Abstract
Heterodimerization domain (HD) mutations in NOTCH1 induce ligand-independent activation of the receptor and contribute to the pathogenesis of one-third of human T-cell lymphoblastic leukemias (T-ALLs). Here we report a novel class of activating mutations in NOTCH1 leading to aberrant activation of NOTCH1 signaling in T-cell lymphoblasts. These so-called juxtamembrane expansion (JME) alleles consist of internal duplication insertions in the vicinity of exon 28 of the NOTCH1 gene encoding the extracellular juxtamembrane region of the receptor. Notably, structure-function analysis of leukemia-derived and synthetic JME mutants demonstrated that the aberrant activation of NOTCH1 signaling is dependent on the number of residues introduced in the extracellular juxtamembrane region of the receptor and not on the specific amino acid sequence of these insertions. JME NOTCH1 mutants are effectively blocked by γ-secretase inhibitors and require an intact metalloprotease cleavage site for activation. Overall, these results show a novel mechanism of NOTCH1 activation in T-ALL and provide further insight on the mechanisms that control the activation of NOTCH1 signaling.
Introduction
NOTCH signaling plays a critical role in lineage specification decisions that enable multipotential precursor cells to become committed to specific cell lineages during development, and therefore has important roles in cell differentiation, proliferation, and apoptosis (reviewed in Greenwald1 and Maillard and Pear2 ). The fundamental components of the NOTCH pathway include the Delta and Serrate family of ligands (Delta-like 1, 3, and 4; and Jagged 1 and 2), the NOTCH receptors (NOTCH1-4), and the CSL (CBF1/Su(H)/LAG-1) DNA-binding protein, which together mediate the conversion of NOTCH-activating signals at the cell surface to changes in gene expression in the nucleus.3
The mature NOTCH1 receptor is a heterodimeric class I transmembrane glycoprotein generated by proteolytic processing of a precursor polypeptide (proNOTCH1) in the trans-Golgi network.4 This first protease cleavage (S1) is catalyzed by a furin protease that cuts the NOTCH1 precursor protein approximately 70 amino acids external to the transmembrane domain to generate an extracellular (NEC) and a transmembrane-intracellular (NTM) NOTCH1 subunit.4 These 2 polypeptides remain noncovalently associated in the resting receptor through the interaction of the sequences flanking the S1 furin cleavage site (C-terminus of NEC and N-terminus of NTM), which constitute the heterodimerization (HD) domain.3 In addition, the extracellular subunit of NOTCH1 contains 36 epidermal growth factor (EGF)–like repeats involved in ligand-receptor interaction, followed by 3 LIN-12/NOTCH repeats (LNRs), which stabilize the interaction between the extracellular and transmembrane subunits and help keep the receptor in a resting state in the absence of ligand.3 The NTM subunit of NOTCH1 consists of a short extracellular juxtamembrane peptide followed by a transmembrane sequence and a series of cytoplasmic domains including a RAM domain, a membrane proximal nuclear localization signal, a series of ankyrin repeats, a distal nuclear localization signal, a transactivation domain, and a carboxy-terminal PEST sequence, which together function as a ligand-activated transcription factor.3
Current models on the mechanism of NOTCH1 activation support that ligand interaction is followed by a conformational change in the LNR repeats-HD domain complex, which leads to the proteolytic cleavage of the NTM subunit, first by an ADAM metalloprotease, which cuts the extracellular juxtamembrane just 12 amino acids proximal to the membrane (S2 site), and subsequently, by the γ-secretase complex, an aspartyl protease multiprotein complex, which cuts the receptor at several different positions within the transmembrane domain.5-8 The final cleavage catalyzed by the γ-secretase complex at position Val1744 (S3 site) releases the intracellular fraction of NOTCH1 (ICN1) from the membrane, allowing it to translocate to the nucleus, where it activates the transcription of target genes in complex with the DNA-binding factor CSL and members of the Mastermind family of coactivators.
The NOTCH signaling pathway plays a critical role in the hematopoietic system by maintaining stem cell homeostasis9 and participating in multiple stages of T-cell development. During early hematopoiesis, NOTCH signaling is required for the commitment of multipotent hematopoietic progenitors to the T-cell lineage.10-13 In addition, NOTCH1 is required later on in T-cell development for progression through the early DN1, DN2, and DN3 stages of thymocyte maturation14 ; participates in the regulation of TCRB gene rearrangement15 ; and regulates lineage decisions between αβ versus γδ lineages16 and, at least in some systems, between CD4 versus CD8 lineages.17-20
Aberrant activation of NOTCH1 signaling induces transformation of T-cell progenitors and plays a prominent role in the pathogenesis of T-cell lymphoblastic leukemia (T-ALL).21 In human leukemias, NOTCH1 activation was first demonstrated in T-ALL cases harboring the t(7;9)(q34;q34.3), a rare chromosomal translocation that juxtaposes a truncated NOTCH1 gene next to the TCRB locus, leading to the aberrant expression of a truncated and constitutively active form of NOTCH1.22,23 More recent studies have demonstrated the major role of NOTCH1 in the pathogenesis of human leukemias by showing the presence of activating mutations in NOTCH1 in more than 50% of human T-ALLs.24,25 Activating mutations in NOTCH1 in human T-ALL are concentrated in exons 26 and 27, which encode the heterodimerization domain, and in exon 34, which encodes the PEST domain in the C-terminal region of the protein.25 HD mutations are typically single amino acid substitutions and small in-frame deletions and insertions and most likely operate by inducing ligand-independent activation of NOTCH1.25,26 In contrast, PEST mutations result in premature stop codons, the loss of the PEST domain, and increased levels of ICN1 due to impaired degradation of the activated receptor by the proteasome.25,27 Importantly, HD and PEST mutations are often found in cis in the same NOTCH1 transcript.25 These HD-PEST double-mutant alleles result in synergistic activation of NOTCH1 signaling and are 10 times more active than NOTCH1 alleles containing an HD or a PEST mutation alone.25
Here, we report the identification and functional characterization of a new class of activating mutations in NOTCH1 in human T-ALL. These novel mutations consist of internal tandem duplications of exon 28 and adjacent intronic sequences in the NOTCH1 gene, which result in expansions of the extracellular juxtamembrane region of the NOTCH1 receptor. Functional characterization of these so-called juxtamembrane expansion (JME) mutants demonstrates that spacing of the HD domain of NOTCH1 from the membrane triggers an aberrant proteolytic processing of NOTCH1 at the canonical S2 site, followed by endomembrane cleavage by the γ-secretase complex. This is in contrast with previously described class 1 and class 2 HD mutations, in which aberrant NOTCH1 signaling results from decreased stability of the HD domain or the displacement of the S2 site out of reach from the protective LNR domains, respectively. From these studies, we conclude that NOTCH1 JME mutant alleles constitute a novel class of oncogenic mutations responsible for aberrant constitutively active NOTCH1 signaling in T-ALL.
