of GFP-positive cells 4 days after infection for growth, apoptosis, and polysome assays and 6 days after infection for Northern analysis and metabolic rRNA labeling. Additional details are given in Document S1.
Cell proliferation and apoptosis
A total of 103 sorted lentivirus-infected cells were plated per well in 96-well plates in 5 replicates for each time-point. Cells were cultured for the indicated times and assayed using CyQUANT NF Cell Proliferation Assay (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To correct for plating variance, the mean fluorescence intensity at each time point was LOG2-transformed and adjusted by the LOG2-transformed intensity on the initial day of plating. Apoptosis was monitored by flow cytometric analysis after staining of shRNA-infected cells with APC-annexin V (Invitrogen) according to the manufacturer's instructions.
Sucrose density gradient fractionation
Extracts of UT-7/Epo cells for polysome analysis were prepared as described.13 Extracts were layered on 16 mL 15% to 55% sucrose gradients and centrifuged in a SW28.1 rotor (Beckman Coulter, Fullerton, CA) for 5 hours at 67 000g. Gradients were fractionated and absorbance at 254 nm monitored on an ISCO model 185 gradient fraction collector and a UA-6 absorbance detector (Teledyne ISCO, Lincoln, NE). Chart records were digitized using Adobe Illustrator (Adobe Systems, San Jose, CA).
Nascent ribosomal RNA metabolic labeling
UT-7/Epo cells infected with lentivirus shRNA constructs were sorted for GFP expression 6 days after infection and plated in 6-well plates. After 24 hours, the media was replaced with phosphate-free RPMI 1640/10% FBS for 2 hours before labeling. For in vivo labeling, cells were incubated in 40 μCi 32P-orthophosphate for 1 hour. The metabolic labeling media was removed and chased with complete RPMI 1640/10% FBS for 4 hours. Total RNA was isolated using Trizol (Invitrogen). A total of 4 μg RNA from each sample was fractionated on 1.3% agarose/formaldehyde gels and dried on Whatman 3M paper. Labeled RNA was visualized by phosphorimage analysis (GE Healthcare, Piscataway, NJ).
Statistical methods
Confidence intervals were calculated using the extended Wald method. Mean proliferation and RPL35A expression levels were compared using the unpaired t test. Annexin binding intensities were compared using the Kolmogorov-Smirnov statistic. In all analyses, a 2-tailed P value less than .05 was considered significant.
GenBank accession numbers
Additional methods
Detailed methods used for gene expression analysis, comparative genomic hybridization (CGH), representational oligonucleotide microarray analysis, Lentivirus production, Southern and Northern blot analysis, inverse and quantitative PCR are available in Document S1.
The microarray data were submitted to MIAMExpress22 at the EMBL-EBI. The accession number is E-MEXP-1648.
Results
Identification of new genetic locus for DBA
The index case (deletion 1) was a 22-month-old male child with a normochromic, macrocytic anemia and congenital abnormalities, including hypertelorism, a ventricular septal defect, hypospadias, low-set ears, and developmental delay. DBA was diagnosed based on elevated eADA activity, hypercellular marrow with an absence of erythroid precursors, and normal chromosome breakage studies. The anemia responded to treatment with corticosteroids. The patient had no family history of DBA and a normal RPS19 gene sequence. At 4 years of age, he developed neutropenia without evidence for antineutrophil antibodies. A repeat bone marrow evaluation revealed decreased myeloid maturation with mild dysplasia. Cytogenetic analysis showed a constitutional deletion of 3q28-qter in both lymphocytes and buccal fibroblasts in the patient that was not present in either parent. This patient is alive at 9 years of age with developmental delay. He remains steroid-dependent for his anemia and has persistent leukopenia with mild dysplasia and normal platelet counts. A second patient with DBA and a 3q deletion from a different family was subsequently identified (deletion 2). This female presented at 7 weeks of age with a low hemoglobin of 59 g/L (5.9 g/dL), reticulocytopenia, an elevated MCV of 105 fL, as well as increased eADA activity and fetal hemoglobin. Other hematologic lineages were unaffected. Physical abnormalities were limited to a prominent pectus excavatum and inguinal hernia. The anemia was also responsive to treatment with steroids. A karyotype was normal, but subtelomeric fluorescence in situ hybridization demonstrated a de novo constitutional 3q29 deletion.
Identification of a limited candidate gene subset by microarray RNA expression analysis
Because haploinsufficiency of the RPS19 gene product is sufficient to cause DBA,9 we hypothesized that deletion of a critical gene in the 3q28-ter interval might lead to haploinsufficiency of a novel DBA disease allele, even in nonhematopoietic tissues. Because of the large number of genes in the deletion 1 interval, we used gene expression array analysis to evaluate expression of these candidates using a novel application of the hypothesis-based analysis of microarrays23 to identify genes with haploinsufficient expression located in the deleted region. To test the event of under-expression in the deletion 1 sample compared with controls, the hypothesis-based analysis of microarray method was applied to probe intensities from each experiment among (1) all probes on the array, (2) to the subset of probes identified as located on chromosome 3, and (3) to those probes lying within the interval, 3q28-qter. The intersection of the candidate gene sets formed a final set of 64 under-expressed probes, which were further examined (Figure S2; Tables S2,S3). Notably, RPL35A, a gene encoding a large ribosomal subunit protein and located at the telomeric end of chromosome 3q, was identified in each of these analyses. We subsequently confirmed decreased expression of RPL35A transcripts from both DBA patient cell lines by quantitative RT-PCR (Figure S3). A detailed discussion of the gene expression microarray analysis is available in the supplemental data.
Deletion breakpoint mapping
CGH and representational oligonucleotide microarray analyses were used to identify the deletion of an approximately 10-megabase region of chromosome 3q (Figure 1A) in deletion 1 that begins within the LPP transcription unit. Southern blotting demonstrated an aberrant band reactive to LPP intron 7 (Figure S4A). This region was amplified and cloned using inverse PCR to identify normal sequence from intron 7 of the LPP gene (to nucleotide 58 874, NT_005612.15) juxtaposed to β-satellite repeat sequence (Figure S4B,C,E). Deletion 2 was analyzed in a similar fashion on a higher density Agilent Technologies 105 000 feature CGH oligonucleotide microarray (Palo Alto, CA). High-density CGH (Figure 1) and confirmatory Southern blot analysis (Figure S4D,E) demonstrated a breakpoint telomeric to HES1.
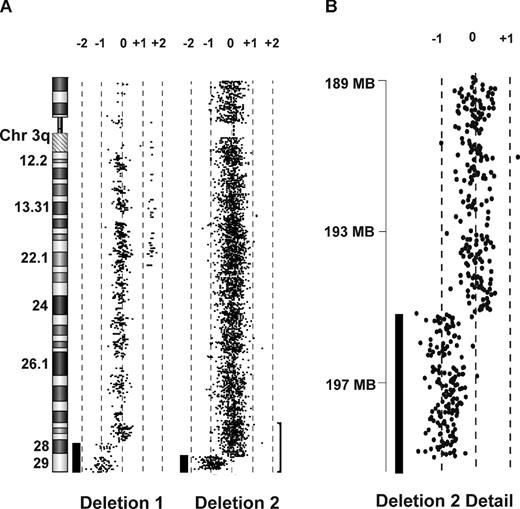
Array CGH of 2 DBA patients with 3q deletions. (A) CGH performed on genomic DNA of EBV lines derived from 2 patients with DBA and chromosome 3q terminal deletions demonstrates single copy terminal deletions of chromosome 3q. The region (bracket) encompassing the smaller deletion (195.46Mb-qter) is enlarged in panel B. Bars to the left of data plots represent segments of copy number loss.
Array CGH of 2 DBA patients with 3q deletions. (A) CGH performed on genomic DNA of EBV lines derived from 2 patients with DBA and chromosome 3q terminal deletions demonstrates single copy terminal deletions of chromosome 3q. The region (bracket) encompassing the smaller deletion (195.46Mb-qter) is enlarged in panel B. Bars to the left of data plots represent segments of copy number loss.
Composite analyses of RNA expression and deletion mapping identify RPL35A, encoding a large ribosomal subunit protein, as a candidate DBA gene
Based on the hypothesis that the critical gene should lie in the deletion interval common to both 3q deletions, we combined the deletion mapping from both DBA patients with 3q deletions and the RNA microarray expression analysis from deletion 1 to develop a restricted list of potential candidate genes. This analysis was further facilitated by the inclusion of mapping data derived from a reported 3q microdeletion syndrome, characterized by skeletal abnormalities and mental retardation but not by hematopoietic deficiencies, and, of note, not deleting RPL35A (Figure 2).24 Based on their roles in erythropoiesis, 2 genes in the initial deletion region considered as potential candidates, TFRC, the transferrin receptor 1, and HES1, a Notch1 signaling protein that regulates erythropoiesis via interaction with GATA1, were considered unlikely gene candidates because they were not included in both deletions (HES1), failed to show a difference in expression level in deletion 1 LCL by microarray (HES1), or fell within the region of the 3q microdeletion syndrome (TFRC).
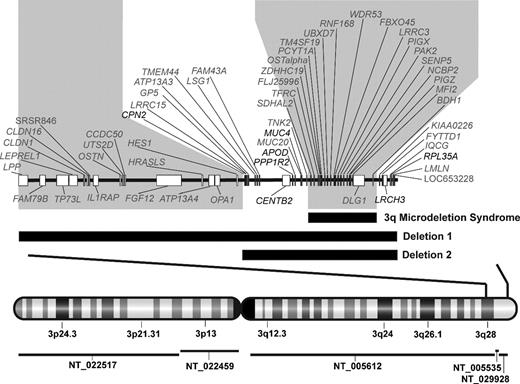
Chromosome 3q deletion mapping in 2 DBA probands. Genomic map of chromosome 3q deletions with combined deletion mapping and RNA expression microarray identifies 3q candidate genes for DBA. Vertical bars through the chromosome 3 map with G-banding pattern indicate the deleted region, which is enlarged above. Contig coverage is shown by horizontal lines below the chromosome ideogram. Black horizontal bars indicate the position of each deletion in relation to known genes on chromosome 3q28-ter. The larger deletion begins in intron 7 of LPP and spans approximately 10 megabases, including 3 contigs (NT_005612, NT_005535, NT_029928) with 2 gaps totaling an estimated 47 kb (NCBI Build 36.1). The smaller deletion begins in the intergenic region between HES1 and CPN2 and spans over 4 megabases. The position of a previously described 3q microdeletion syndrome, which was not associated with DBA or hematologic abnormalities, is also indicated.24 Regions of 3q which were either not involved in both DBA deletions or include the 3q microdeletion syndrome region (shaded gray) were considered improbable for candidate genes. Seven genes (noted in black), which lie in the intervals defined by these deletions, also demonstrated haploinsufficient expression in deletion 1 EBV LCL. RPL35A, a gene encoding a structural component of the large ribosomal subunit, is located within the extreme terminal region of the chromosome 3q deletions.
Chromosome 3q deletion mapping in 2 DBA probands. Genomic map of chromosome 3q deletions with combined deletion mapping and RNA expression microarray identifies 3q candidate genes for DBA. Vertical bars through the chromosome 3 map with G-banding pattern indicate the deleted region, which is enlarged above. Contig coverage is shown by horizontal lines below the chromosome ideogram. Black horizontal bars indicate the position of each deletion in relation to known genes on chromosome 3q28-ter. The larger deletion begins in intron 7 of LPP and spans approximately 10 megabases, including 3 contigs (NT_005612, NT_005535, NT_029928) with 2 gaps totaling an estimated 47 kb (NCBI Build 36.1). The smaller deletion begins in the intergenic region between HES1 and CPN2 and spans over 4 megabases. The position of a previously described 3q microdeletion syndrome, which was not associated with DBA or hematologic abnormalities, is also indicated.24 Regions of 3q which were either not involved in both DBA deletions or include the 3q microdeletion syndrome region (shaded gray) were considered improbable for candidate genes. Seven genes (noted in black), which lie in the intervals defined by these deletions, also demonstrated haploinsufficient expression in deletion 1 EBV LCL. RPL35A, a gene encoding a structural component of the large ribosomal subunit, is located within the extreme terminal region of the chromosome 3q deletions.
RPL35A mutations in DBA probands
To determine whether nondeletion RPL35A abnormalities occur in DBA, we screened genomic DNA from an additional 148 probands with DBA. This analysis revealed 3 mutations of RPL35A (Table 1), yielding an estimated 3.3% rate of RPL35A abnormalities in DBA probands (5 of 150, 95% confidence interval, 0.0122-0.0777). Both deletion patients had wild-type RPL35A sequence from the unaffected allele. An evaluation of polymorphic sequence variation of RPL35A in 180 normal control patients did not identify these sequence changes or other deviations from reference sequence in the coding region of RPL35A. However, 3 putative promoter polymorphisms were identified in both DBA and control samples: a linked T>G transversion at −110 and a G>T transversion at −68 as well as a G>A transition at −99 base pairs upstream from the transcriptional start site were seen in approximately 10% and 2% of cases, respectively.