Methods
These studies were supervised by the Columbia University Medical Center (CUMC) Institutional Review Board. All clinical samples were collected in the context of clinical trials approved by the IRB committee of the participating institutions (see “Clinical samples”).
NOTCH1 expression plasmids
The pcDNA3 NOTCH1 expression plasmid is a full-length NOTCH1 expression construct containing codons 1 to 2555 of NOTCH1 followed by a FLAG tag sequence. pcDNA3 NOTCH1 L1600P encodes an HD (substitution of L to P at position 1600) mutant form of NOTCH1 tagged with a Flag tag epitope in the C-terminus. pcDNA3 NOTCH1 L1601P-ΔPEST encodes a double HD (substitution of L to P at position 1600) plus ΔPEST (truncation at position 2473) mutant form of NOTCH1 tagged with a FLAG tag epitope in the C-terminus. pcDNA3 NOTCH1, pcDNA3 NOTCH1 L1600P, and pcDNA3 NOTCH1 L1600P-ΔPEST constructs were a gift from Dr Iannis Aifantis at New York University. The pcDNA3 NOTCH1 Jurkat JME17 mutant was generated by cloning a partial NOTCH1 transcript (exons 19 to 29) amplified by polymerase chain reaction (PCR) from Jurkat cells, which contain an internal tandem duplication of 51 bases within exon 28 of the NOTCH1 gene, in the unique BamH1 and NotI restriction sites of pcDNA3 NOTCH1. A series of artificial NOTCH1 alleles (pcDNA3 NOTCH1 JME VSV-G 5, 8, 11-14) encoding insertions of 5 amino acids (NRLGK), 8 amino acids (IEMNRLGK), 11 amino acids (YTDIEMNRLGK), 12 amino acids (QYTDIEMNRLGK), 13 amino acids (EQYTDIEMNRLGK), and 14 amino acids (KEQYTDIEMNRLGK) in the extracellular juxtamembrane region of NOTCH1 at position 1740 were generated by cloning synthetic oligonucleotides containing the corresponding codons in the unique NotI restriction site of pcDNA3 NOTCH1. Mutant forms of NOTCH1 at the S2 cleavage site (AV 1720-1721 VH and AV 1720-1721 ED) were generated using the Quick Change II site-directed mutagenesis kit (Stratagene, La Jolla, CA). NOTCH1 amino acid sequence NP_060087.328 was used as reference for the annotation of all JME constructs and mutations.
Clinical samples
Samples of cryopreserved lymphoblasts from 210 children and young adults with T-ALL collected in the context of the DCOG and COALL clinical trials or treated at St Jude Children's Research Hospital (Memphis, TN) and Dana-Farber Cancer Institute (Boston, MA) were obtained with informed consent in accordance with the Declaration of Helsinki at the time of diagnosis. Genomic DNA from each sample was extracted with a commercial kit (GENTRA, Minneapolis, MN).
Western blot
Antibodies against activated NOTCH1 (NOTCH1 Val 1774; Cell Signaling, Beverly, MA), Renilla luciferase (Chemicon, Temecula, CA), FLAG (M2; Sigma-Aldrich, St Louis, MO) and tubulin (SC-8035; Santa Cruz Biotechnology, Santa Cruz, CA) were used in immunoblot assays in 293T cells following the manufacturers' instructions.
NOTCH1 mutation analysis
NOTCH1 mutation analysis in the Jurkat cell line was performed by direct sequencing of reverse transcription (RT)–PCR–amplified NOTCH1 transcripts. Primary T-ALL samples were analyzed by direct sequencing of PCR products expanding NOTCH1 exons 26, 27, and 28 and the distal part of exon 34 encompassing the sequences encoding the TAD and PEST domains. Primer sequences were as follows: exon 26 FW: GCTGAGGGAGGACCTGAACTTGG; exon 26 RV: CCTGAGCTGGAATGCTGCCTCTA; exon 27 FW: CATGGGCCTCAGTGTCCT; exon 27 RV: TAGCAACT-GGCACAAACAGC; exon 28 FW: GCGTAGCCGCTGCCTGAT; exon 28 RV: CAGACTCCCGGTGAGGATGC; exon 34 FW1: GCTGGCCT-TTGAGACTGG; exon 34 RV1: CTCCTGGGGCAGAATAGTGT; exon 34 FW2: ACAGATGCAGCAGCAGAACC; and exon 34 RV2: CCTGG-GGCCAGATAAAACAGTACA.
Reporter gene assays
pcDNA3 NOTCH1 expression plasmids were transiently transfected in HeLa cells using FuGene (Roche, Indianapolis, IN) transfection reagent together with the pGaLUC artificial luciferase reporter construct (a gift from Dr Honjo at Kyoto University, Kyoto, Japan), which contains 6 tandem CSL-binding sites; pRL, a plasmid expressing the Renilla luciferase gene under the control of the cytomegalovirus (CMV) promoter, was used as an internal control. Total DNA was kept constant by adding empty vector as needed. All transfections were carried out in triplicate. Cell lysates were harvested 48 hours after transfection, and luciferase assays were carried out using the Dual Luciferase Assay System (Promega, Madison, WI) on a LUMAT LB 9507 luminometer (Berthold Technologies, Oak Ridge, TN).