Summary of RPL35A mutations
| Patient . | Inheritance . | DNA change . | Exon . | RNA change . | Predicted protein change . | Sex . | Presenting age, mo . | eADA . | Steroid responsive . | Associated anomalies . |
|---|---|---|---|---|---|---|---|---|---|---|
| Del 1 | Sporadic | Allelic deletion | — | ∼50% transcript | Protein haploinsufficiency | M | 2 | 2.8 | Yes | Hypertelorism, VSD, hypospadias, low-set ears, developmental delay |
| Del 2 | Sporadic | Allelic deletion | — | ∼50% transcript | Protein haploinsufficiency | F | 2 | 3.6 | Yes | Inguinal hernia, pectus excavatum |
| D9 | Sporadic | c.82_84delCTT | 3 | Del 82–84CTT | Del L27 | M | 14 | 1.04 | Yes | None |
| E2 | Sporadic | c.304 C>T | 4 | 304 C>U | R102stop | M | 2 | ND | No | Bilateral duplicated ureters, bilateral duplicated 11th ribs |
| 9 aa C-terminal truncation | ||||||||||
| 019 | Familial | c.97 G>A | 3 | 97 G>A; cryptic exon 3 splice donor | V33I; frameshift following E31 | M | 4 | 0.46 | Yes | Hypospadias, short stature |
| Patient . | Inheritance . | DNA change . | Exon . | RNA change . | Predicted protein change . | Sex . | Presenting age, mo . | eADA . | Steroid responsive . | Associated anomalies . |
|---|---|---|---|---|---|---|---|---|---|---|
| Del 1 | Sporadic | Allelic deletion | — | ∼50% transcript | Protein haploinsufficiency | M | 2 | 2.8 | Yes | Hypertelorism, VSD, hypospadias, low-set ears, developmental delay |
| Del 2 | Sporadic | Allelic deletion | — | ∼50% transcript | Protein haploinsufficiency | F | 2 | 3.6 | Yes | Inguinal hernia, pectus excavatum |
| D9 | Sporadic | c.82_84delCTT | 3 | Del 82–84CTT | Del L27 | M | 14 | 1.04 | Yes | None |
| E2 | Sporadic | c.304 C>T | 4 | 304 C>U | R102stop | M | 2 | ND | No | Bilateral duplicated ureters, bilateral duplicated 11th ribs |
| 9 aa C-terminal truncation | ||||||||||
| 019 | Familial | c.97 G>A | 3 | 97 G>A; cryptic exon 3 splice donor | V33I; frameshift following E31 | M | 4 | 0.46 | Yes | Hypospadias, short stature |
Del indicates deletion; aa, amino acid; eADA, erythrocyte adenosine deaminase activity (IU/g hemoglobin); VSD, ventricular septal defect; ND, not done; and —, not applicable.
DNA from family members was available for cases D9 and 019. The D9 delCTT mutation was not found in 2 unaffected siblings of the proband (data not shown). The 97 G>A mutation in patient 019 segregated with 2 family members with clinical features characteristic of silent DBA carriers but not with family members with normal hematologic parameters (Figure 3A). Although the valine 33 and leucine 27 positions are conserved among eukaryotes, the predicted V33I change results in a conservative amino acid substitution. However, a consistent doublet in the full-length RT-PCR product of RPL35A was observed in this sample that was not seen in other normal or DBA samples. Sequencing of both products demonstrated that the shorter cDNA resulted from aberrant splicing causing a nonsense mutation after amino acid position 31 (Figure 3B). The full-length band was found to contain both wild-type and 97 G>A sequence with the wild-type splicing pattern. Thus, this mutation results in haploinsufficiency of RPL35A mRNA as a result of the formation of a cryptic splice donor site.
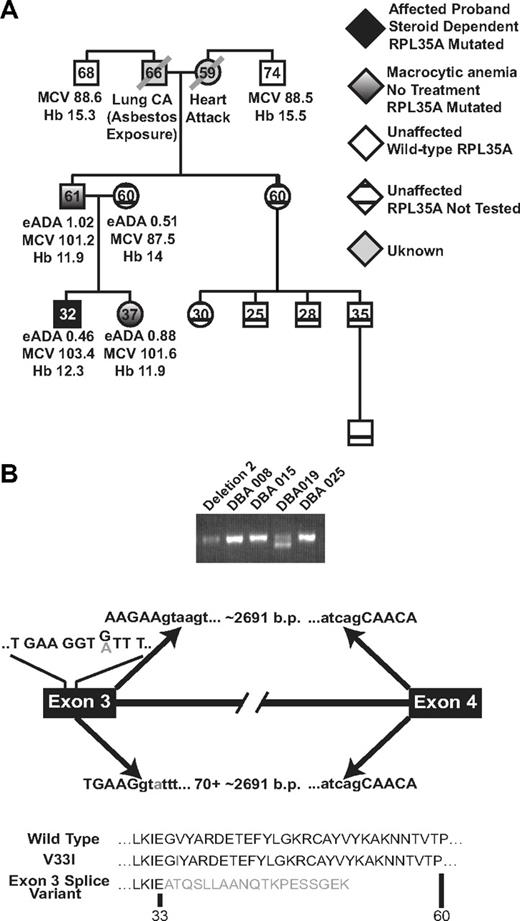
Genotype/phenotype analysis in patient DBA019. (A) RPL35A mutational analysis in this family showed 2 first-degree relatives, a father and sister, with the heterozygous RPL35A 97 G>A mutation. Both of these patients had macrocytic anemia suggestive of subclinical DBA carriers. Hemoglobin (Hb), mean red cell volume (MCV), and eADA activity are indicated; the hemoglobin value indicated for the proband is during treatment with steroids. Erythrocyte ADA was normal in all tested members of this pedigree. Black shading represents the DBA proband; white shading, hematologically unaffected members of the paternal lineage with normal RPL35A sequence; diagonal lines, deceased members with unknown RPL35A status; cross-hatched symbols, clinically normal persons who were not tested for RPL35A mutations. (B) 2 RPL35A RNA products were amplified from patient DBA019 using RT-PCR with primers designed to amplify the full-length RPL35A message. The shorter product results from an alternative splicing event between exons 3 and 4, leading to a truncated protein. The arrows above the transcript diagram show the normal splicing event, leading to wild-type RPL35A and removal of the approximately 2691 nucleotide intron. The lines above exon 3 indicate the wild-type codon sequence within exon 3; the 97 G/A mutation is indicated in gray. The arrows below the line demonstrate the abnormal splicing event. The 97 G>A mutation results in selection of a cryptic splice donor site within exon 3 immediately upstream of the change, causing removal of 70 base pairs of 3′ exon 3 coding sequence in addition to the intron. The predicted amino acid sequence of wild-type, simple amino-acid substitution, and the splicing variant are shown below.
Genotype/phenotype analysis in patient DBA019. (A) RPL35A mutational analysis in this family showed 2 first-degree relatives, a father and sister, with the heterozygous RPL35A 97 G>A mutation. Both of these patients had macrocytic anemia suggestive of subclinical DBA carriers. Hemoglobin (Hb), mean red cell volume (MCV), and eADA activity are indicated; the hemoglobin value indicated for the proband is during treatment with steroids. Erythrocyte ADA was normal in all tested members of this pedigree. Black shading represents the DBA proband; white shading, hematologically unaffected members of the paternal lineage with normal RPL35A sequence; diagonal lines, deceased members with unknown RPL35A status; cross-hatched symbols, clinically normal persons who were not tested for RPL35A mutations. (B) 2 RPL35A RNA products were amplified from patient DBA019 using RT-PCR with primers designed to amplify the full-length RPL35A message. The shorter product results from an alternative splicing event between exons 3 and 4, leading to a truncated protein. The arrows above the transcript diagram show the normal splicing event, leading to wild-type RPL35A and removal of the approximately 2691 nucleotide intron. The lines above exon 3 indicate the wild-type codon sequence within exon 3; the 97 G/A mutation is indicated in gray. The arrows below the line demonstrate the abnormal splicing event. The 97 G>A mutation results in selection of a cryptic splice donor site within exon 3 immediately upstream of the change, causing removal of 70 base pairs of 3′ exon 3 coding sequence in addition to the intron. The predicted amino acid sequence of wild-type, simple amino-acid substitution, and the splicing variant are shown below.
Deficient RPL35A expression results in reduced proliferation and increased apoptosis in hematopoietic cell lines
Decreased expression of RPS19 is associated with decreased proliferation and increased apoptosis in erythroid precursor cell lines and bone marrow progenitors, with preferential effects on erythroid development.25-27 To test whether decreased expression of RPL35A would have similar effects on cell growth and/or apoptosis, we tested 4 small hairpin RNAs (shRNA) that target RPL35A mRNA (Figure S1) and a control shRNA targeting firefly luciferase19 using a lentiviral system.20 The TF-1 and UT-7 cell lines are multipotential leukemic cell lines derived from human erythroleukemia and megakaryoblastic leukemia, respectively. UT-7/Epo is an erythropoietin-dependent UT-7 subclone with a committed erythroid progenitor phenotype.28 These cell lines have proven to be effective model systems for DBA with RPS19 mutations in that shRNA inhibition of RPS19 recapitulates the decreased proliferation, increased apoptosis, and 40S assembly defects seen in the bone marrow from DBA patients with mutations in RPS19.13,25,27 UT-7/Epo and TF-1 cells were infected with shRNA lentivirus and sorted for expression of the GFP marker. Real-time quantitative PCR demonstrated an 80% to 90% reduction in RPL35A message in these cell lines with 3 of the 4 shRNAs, each of which targets a different region in the coding sequence. A fourth shRNA (sh-2), targeting the 3′ untranslated region, led to RPL35A transcript levels of 50% to 80% of the control infected cells (Figure 4A). Cellular proliferation was monitored after cell sorting using a fluorescent DNA binding dye assay. The 3 effective RPL35A knockdown shRNAs caused significantly decreased proliferation in the UT-7/Epo erythroid precursor cell line compared with control or sh-2–transduced cells (Figure 4B-E). Similar reductions in proliferation were also observed in TF-1 cells (data not shown). Annexin V staining after RPL35A knockdown was increased by 45% to 55% over control-infected UT-7/Epo cells, suggesting that increased apoptosis was responsible at least in part for the proliferative defect in these cells (Figure 4F).
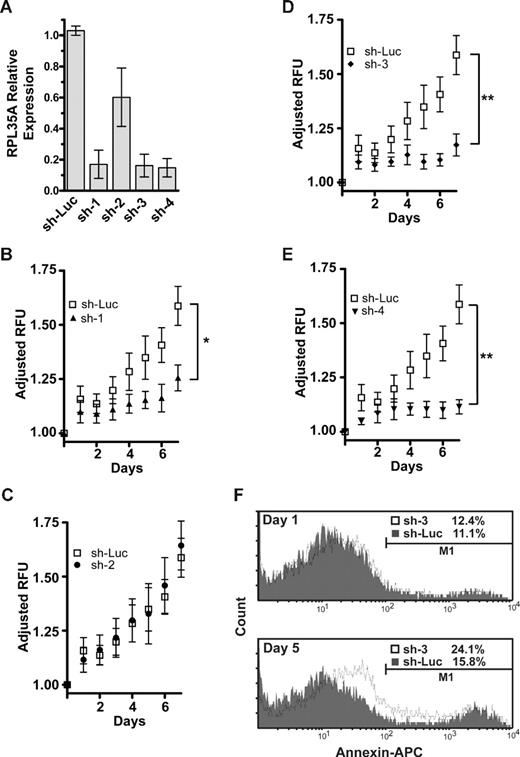
shRNA directed against RPL35A mRNA causes decreased proliferation and apoptosis. (A) The efficacy of expression knockdown was assessed by real-time quantitative RT-PCR 4 days after transduction of UT-7/Epo or TF-1 cells with the lentiviral siRNA construct. Bar graph represents expression of RPL35A transcripts after normalization to GAPDH in each of 4 shRNA constructs compared with cells infected with a control shRNA-lentiviral construct targeting firefly luciferase. Bars represent the aggregated mean expression level from 5 independent experiments, 3 in TF-1 and 2 in UT-7/Epo. Error bars indicate SD. (B-E) UT-7/epo cell proliferation was assessed by quantitation of fluorescent dye DNA binding. GFP-positive UT-7/epo cells were sorted 3 days after lentiviral infection, subsequently plated (day 0) in 96-well plates in 5 replicates at 103 cells/well, and assayed at the indicated times after sorting. Plating variation was corrected by adjusting the LOG2-transformed intensity at each day by the LOG2-transformed intensity on the initial day of plating. Curves represent the average of 4 platings from 2 independent experiments. Error bars indicate SDs. Differences between shLuc control-infected cells and RPL35A sh-2 were not significant. *sh-1, P < .05, compared with shLuc control cells. **sh-3 and 4, P < .01, compared with shLuc control cells. (F) Apoptosis was quantified by flow cytometric analysis after annexin V staining. GFP-positive UT-7/epo cells were sorted 3 days after infection and returned to culture for 24 hours. Cells were subsequently assessed on day 1 (top panel) and day 5 (bottom panel) after sorting. The histogram plots show cell counts versus annexin V intensity in shLuc (gray shading) and sh-3 (solid line) with apoptotic cells from each group enumerated above the gating threshold (M1). After sorting, no significant increase in annexin V–positive cells was discernable between control and sh-3–infected cells (K-S, P > .1). After adjustment for the proportion of annexin V–positive cells on day 1 (treatment group day 5 − treatment group day 1), RPL35A knockdown by day 5 resulted in a nearly 2.5-fold increase in annexin V–positive cells compared with controls (4.7% vs 11.7%; K-S, P < .001). Of note, a significant shift in the overall intensity of annexin V staining of the entire population of cells was also observed, suggesting that the overall effect on apoptosis was greater. Similar results were observed in sh-1– and sh-4–infected cells (not shown). Sh-Luc indicates Luciferase-control transduced cells; sh-1, -2, -3, and -4, respective RPL35A shRNAs.