Cell lines
Jurkat cells were obtained from the ATCC (Manassas, VA). LOUCY, MOLT16, SUPT11, SUPT13, and P12-Ichikawa cells were a gift from Dr Thomas Look (Dana-Farber Cancer Institute). T-ALL cell lines were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere under 5% CO2. HeLa and 293T cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37°C in a humidified atmosphere under 5% CO2.
Heterodimer stability assays
We transfected 293T cells in 60-mm dishes with 2 μg empty pcDNA3 plasmid or with pcDNA3.1 plasmids encoding soluble N-terminal FLAG and C-terminal HA tags NOTCH1 minireceptors corresponding to wild-type NOTCH1 (amino acids 1446 to 1743) and Jurkat JME17 and VSV-G JME 14 NOTCH1 mutant alleles. Three days after transfection, the conditioned media were collected, centrifuged at 500g for 5 minutes, and divided in 2 aliquots that were incubated with anti-FLAG and anti-HA beads at 4°C overnight. Following 3 washes with wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% NP40), the precipitated proteins were eluted with FLAG or HA peptide and analyzed by Western blotting with antibodies against the epitope tags. For the experiment in the presence of urea, HA immunoprecipitates were prepared as above and following 3 washes with wash buffer, were incubated with 1 mL wash buffer with 0 to 4 M urea for 30 minutes at 24°C. Following 2 more washes, immunoprecipitated proteins were eluted and analyzed by Western blot as above.
Results
Internal tandem duplications in exon 28 of NOTCH1 in T-ALL
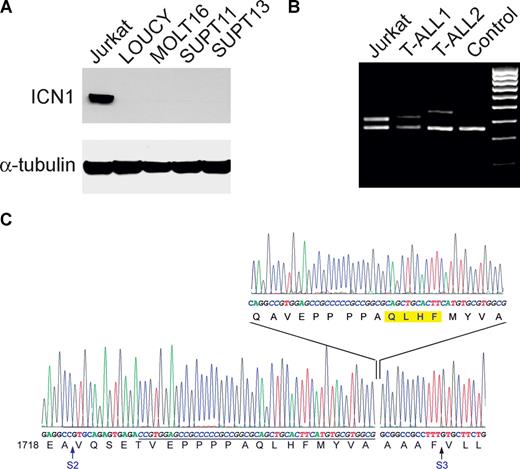
Activating mutations in the HD and PEST domains of NOTCH1 have been detected in more than 70% of T-ALL cell lines and are associated with the expression of high levels of ICN1. To analyze the possible role of NOTCH1 signaling in T-ALL lines lacking known activating mutations in the NOTCH1 gene, we analyzed the levels of ICN1 protein by Western blot using the Val1744 antibody, which specifically recognizes the γ-secretase cleaved activated form of NOTCH1, in a panel of T-ALL lines lacking mutations in exons 26, 27, and 34 of the NOTCH1 gene. This analysis demonstrated high levels of ICN1 protein in the Jurkat T-ALL cell line but not in LOUCY, MOLT16, SUPT11, or SUPT13 cells, suggesting the presence of an as-yet-unidentified activating mutation in the NOTCH1 gene in this cell line (Figure 1A). To screen for further activating mutations in NOTCH1, we performed direct sequencing analysis of RT-PCR products encompassing the complete coding sequence of the NOTCH1 gene in Jurkat cells. Sequence analysis revealed an in-frame insertion of the CAGG tetranucleotide followed by an internal tandem duplication of 47 bp in exon 28 of NOTCH1, resulting in the insertion of 17 amino acids (QAVEPPPPAQLHFMYVA) at position 1740 in the extracellular juxtamembrane region of the NOTCH1 receptor (Figure 1B). No further mutations in the entire NOTCH1 cDNA sequence from this cell line were detected. PCR amplification of exon 28 sequences from genomic DNA from 210 primary T-ALL samples identified 7 patients showing an extra band of increased size in addition to the normal exon 28 PCR product (Figure 1C). Sequence analysis of these aberrant PCR products demonstrated the presence of internal tandem duplications ranging from 33 to 108 bases in the region extending from the distal part of intron 27 to the proximal segment of exon 28 resulting in the in-frame insertion of 11 to 36 amino acids in the extracellular juxtamembrane segment of NOTCH1 (Table 1). Direct sequence analysis of exon 28 and flanking intronic sequences in 50 T-ALL samples showing normal sized PCR products failed to detect any additional NOTCH1 mutations in this region.
Identification of internal tandem duplications in the extracellular juxtamembrane domain of NOTCH1 in T-ALL. (A) Western blot analysis of T-ALL cell lines lacking HD and PEST domain mutations in NOTCH1 demonstrating high levels of activated NOTCH1 in the Jurkat cell line. ICN1 levels were detected with the NOTCH1 Val1744 antibody. Tubulin is shown as loading control. (B) Sequence analysis of the NOTCH1 transcripts showing the mutation identified in the Jurkat cell line. The wild-type NOTCH1 transcript sequence is shown at the bottom and inserted sequences on top. S2 and S3 indicate the sites for ADAM metalloprotease and γ-secretase cleavage, respectively. Duplicated nucleotides are shown in italics and underlined in the wild-type sequence. The common 4 amino acids (QLHF) present in all JME mutants identified in this series are highlighted in yellow. (C) PCR amplification of exon 28 of NOTCH1 and adjacent intronic sequences. Three samples corresponding to Jurkat cells and 2 independent primary T-ALL cases show bands of increased size compared with the control genomic DNA indicating the presence of insertions in the extracellular juxtamembrane domain of NOTCH1.
Identification of internal tandem duplications in the extracellular juxtamembrane domain of NOTCH1 in T-ALL. (A) Western blot analysis of T-ALL cell lines lacking HD and PEST domain mutations in NOTCH1 demonstrating high levels of activated NOTCH1 in the Jurkat cell line. ICN1 levels were detected with the NOTCH1 Val1744 antibody. Tubulin is shown as loading control. (B) Sequence analysis of the NOTCH1 transcripts showing the mutation identified in the Jurkat cell line. The wild-type NOTCH1 transcript sequence is shown at the bottom and inserted sequences on top. S2 and S3 indicate the sites for ADAM metalloprotease and γ-secretase cleavage, respectively. Duplicated nucleotides are shown in italics and underlined in the wild-type sequence. The common 4 amino acids (QLHF) present in all JME mutants identified in this series are highlighted in yellow. (C) PCR amplification of exon 28 of NOTCH1 and adjacent intronic sequences. Three samples corresponding to Jurkat cells and 2 independent primary T-ALL cases show bands of increased size compared with the control genomic DNA indicating the presence of insertions in the extracellular juxtamembrane domain of NOTCH1.
NOTCH1 JME mutations in T-ALL
| Sample . | NOTCH1 sequence (NM_017617.2)28 . | Predicted amino acid change . |
|---|---|---|
| Jurkat | 5220 Ins CAGGCCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGGCG | 1740 Ins QAVEPPPPAQLHFMYVA |
| T-ALL 1 | 5212 Ins TGAAGGGGAGCCGCTGCCTGATGTCCGGGCACCTGCCCCTGGCCCCCGTGCCCGCAGGTGA-GACCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGT | 1738 InsLKGSRCLMSGHLPLAPVPAGETVEPPPPAQLHFMY |
| T-ALL 2 | 5221 Ins TGCGTAGCCGCTGCCTGATGTCCGGGCACCTGCCCCTGGCCCCCGTGCCCGCAGGTGAGAC-CGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGGCGG | 1741 Ins VRSRCLMSGHLPLAPVPAGETVEPPPPAQLHFMTWR |
| T-ALL 3 | 5214 Ins CCGCCCCCGCCGGCGCAGCTGCACTTCATGTAC | 1738 Ins PPPPAQLHFMY |
| T-ALL 4 | 5209 Ins GGACCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCA | 1737 Ins RTVEPPPPAQLHF |
| T-ALL 5 | 5221 Ins AGGCCCGGCAGCTGCACTTCATGTACGTGGCGG | 1741 Ins EARQLHFMYVA |
| T-ALL 6 | 5218 Ins GGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGG | 1740 Ins GEPPPPAQLHFMYV |
| T-ALL 7 | 5218 Ins CCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGG | 1740 Ins AVEPPPPAQLHFMYV |
| Sample . | NOTCH1 sequence (NM_017617.2)28 . | Predicted amino acid change . |
|---|---|---|
| Jurkat | 5220 Ins CAGGCCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGGCG | 1740 Ins QAVEPPPPAQLHFMYVA |
| T-ALL 1 | 5212 Ins TGAAGGGGAGCCGCTGCCTGATGTCCGGGCACCTGCCCCTGGCCCCCGTGCCCGCAGGTGA-GACCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGT | 1738 InsLKGSRCLMSGHLPLAPVPAGETVEPPPPAQLHFMY |
| T-ALL 2 | 5221 Ins TGCGTAGCCGCTGCCTGATGTCCGGGCACCTGCCCCTGGCCCCCGTGCCCGCAGGTGAGAC-CGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGGCGG | 1741 Ins VRSRCLMSGHLPLAPVPAGETVEPPPPAQLHFMTWR |
| T-ALL 3 | 5214 Ins CCGCCCCCGCCGGCGCAGCTGCACTTCATGTAC | 1738 Ins PPPPAQLHFMY |
| T-ALL 4 | 5209 Ins GGACCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCA | 1737 Ins RTVEPPPPAQLHF |
| T-ALL 5 | 5221 Ins AGGCCCGGCAGCTGCACTTCATGTACGTGGCGG | 1741 Ins EARQLHFMYVA |
| T-ALL 6 | 5218 Ins GGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGG | 1740 Ins GEPPPPAQLHFMYV |
| T-ALL 7 | 5218 Ins CCGTGGAGCCGCCCCCGCCGGCGCAGCTGCACTTCATGTACGTGG | 1740 Ins AVEPPPPAQLHFMYV |
Expansion of the extracellular juxtamembrane region of NOTCH1 induces strong receptor activation
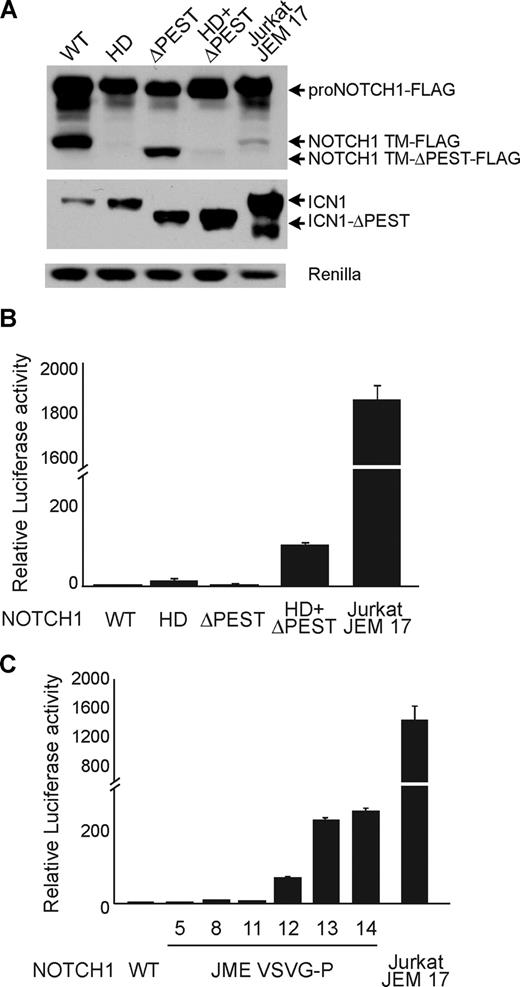
To analyze the functional relevance of NOTCH1 JME mutants, we analyzed the levels of activated ICN1 in cells transfected with wild-type NOTCH1, the JME17 mutation isolated from Jurkat cells, and a series of prototypical mutant forms of NOTCH1 including the HD (L1600P), ΔPEST, and HD (L1600P) ΔPEST NOTCH1 mutant alleles. These experiments showed that wild-type NOTCH1 and each NOTCH1 mutant allele result in high levels of NOTCH1 precursor protein (proNOTCH1). In contrast, ICN1 levels were markedly increased in cells expressing mutant NOTCH1 alleles compared with control wild-type NOTCH1 transfected cells. Notably, the NOTCH1 Jurkat JME17 mutant allele induced ICN1 levels higher than previously described HD, ΔPEST, and HD-ΔPEST NOTCH1 mutant alleles (Figure 2A). Similarly, luciferase reporter assays showed that expression of NOTCH1 HD, ΔPEST, and HD-ΔPEST NOTCH1 mutant alleles resulted in 12-, 2.5-, and 105-fold activation of NOTCH1 signaling over basal levels, respectively; while expression of the NOTCH1 Jurkat JME17 mutant allele induced almost 2000-fold activation of the NOTCH1 reporter compared with controls (Figure 2B).