shRNA directed against RPL35A mRNA causes decreased proliferation and apoptosis. (A) The efficacy of expression knockdown was assessed by real-time quantitative RT-PCR 4 days after transduction of UT-7/Epo or TF-1 cells with the lentiviral siRNA construct. Bar graph represents expression of RPL35A transcripts after normalization to GAPDH in each of 4 shRNA constructs compared with cells infected with a control shRNA-lentiviral construct targeting firefly luciferase. Bars represent the aggregated mean expression level from 5 independent experiments, 3 in TF-1 and 2 in UT-7/Epo. Error bars indicate SD. (B-E) UT-7/epo cell proliferation was assessed by quantitation of fluorescent dye DNA binding. GFP-positive UT-7/epo cells were sorted 3 days after lentiviral infection, subsequently plated (day 0) in 96-well plates in 5 replicates at 103 cells/well, and assayed at the indicated times after sorting. Plating variation was corrected by adjusting the LOG2-transformed intensity at each day by the LOG2-transformed intensity on the initial day of plating. Curves represent the average of 4 platings from 2 independent experiments. Error bars indicate SDs. Differences between shLuc control-infected cells and RPL35A sh-2 were not significant. *sh-1, P < .05, compared with shLuc control cells. **sh-3 and 4, P < .01, compared with shLuc control cells. (F) Apoptosis was quantified by flow cytometric analysis after annexin V staining. GFP-positive UT-7/epo cells were sorted 3 days after infection and returned to culture for 24 hours. Cells were subsequently assessed on day 1 (top panel) and day 5 (bottom panel) after sorting. The histogram plots show cell counts versus annexin V intensity in shLuc (gray shading) and sh-3 (solid line) with apoptotic cells from each group enumerated above the gating threshold (M1). After sorting, no significant increase in annexin V–positive cells was discernable between control and sh-3–infected cells (K-S, P > .1). After adjustment for the proportion of annexin V–positive cells on day 1 (treatment group day 5 − treatment group day 1), RPL35A knockdown by day 5 resulted in a nearly 2.5-fold increase in annexin V–positive cells compared with controls (4.7% vs 11.7%; K-S, P < .001). Of note, a significant shift in the overall intensity of annexin V staining of the entire population of cells was also observed, suggesting that the overall effect on apoptosis was greater. Similar results were observed in sh-1– and sh-4–infected cells (not shown). Sh-Luc indicates Luciferase-control transduced cells; sh-1, -2, -3, and -4, respective RPL35A shRNAs.
Deficient RPL35A expression is associated with decreased biogenesis of mature 60S ribosomal subunits
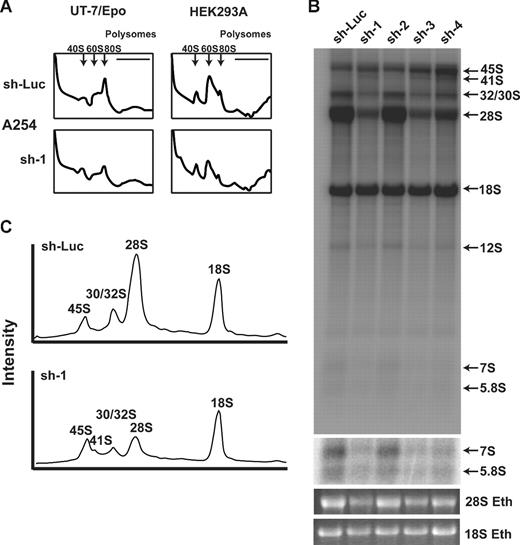
Deficiency of RPS19 in yeast and hematopoietic cell line knockdown experiments has been shown to result in a reduction of free 40S ribosomal subunits.11-13 We hypothesized that disruption of RPL35A would adversely affect 60S subunit biogenesis. Compared with UT-7/Epo cells transduced with control shRNA, cells transduced with RPL35A sh-1 showed a marked increase in the 40S:60S subunit ratio and decreased 80S complexes, suggesting that the apoptotic growth defect observed in these cells is the result of failure to produce 60S ribosomal subunits (Figure 5A). A similar, though less pronounced, effect on 60S formation was observed in the nonhematopoietic cell line, HEK293A.
Rpl35A is required for large ribosomal subunit assembly and pre-rRNA processing. (A) To identify potential sources of altered proliferation, polysome analysis was performed on UT-7/Epo (left column) and HEK293A (right column) cells infected with control (top row) or RPL35A (bottom row) sh-1. Cells were sorted 3 days after lentiviral infection and returned to media for 24 hours before treatment with cycloheximide and fractionation of lysates on a sucrose gradient. The 40S, 60S, and 80S peaks are indicated by; the polysome fraction lies below the horizontal line. In comparison to control shRNA-infected cells, cells infected with RPL35A shRNA demonstrated a decreased 40S:60/80S ratio, indicating a relative reduction of free 60S subunits. Similar results were seen in UT-7/Epo and HEK293A cells transduced with RPL35A sh-3 (not shown). Experiments were performed once with 2 different RPL35A shRNA for HEK293A cells and twice with 2 RPL35A shRNAs in UT-7/Epo cells. (B) Metabolic labeling of nascent RNA with 32P was used to identify abnormalities of pre-rRNA processing. Lentivirus-infected cells sorted 6 days after infection were plated in phosphate-free media in 6-well plates for 2 hours before the addition of 32P orthophosphate for 1 hour, washed, and then incubated in complete media for 4 hours. RNA was fractionated on 1.3% agarose/formaldehyde gels, dried, and autoradiographed. denotes the indicated mature and pre-rRNA species. RPL35A knockdown resulted in marked decrease of 32S, 28S, and 12S labeling without affecting mature 18S labeling. An increased exposure of the gel demonstrates reduced 7S and 5.8S rRNA. An ethidium bromide stain of the gel is also shown. (C) Intensity profile of lanes 1 and 2 demonstrates the reduction of 28S, preservation of 18S, and the appearance of a 41S band (shoulder adjacent to the 45S peak) not seen in control cells. Eth indicates ethidium bromide; sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, and -4, respective RPL35A shRNAs.