NOTCH1 JME mutants show high levels of NOTCH1 signaling. (A) Western blot analysis of 293T cells expressing FLAG-tagged forms of wild-type (WT) and NOTCH1 mutant alleles. NOTCH1 HD L1600P is class 1 HD mutant; ΔPEST encodes a C-terminal truncation of NOTCH1 lacking a PEST domain; L1600P+ΔPEST is a double-mutant allele of NOTCH1 harboring both an HD mutation and a PEST domain truncation; and Jurkat JME 17 is the NOTCH1 exon 28 JME mutant allele identified in Jurkat cells. Total NOTCH1 levels were detected with anti-FLAG antibodies (top panel). ICN1 protein was detected with NOTCH1 Val1744 antibody, which specifically recognizes the γ-secretase cleaved activated form of NOTCH1. Luciferase is shown as transfection/loading control. (B) Transactivation activity of HD, ΔPEST, HD-ΔPEST, and Jurkat JME 17 NOTCH1 alleles in a CSL luciferase reporter assay. Reporter activity is shown as fold change compared with wild-type NOTCH1 (WT). (C) Transactivation activity of artificially generated NOTCH1 JME alleles. NOTCH1 constructs expressing juxtamembrane insertions of 5 to 14 amino acids corresponding to the VSV-G epitope as well as the Jurkat JME 17 allele were tested in a CSL luciferase reporter assay. Reporter activity is shown as fold change compared with wild-type NOTCH1 (WT). Error bars represent SD.
NOTCH1 JME mutants show high levels of NOTCH1 signaling. (A) Western blot analysis of 293T cells expressing FLAG-tagged forms of wild-type (WT) and NOTCH1 mutant alleles. NOTCH1 HD L1600P is class 1 HD mutant; ΔPEST encodes a C-terminal truncation of NOTCH1 lacking a PEST domain; L1600P+ΔPEST is a double-mutant allele of NOTCH1 harboring both an HD mutation and a PEST domain truncation; and Jurkat JME 17 is the NOTCH1 exon 28 JME mutant allele identified in Jurkat cells. Total NOTCH1 levels were detected with anti-FLAG antibodies (top panel). ICN1 protein was detected with NOTCH1 Val1744 antibody, which specifically recognizes the γ-secretase cleaved activated form of NOTCH1. Luciferase is shown as transfection/loading control. (B) Transactivation activity of HD, ΔPEST, HD-ΔPEST, and Jurkat JME 17 NOTCH1 alleles in a CSL luciferase reporter assay. Reporter activity is shown as fold change compared with wild-type NOTCH1 (WT). (C) Transactivation activity of artificially generated NOTCH1 JME alleles. NOTCH1 constructs expressing juxtamembrane insertions of 5 to 14 amino acids corresponding to the VSV-G epitope as well as the Jurkat JME 17 allele were tested in a CSL luciferase reporter assay. Reporter activity is shown as fold change compared with wild-type NOTCH1 (WT). Error bars represent SD.
Three outstanding features were in common in all 8 JME mutant alleles identified in our series. First, they all encoded NOTCH1 receptors with significantly extended extracellular juxtamembrane regions that would presumably leave intact, but displaced from the membrane, the ADAM protease S2 cleavage site and the complex formed by the 2 subunits of the HD domain and the LNR repeats. Second, they all clustered in a specific location around amino acid 1740 of NOTCH1. And finally, they all shared 4 common amino acids (QLHF) in each peptide inserted in the extracellular juxtamembrane region of the receptor.
Based on these observations, we hypothesized that NOTCH1 JME mutations could be related to rare NOTCH1 HD mutant alleles previously identified in the distal region of exon 27 in the P12-ICHIKAWA cell line and in primary T-ALL patient samples,24,25 which consisted of in-frame internal tandem duplications resulting in the displacement of the NOTCH1 S2 cleavage site away from the inhibitory LNR-HD complex. Thus, NOTCH1 JME mutations might result in aberrant proteolytic processing of the extracellular juxtamembrane region of NOTCH1 by creating an alternative S2-like site, possibly located within their common QLHF sequence. Alternatively, aberrant activation of NOTCH1 might result from conformational changes in the HD-LNR complex upon its displacement away from the membrane. To test these possibilities, we generated a series of artificial NOTCH1 JME mutants consisting of insertions of 5, 8, 11, 12, 13, and 14 amino acids derived from the VSV-G epitope in position 1740 of NOTCH1 and tested their ability to induce activation of a CSL-driven promoter in a luciferase reporter assay. These amino acid sequences lack any significant similarities with the NOTCH1 JME insertions and are devoid of any valine or alanine residues to avoid introducing an alternative S2-like cleavage site (“NOTCH1 expression plasmids”). These experiments demonstrated moderate (1.5- to 12-fold) gains in NOTCH1 signaling for JME mutants with insertions of 5, 8, or 11 amino acids. In contrast, the introduction of 12 to 14 amino acids resulted in marked increases in NOTCH1 signaling, ranging from 200- to 700-fold over baseline levels (Figure 2C). These results strongly suggest that the activation of NOTCH1 in JME mutants depends on the length of the spacing inserted between the HD-LNR repeat complex and the membrane, and is independent of the specific amino acid sequence introduced in the juxtamembrane region of the receptor by these mutations.
NOTCH1 JME mutations require γ-secretase and S2 processing for activation
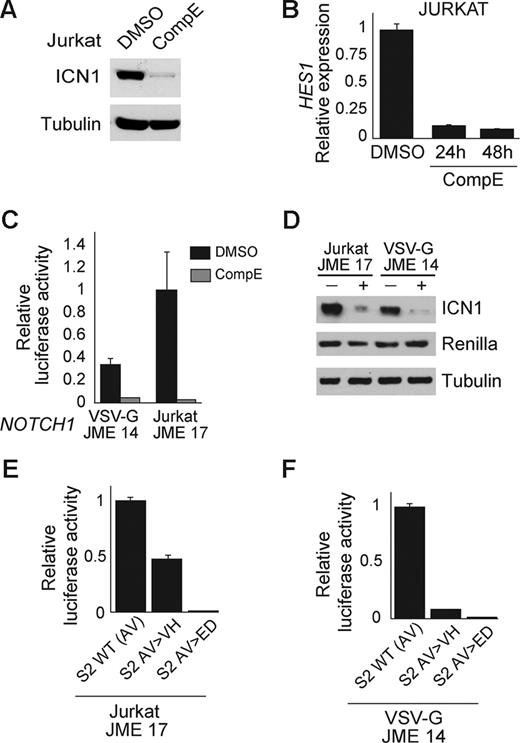
Activating mutations in the HD domain of NOTCH1 typically work by facilitating the S2 processing of the receptor and the subsequent S3 cleavage catalyzed by the γ-secretase complex. As a result, inhibition of S3 processing by small molecule inhibitors of the γ-secretase complex effectively abrogate the activity of these mutants. Treatment of Jurkat T-ALL cells with CompE, a highly active γ-secretase inhibitor, resulted in almost complete clearance of ICN1 at 24 hours as determined by Western blot analysis with the NOTCH1 Val 1744 antibody, and in marked down-regulation of NOTCH1 transcriptional activity evidenced by a down-regulation of HES1, a direct target of NOTCH1 (Figure 3A,B). To further analyze the requirement of the γ-secretase complex for the activity of NOTCH1 JME alleles, we performed luciferase reporter assays in the presence and absence of CompE. Incubation of HeLa cells transfected with the NOTCH1 Jurkat JME17 or with the artificial NOTCH1 JME VSV-G 14 mutant allele with CompE resulted in complete abrogation of NOTCH1 signaling (Figure 3C) and marked reduction of ICN1 expression (Figure 3D). Overall, these results demonstrate a strict requirement of the γ-secretase complex for the activity of NOTCH1 JME mutant alleles.
JME mutants are sensitive to inhibition of the γ-secretase complex and require S2 cleavage for activation. (A) Western blot analysis of activated NOTCH1 (ICN1) levels in Jurkat cells treated with CompE (100 nM) for 24 hours shows inhibition of NOTCH1 processing and clearance of activated intracellular NOTCH1. (B) Quantitative RT-PCR analysis of HES1 expression in Jurkat cells treated with CompE (100 nM) or vehicle only (DMSO). (C,D) CSL luciferase reporter assay (C) and Western blot analysis (D) of ICN1 activity and expression in cells expressing NOTCH1 JME mutants treated with vehicle (DMSO) or a γ-secretase inhibitor (CompE, 100 nM). (E,F) Luciferase reporter assay with a CSL reporter construct shows effective inhibition of NOTCH1 signaling from the NOTCH1 JURKAT JME 17 allele (E) and the NOTCH1 VSV-G JME 14 allele (F) upon mutation of the S2 cleavage site. Error bars represent SD.
JME mutants are sensitive to inhibition of the γ-secretase complex and require S2 cleavage for activation. (A) Western blot analysis of activated NOTCH1 (ICN1) levels in Jurkat cells treated with CompE (100 nM) for 24 hours shows inhibition of NOTCH1 processing and clearance of activated intracellular NOTCH1. (B) Quantitative RT-PCR analysis of HES1 expression in Jurkat cells treated with CompE (100 nM) or vehicle only (DMSO). (C,D) CSL luciferase reporter assay (C) and Western blot analysis (D) of ICN1 activity and expression in cells expressing NOTCH1 JME mutants treated with vehicle (DMSO) or a γ-secretase inhibitor (CompE, 100 nM). (E,F) Luciferase reporter assay with a CSL reporter construct shows effective inhibition of NOTCH1 signaling from the NOTCH1 JURKAT JME 17 allele (E) and the NOTCH1 VSV-G JME 14 allele (F) upon mutation of the S2 cleavage site. Error bars represent SD.
However, perhaps an even more intriguing question is whether S2 cleavage is required for the activity of these mutants. To test whether NOTCH1 JME mutants are dependent on a proteolytic processing at the prototypical S2 cleavage site, we generated 2 different targeted mutations at this position (AV 1720-1721 VH and AV 1720-1721 ED) in the context of the NOTCH1 Jurkat JME17 and VSV-G 14 alleles (Figure 3E,F). These mutations have been reported to partially (AV>VH) and completely (AV>ED) abrogate S2 cleavage and activation of NOTCH1 signaling.29 Functional analysis of NOTCH1 Jurkat JME17 and VSV-G 14 alleles with S2 cleavage site mutations showed abrogation of NOTCH1 activity in luciferase reporter assays. These results demonstrate that activation of NOTCH1 by JME mutations requires cleavage at the canonical S2 site and does not support that these mutations generate alternative S2-like cleavage motifs in the juxtamembrane extracellular region of the receptor.