Rpl35A is required for large ribosomal subunit assembly and pre-rRNA processing. (A) To identify potential sources of altered proliferation, polysome analysis was performed on UT-7/Epo (left column) and HEK293A (right column) cells infected with control (top row) or RPL35A (bottom row) sh-1. Cells were sorted 3 days after lentiviral infection and returned to media for 24 hours before treatment with cycloheximide and fractionation of lysates on a sucrose gradient. The 40S, 60S, and 80S peaks are indicated by; the polysome fraction lies below the horizontal line. In comparison to control shRNA-infected cells, cells infected with RPL35A shRNA demonstrated a decreased 40S:60/80S ratio, indicating a relative reduction of free 60S subunits. Similar results were seen in UT-7/Epo and HEK293A cells transduced with RPL35A sh-3 (not shown). Experiments were performed once with 2 different RPL35A shRNA for HEK293A cells and twice with 2 RPL35A shRNAs in UT-7/Epo cells. (B) Metabolic labeling of nascent RNA with 32P was used to identify abnormalities of pre-rRNA processing. Lentivirus-infected cells sorted 6 days after infection were plated in phosphate-free media in 6-well plates for 2 hours before the addition of 32P orthophosphate for 1 hour, washed, and then incubated in complete media for 4 hours. RNA was fractionated on 1.3% agarose/formaldehyde gels, dried, and autoradiographed. denotes the indicated mature and pre-rRNA species. RPL35A knockdown resulted in marked decrease of 32S, 28S, and 12S labeling without affecting mature 18S labeling. An increased exposure of the gel demonstrates reduced 7S and 5.8S rRNA. An ethidium bromide stain of the gel is also shown. (C) Intensity profile of lanes 1 and 2 demonstrates the reduction of 28S, preservation of 18S, and the appearance of a 41S band (shoulder adjacent to the 45S peak) not seen in control cells. Eth indicates ethidium bromide; sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, and -4, respective RPL35A shRNAs.
Deficient RPL35A expression is associated with abnormal large ribosomal subunit rRNA processing
Decreased expression of RPS19 in human cells and in yeast is associated with a defect in rRNA processing characterized by a deficiency of mature 18S rRNA with accumulation of the 21S rRNA precursor.11-14 This rRNA processing abnormality underlies the failure to synthesize an adequate pool of 40S ribosomal precursors. To test whether decreased levels of RPL35A resulted in similar alterations in large subunit rRNA maturation, we used 32P-orthophosphate in vivo metabolic labeling of UT-7/Epo cells to study pre-rRNA processing after RPL35A knock-down. RPL35A knock-down resulted in marked reduction in the labeling of mature 28S and 5.8S rRNAs. Labeling of the 32S, 12S, and 7S pre-rRNA species was also significantly decreased. In contrast, the levels of 45S and 41S were increased relative to controls, suggesting a delay in processing of these precursors. There was no significant change in mature 18S rRNA associated with RPL35A knockdown (Figure 5B,C).
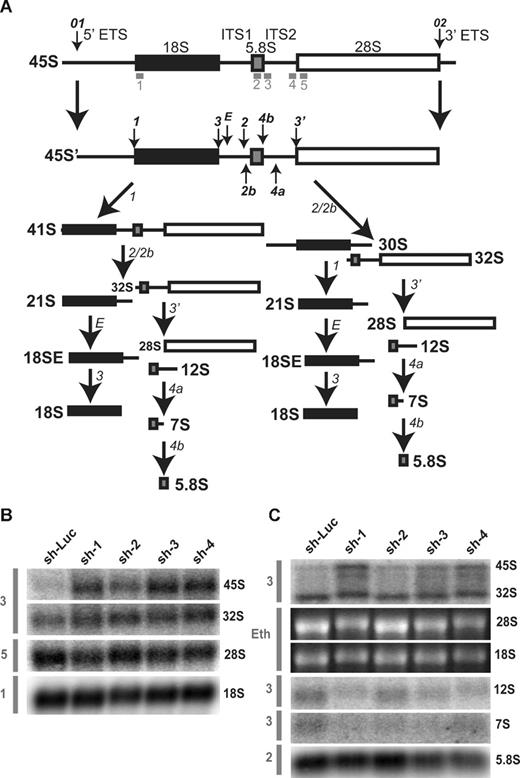
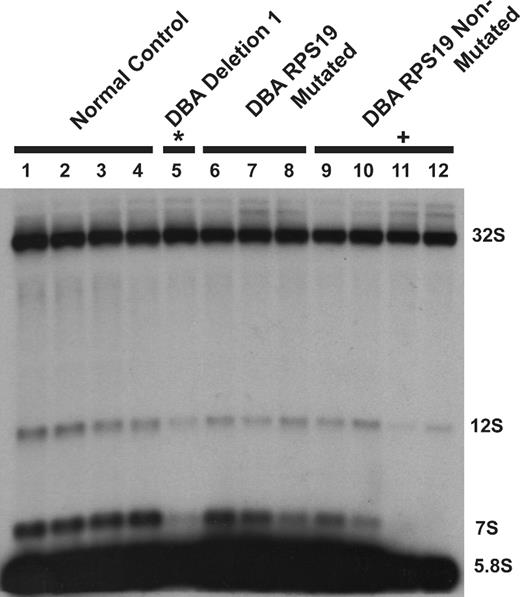
To confirm steady-state reductions of mature and precursor rRNA, Northern blot analysis was performed using oligonucleotide probes to detect the presence of mature and intermediate rRNA species after knockdown in both TF-1 and UT-7/Epo cell lines. A schematic of human ribosomal RNA processing pathways and the probes used for Northern blot analysis are shown in Figure 6A. Compared with control samples, knockdown of RPL35A transcript expression results in increased steady-state levels of the very early rRNA precursors 45S and 41S with a marked increase in 45S:32S ratios (Figure 6B), indicating an early processing defect affecting cleavages 1 or 2. In addition, 32S:12S ratios were increased after inhibition of RPL35A expression (Figure 6C), demonstrating that Rpl35A also functions in rRNA maturation downstream of 32S pre-rRNA maturation. In accordance with the in vivo labeling experiments and data from yeast with mutations and deletions of the RPL35A ortholog, RPL33A29 (J. Moore, J.E.F., R.J.A., J. Liu, S.R.E., manuscript submitted June 2008), we observed reductions in the 12S and 7S precursors to 5.8S rRNA as well as mature 5.8S rRNA which, after normalization to 18S levels, was decreased to approximately 60% to 65% of levels observed in cells with control shRNAs (Figure 6C). The steady-state 28S rRNA level was similarly decreased, although to a lesser extent, with 28S:18S ratios ranging from 75% to 85% of those in cells with control shRNAs (Figure 6B,C). Ribosomal RNA processing abnormalities have also been observed in DBA nonhematopoietic tissues12,14 ; we therefore used Northern blot analysis to evaluate a panel of DBA EBV cell lines for large subunit rRNA processing abnormalities (Figure 7). This demonstrated reduced 32S:12S ratios in both the deletion 1 and 019 EBV LCL samples as well as an additional DBA sample with wild-type RPL35A, suggesting the involvement of other genes linked to the large ribosomal subunit in DBA.
Rpl35A is required for pre-rRNA processing in ITS1 and ITs2. (A) A schematic of human pre-rRNA processing. Mature ribosomal RNA species are indicated by shaded boxes: ■, 18S;  , 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al,53 are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.
, 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al,53 are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.
Rpl35A is required for pre-rRNA processing in ITS1 and ITs2. (A) A schematic of human pre-rRNA processing. Mature ribosomal RNA species are indicated by shaded boxes: ■, 18S;  , 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al,53 are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.
, 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al,53 are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.
DBA EBV cell lines display altered large subunit pre-rRNA processing. RNA isolated from DBA EBV cell lines was probed with a 5.8S rRNA probe: lanes 1 to 4, healthy subject LCL; lane 5, deletion 1; lanes 6 to 8, DBA LCL with S19 mutations; lanes 9 to 12, non-S19-mutated DBA LCL. An altered 32S:12S ratio was observed in deletion 1 (*lane 5) and DBA019 EBV (+lane 11) cell lines. The sample in lane 12 also has increased 32S:12S with normal RPL35A transcript levels by qPCR and normal sequence for RPL35A and RPS19, suggesting additional gene defects may impair this pre-rRNA processing pathway in DBA.
DBA EBV cell lines display altered large subunit pre-rRNA processing. RNA isolated from DBA EBV cell lines was probed with a 5.8S rRNA probe: lanes 1 to 4, healthy subject LCL; lane 5, deletion 1; lanes 6 to 8, DBA LCL with S19 mutations; lanes 9 to 12, non-S19-mutated DBA LCL. An altered 32S:12S ratio was observed in deletion 1 (*lane 5) and DBA019 EBV (+lane 11) cell lines. The sample in lane 12 also has increased 32S:12S with normal RPL35A transcript levels by qPCR and normal sequence for RPL35A and RPS19, suggesting additional gene defects may impair this pre-rRNA processing pathway in DBA.
Discussion
A central and unresolved question since the identification of RPS19 mutations in DBA has been how to explain the limited clinical manifestations resulting from mutation and/or haploinsufficiency of a widely expressed ribosomal protein gene. Extra-ribosomal functions have been identified for a number of ribosomal protein genes,30 including RPS19,10 providing one plausible explanation for tissue specificity. However, the discovery of 2 additional small subunit ribosomal proteins responsible for the disorder suggests that the ribosome itself may be the site of the central defect in DBA. In the present study, we identified functional abnormalities linked to alterations of RPL35A in patients with DBA, thus establishing this as the first structural large ribosomal subunit protein defect directly implicated in human disease.
As with RPS19 mutations, a variety of mutation types were identified, including amino acid deletions, splicing defects, and genomic deletions. Haploinsufficiency of the RPL35A gene product is inferred in 3 of these cases based on genomic deletions or a nonsense splicing defect. These changes were not seen in the evaluation of 180 normal controls (360 normal chromosomes), a sample size sufficient to detect polymorphisms occurring at a frequency of 1% with 95% power.31 Although the size of normal controls is not sufficient to exclude very rare polymorphisms, the remaining 2 mutations delete either one or several highly conserved amino acids in regions predicted to form β-sheets,29 suggesting they may result in loss of protein function and thus are likely pathogenic. An autosomal dominant familial case of DBA with an RPL35A mutation was also identified in this study. The proband in this multiplex family demonstrated classic features of DBA: macrocytic anemia, reticulocytopenia, and a normocellular bone marrow with a paucity of erythroid precursors. The proband's father and sister demonstrate nonclassic DBA features of mild anemia and otherwise unexplained macrocytosis. Nonclassic DBA phenotypes32 and even nonexpressing phenotypes33 have been previously demonstrated in RPS19 mutated and nonmutated multiplex DBA families. Hence, the variable expressivity generally observed in DBA appears a feature of DBA resulting from RPL35A alterations as well. An important future question will be to determine whether defects in genes encoding large ribosomal subunit proteins lead to distinct differences in the types or frequencies of congenital anomalies or malignancies in DBA compared with defects of the small subunit.
Rpl35A, a 110-amino-acid ribosomal protein, is one of 47 structural components of the 60S ribosomal subunit. Like all small subunit proteins identified as mutated in DBA to date, Rpl35A is highly conserved in eukaryotes34 with archaeal but not bacterial orthologs; like Rps19 and Rps17, it is an essential ribosomal protein in yeast.35 In vitro studies have demonstrated that Rpl35A is capable of binding both 5.8S as well as initiator and elongator tRNA36,37 ; cross-linking studies in rats38 have suggested localization of Rpl35a in the ribosome near the A and/or P sites, although no more definitive localization has been made.39 The first mammalian RPL35A gene was cloned from murine erythroleukemia cells where it was found at high levels that decreased significantly on differentiation40 in a pattern reminiscent of RPS19 expression changes during terminal erythroid differentiation.41
We used shRNA-mediated knockdown in 2 hematopoietic cell lines to demonstrate a requirement for Rpl35A in the proliferation and viability of these cells as well as to identify a functional role for Rpl35A in 60S ribosomal subunit formation. Three of the 4 RPL35A shRNAs led to marked reduction of RPL35A mRNA; cells expressing each of these 3 shRNA constructs are nearly indistinguishable with respect to phenotypic abnormalities observed. Although the studies presented herein rely on mRNA estimates of effective knockdown of expression, we would predict significant reductions in Rpl35A protein given the extent of mRNA knockdown in the 3 effective shRNA constructs. In contrast, shRNA targeting the 3′ untranslated region was only modestly effective, leaving approximately 75% of RPL35A mRNA intact as assessed by quantitative PCR (qPCR), and did not lead to statistically significant changes in proliferation or to marked abnormalities in rRNA maturation compared with control cells. It is possible that this level of mRNA expression is insufficient to significantly alter protein levels. Alternatively, small reductions of Rpl35a protein may be insufficient to cause a phenotypic change in this system or posttranscriptional regulatory mechanisms may compensate for modest decreases in transcript level. Ribosomal protein expression is subject to translational control,42 and tissue-specific translational regulation is one factor that has been proposed to explain the lack of a more generalized phenotype in DBA.9
Knockdown of RPL35A with each of the 3 effective shRNAs resulted in significant reductions in proliferation as well as increased apoptosis, suggesting an important role for Rpl35A in promoting cellular proliferation and/or preventing apoptosis. Similarly, deletions of Rpl33A, the yeast RPL35A ortholog, result in a severe slow growth phenotype.29 In contrast to the 40S deficiency observed after RPS19 knockdown,13 polysome analysis after RPL35A knockdown demonstrated a reduction of free 60S subunits, a finding showing the requirement for Rpl35A in normal 60S subunit biogenesis, and consistent with marked 60S reductions observed in yeast with Rpl33A deletions.29
Northern blot analysis and metabolic rRNA labeling were used to further define the underlying defect in 60S subunit synthesis. Metabolic labeling showed reductions of 32S and subsequent rRNA species without significant changes in mature 18S levels. Northern analysis showed increased 45S:32S pre-rRNA ratios, suggesting an early rRNA processing defect, as well as increased 32S:12S ratios, indicating a processing defect downstream of 32S pre-rRNA. As shown in Figure 6, a processing defect in cleavages 1 or 2 should result in an accumulation of 45S pre-rRNA and lead to decreased levels of all 3 mature rRNA species derived from this transcript. However, levels of 18S rRNA are relatively unaffected in RPL35A knockdown cells. This implies an additional cleavage site(s) in ITS1 (cleavage 2b depicted in Figure 6A), analogous to the yeast A3 site, which would result in a selective reduction in 60S relative to 40S subunit rRNAs. Based on these findings, we would argue that the primary defect resulting from RPL35A knockdown is on processing of 32S pre-rRNA but not directly affecting earlier cleavages in ITS1. This primary cleavage defect is likely accompanied by a secondary effect on earlier processing, which results in the inhibition of cleavages at sites 1 and 2, leading to the observed elevation in 45S:32S ratios. A similar situation occurs in yeast where mutations that cause decreased 60S subunit production lead to secondary inhibition of cleavage at sites A0, A1, and A2 (similar to sites 1 and 2 in humans).43 Taken together, these metabolic rRNA labeling and Northern blot hybridization data indicate a role for RPL35A in efficient maturation of the 5′ end of the 32S pre-rRNA and suggest that it may act across a broader range of pre-rRNA processing, including maturation of the 12S rRNA.