NOTCH1 JME mutations do not affect the stability of the HD-LNR repeat complex
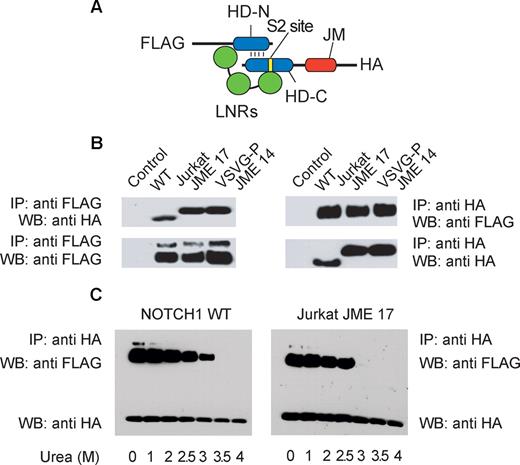
The strict requirement of a prototypical S2 cleavage site for the activation of NOTCH1 JME mutant alleles suggested that these insertions may work similarly to class 1 NOTCH1 HD mutations,26 by severely destabilizing the HD-LNR repeat complex, which normally protects the S2 site from the activity of ADAM proteases. To test this hypothesis, we generated a construct encoding a secreted LNR-HD juxtamembrane segment of NOTCH1 (amino acids 1446 to 1743) tagged with a FLAG epitope in the N-terminus and an HA epitope in the C-terminus (Figure 4A). Transfection of this construct in mammalian cells generates a soluble HD-LNR heterodimer that can be immunoprecipitated from the conditioned media with a FLAG antibody and detected by Western blot using the anti-HA tag antibody.25,26 In addition to a wild-type LNR-HD construct, we generated versions of this NOTCH1 minigene containing the NOTCH1 Jurkat JME17 and NOTCH1 VSV-G JME14 alleles. In agreement with previous reports,26 immunoprecipitation of the wild-type protein using the FLAG antibody recovered a 25 kDa N-terminus fragment containing the FLAG epitope and a 14 kDa C-terminus fragment with the HA tag (Figure 4B). This result demonstrates that the wild-type LNR-HD protein was correctly processed by furin proteases and secreted into the media as a stable LNR-HD heterodimer. Notably, FLAG immunoprecipitation of Jurkat and VSV-G JME LNR-HD also recovered both the N-terminus and the C-terminus peptides consistent with the production and secretion of a stable soluble LNR-HD repeat complex in the context of activating JME insertions (Figure 4B). Treatment of NOTCH1 LNR-HD complexes encoded by these NOTCH1 minigenes with increased concentrations of urea showed only a slight increase in urea sensitivity for the dissociation of the LNR-HD repeat complex in Jurkat JME17 mutant compared with wild-type NOTCH1 (Figure 4C). Overall, these results demonstrate that highly active JME mutants do not affect significantly the stability of soluble LNR-HD complexes. This is in contrast with results previously reported for class 1 NOTCH1 HD mutations,26 which typically induce receptor activation by destabilizing the interaction between the 2 HD subunits making the S2 site accessible to metalloprotease cleavage.
JME mutants do not affect the stability of the LNR-HD complex. (A) Schematic representation of the soluble LNR-HD complex encoded by the NOTCH1 minigene. In addition to the LNR repeats (green), the N-terminal (HD-N) and C-terminal (HD-C) subunits of the HD domain (blue), and the extracellular juxtamembrane region (JM), this construct contains an N-terminal FLAG tag and a C-terminal HA tag. (B) Immunoprecipitation immunoblot analysis of NOTCH1 minigene protein products recovered from conditioned media of 293T cells transfected with empty plasmid (control), a NOTCH1 wild-type minigene (WT), a NOTCH1 minigene containing the Jurkat JME 17 amino acid insertion (Jurkat JME 17), and a NOTCH1 minigene containing the artificial VSV-G-P JME 14 amino acid insertion. (C) Immunoprecipitation immunoblot analysis of NOTCH1 minigene protein products from a NOTCH1 wild-type minigene (WT) and NOTCH1 minigene containing the Jurkat JME 17 amino acid insertion (Jurkat JME 17) incubated with increased concentrations of urea to test the stability of the LNR-HD domain complex.
JME mutants do not affect the stability of the LNR-HD complex. (A) Schematic representation of the soluble LNR-HD complex encoded by the NOTCH1 minigene. In addition to the LNR repeats (green), the N-terminal (HD-N) and C-terminal (HD-C) subunits of the HD domain (blue), and the extracellular juxtamembrane region (JM), this construct contains an N-terminal FLAG tag and a C-terminal HA tag. (B) Immunoprecipitation immunoblot analysis of NOTCH1 minigene protein products recovered from conditioned media of 293T cells transfected with empty plasmid (control), a NOTCH1 wild-type minigene (WT), a NOTCH1 minigene containing the Jurkat JME 17 amino acid insertion (Jurkat JME 17), and a NOTCH1 minigene containing the artificial VSV-G-P JME 14 amino acid insertion. (C) Immunoprecipitation immunoblot analysis of NOTCH1 minigene protein products from a NOTCH1 wild-type minigene (WT) and NOTCH1 minigene containing the Jurkat JME 17 amino acid insertion (Jurkat JME 17) incubated with increased concentrations of urea to test the stability of the LNR-HD domain complex.
Discussion
The extracellular HD and LNR repeat domains of NOTCH proteins play a critical role in the stabilization of the receptor in a resting state and are the target of activating mutations that result in aberrant NOTCH1 signaling in T-ALL. The resolution of the crystal structure of the NOTCH2 LNR-HD complex has provided mechanisms that mediate the inhibitory effects of these extracellular domains on the processing and activation of NOTCH receptors.29 These studies have shown that the N-terminal and C-terminal subunits of the HD domain are closely associated, an interaction that is further stabilized by the LNR repeats, which fold over the HD domain and hold the 2 HD subunits together.30 In addition, the third LNR domain buries the S2 cleavage site located in the C-terminal HD subunit protecting it from cleavage by metalloproteases.30 According to this model, physiologic NOTCH1 signaling is initiated by the binding of a Delta-like or Jagged ligand molecule to the EGF repeats in the extracellular subunit of the receptor. This triggers a conformational change in the LNR-HD complex that exposes the S2 site to proteolytic cleavage by ADAM metalloproteases.30 Following S2 cleavage, the short extracellular juxtamembrane stub of the transmembrane-intracellular subunit binds to nicastrin, a membrane protein that works as a receptor for the γ-secretase complex, which ultimately cleaves NOTCH1 at the S3 site located in the transmembrane domain receptor.31 This final proteolytic cleavage activates the receptor by re-leasing the intracellular domains of NOTCH1 from the membrane, which then translocate to the nucleus and activate the expression of target genes.