The finding that DBA results from abnormalities of a large ribosomal subunit has several important implications. With the genetic defect unknown in roughly two-thirds of DBA cases, this finding suggests that mutations in other large subunit ribosomal proteins may also lead to DBA. This possibility is supported by our observation of DBA EBV lines with altered 32S:12S ratios and wild-type RPL35A. Based on the relatively low mutational rates observed in RPS24, RPS17, and RPL35A and the clinical heterogeneity of the disease, it is possible that DBA can result from mutations of many different structural ribosomal proteins or of the accessory factors required for ribosome assembly. It is also possible that these rates underestimate the true frequency of genetic changes at these loci because no detailed analysis of deletions or rearrangements has been reported in DBA. In terms of understanding the molecular pathobiology of DBA, the finding of a large subunit defect adds significant new support to the hypothesis that DBA is fundamentally a disorder of defective ribosome synthesis, rather than of extra-ribosomal functions of ribosomal proteins.
At least 3 nonexclusive consequences of a failure to synthesize adequate ribosomal components might contribute to the phenotype observed in DBA. First, insufficient ribosomes, whether the result of a relative 40S or 60S subunit deficiency, would be predicted to reduce the overall translational output in a quantitative fashion. Globally diminished protein production might then lead to a downstream apoptotic defect in erythroid progenitors or other tissues dependent on very high translational rates. In support of this idea, a quantitative reduction in translation has been reported in DBA patient-derived lymphocytes, irrespective of RPS19 mutational status.44
Second, ribosomal alterations might lead to qualitative differences in translational output. Although this theoretical consideration has not been demonstrated in DBA, a feature of Rpl35A function is intriguing. Martin-Marcos et al demonstrated aberrant translational regulation of GCN4, a transcriptional activator involved in the general amino acid control response, as a result of an Rpl33A yeast mutation (the yeast RPL35A ortholog) and, to a lesser extent, RPL33A deletion.29 Hence, abnormalities of RPL35A, and possibly additional ribosomal proteins, may qualitatively alter protein expression through changes in translational control.
A third potential mechanism whereby ribosomal protein deficiency might lead to a proapoptotic defect comes from nucleolar stress models where defective ribosome assembly signals to increase the steady-state level of p53 and promote a proapoptotic phenotype.45,46 A proapoptotic erythroid progenitor defect might thus be a consequence of abnormal ribosomes or imbalances of free ribosomal constituents interacting with cell-cycle checkpoints.47,48 In a scenario analogous to one proposed to explain the limited phenotype associated with RPL24 haploinsufficiency in the Bst/mouse,49 the predominantly erythroid phenotype could result from an inflexibility of rapidly differentiating erythroid precursors to tolerate cell-cycle prolongation relative to the normal erythroid differentiation program. Alternatively, haploinsufficient expression of ribosomal proteins might limit ribosome assembly because of erythroid specific differences in promoter or 5′ terminal oligopyrimidine (5′TOP) regulatory activity.50,51
The link of altered ribosome biogenesis to cell-cycle regulation and apoptosis may also provide important clues to the cancer predisposition of patients with DBA and other inherited bone marrow failure syndromes associated with ribosomal abnormalities. In this regard, it is notable that haploinsufficiency of several large and small subunit ribosomal proteins was associated with a high incidence of tumor formation in zebrafish.52 Furthermore, haploinsufficiency of Rps14 has been linked to the 5q- syndrome and myelodysplasia.17 One potential explanation of these findings is that alterations of ribosomal precursors, ribosome subunits, or their constituents may trigger alterations in cell-cycle control, inducing apoptotic stress, which can be circumvented by mutation or down-regulation of tumor suppressors. Alternatively, quantitative or qualitative defects in protein synthesis may lead to altered translation of tumor suppressors. Future studies should be able to experimentally test these alternative explanations as contributing pathways leading to cancer predisposition.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and families for their participation in this study, Drs Gita Massey and Nancy Dunn for the identification of the second deletion patient, Dr Joseph Palumbo for caring for the first deletion patient, and Amanda Black for computational microarray assistance.
This work was supported in part by grants from the Children's Cancer Foundation (R.J.A.), the Lyles Parachini Fund (R.J.A.), the Michael Corb Fund (R.J.A.), the Michael Garil Leukemia Survivors Program (R.J.A.), the Pediatric Cancer Foundation (J.M.L.), Alex's Lemonade Stand Foundation (J.E.F.), ASCO Foundation (J.E.F.), the Diamond Blackfan Anemia Foundation (H.T.G.) and National Institutes of Health (NIH) grants HL079583 (S.R.E.), HL079571 (J.M.L.), GCRC Grant MO1RR0918535 (J.M.L., A.V., E.A.), and CA120535 (R.J.A.). J.E.F. also received NIH support under Ruth L. Kirschstein National Research Service Award CA60441.
National Institutes of Health
Authorship
Contribution: E.C., M.A.M., J.K., S.R.E., and H.T.G. designed and performed research, analyzed data, and drafted the manuscript; M.N., C.C.T., P.M., D.E., A.H.B., H.E.S., and A.G. performed research and analyzed data; C.M.T., A.G., S.E.B., E.N., C.A.S., A.V., E.A., J.M.L., and H.T.G. contributed vital clinical samples; J.E.F. and R.J.A. developed the project, designed and performed research, analyzed data, performed statistical analysis, and drafted the manuscript; and R.J.A. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert J. Arceci, 1650 Orleans Street, CRB I Room 208, Baltimore, MD 21231; e-mail: arcecro@jhmi.edu.