Mutations in the heterodimerization domain of NOTCH1 induce ligand-independent activation of the receptor. Aberrant NOTCH1 activation by most HD mutations (class 1) occurs because of reduced heterodimer stability, resulting in either overt heterodimer dissociation (class 1A mutations) or, in some cases, more subtle weakening of the LNR-HD complex inducing a slow heterodimer dissociation and/or an increase in S2 site exposure (class 1B mutations; Figure 5). In rare cases, HD mutations do not affect the stability of the LNR-HD complex but introduce extra amino acids between the distal part of the HD domain and the S2 cleavage site, displacing it away from the protective effects of the LNR-HD complex (class 2 mutations; Figure 5). Common features of both class 1 and class 2 HD mutations are (1) their strict dependence on the integrity of the S2 cleavage site for activation, (2) having ADAM10 as the main metalloprotease involved in S2 processing, and (3) the requirement of γ-secretase activity for NOTCH1 activation. However, they differ in their relative activity, with class 2 mutants being significantly more active than class 1 NOTCH1 alleles.
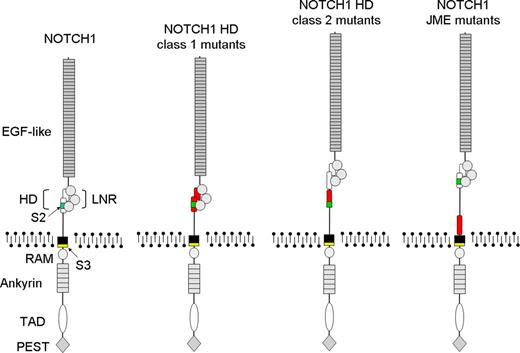
Schematic representation of NOTCH1 mutations leading to increased γ-secretase processing and activation in T-ALL. Functional domains of NOTCH1 are annotated in the structure of wild-type NOTCH1. EGF-like indicates EGF-like repeats; HD, heterodimerization domain; LNR, LNR repeats; RAM, RAM domain; ankyrin, ankyrin repeats; TAD, transactivation domain; PEST, PEST domain; S2, metalloprotease cleavage site; and S3, γ-secretase cleavage site. Sequences altered by the different NOTCH1 mutations are highlighted in red. HD class 1 mutants disrupt the HD domain structure. HD class 2 mutants displace the S2 cleavage site away from the HD-LNR complex. NOTCH1 JME mutants displace the HD-LNR repeat complex away from the cell surface.
Schematic representation of NOTCH1 mutations leading to increased γ-secretase processing and activation in T-ALL. Functional domains of NOTCH1 are annotated in the structure of wild-type NOTCH1. EGF-like indicates EGF-like repeats; HD, heterodimerization domain; LNR, LNR repeats; RAM, RAM domain; ankyrin, ankyrin repeats; TAD, transactivation domain; PEST, PEST domain; S2, metalloprotease cleavage site; and S3, γ-secretase cleavage site. Sequences altered by the different NOTCH1 mutations are highlighted in red. HD class 1 mutants disrupt the HD domain structure. HD class 2 mutants displace the S2 cleavage site away from the HD-LNR complex. NOTCH1 JME mutants displace the HD-LNR repeat complex away from the cell surface.
Here, we describe a new family of activating mutations within the extracellular juxtamembrane region of NOTCH1 in T-ALL (Figure 5). These juxtamembrane expansion mutants or JME NOTCH1 mutant alleles are generated by an internal tandem duplications in the 3′ end of intron 27 and/or in the proximal region of exon 28 and result in the insertion of relatively long peptides around position 1740 of NOTCH1. Interestingly, each of the 8 JME insertions identified in this series introduces a common tetrapeptide sequence (QLHF) proximal to the transmembrane domain of the NOTCH1 receptor. However, activation of NOTCH1 by JME mutants is not dependent on the sequence of this common motif, but on the number of amino acid residues introduced proximal to the membrane domain of the NOTCH1 receptor. Thus, artificially generated JME insertions of 5, 8, and 11 amino acids failed to induce significant activation of the NOTCH1 receptor, while the insertion of 14 amino acids induced marked increase in NOTCH1 signaling. Importantly, the amino acid sequences introduced in the extracellular juxtamembrane region of NOTCH1 by these constitutively active artificial JME alleles were unrelated to the insertions found in leukemia-derived JME mutants.
Based on these results, we proposed that JME mutations could result in aberrant processing of NOTCH1 at the typical S2 metalloprotease cleavage site, or alternatively, in proteolytic cleavage of the receptor at an alternative S2-like cleavage site located in the inserted peptide sequences encoded by these NOTCH1 alleles, followed by γ-secretase cleavage. Our finding that the activity of JME mutants seems to be dependent on the length of the inserted sequences and not on the specific amino acid sequence of the insertion suggested that these mutants probably work by inducing a conformational change in the LNR-HD complex that facilitates constitutive cleavage at the classic S2 site. Consistent with this hypothesis, activation of NOTCH1 signaling is strictly dependent on the integrity of the standard S2 cleavage site and is also effectively blocked by small molecule inhibitors of the γ-secretase complex.
NOTCH1 JME alleles and class 2 HD mutants have several features in common: (1) both are generated by relatively long insertions resulting from internal tandem duplications in the NOTCH1 gene; (2) they result in high levels of NOTCH1 activity compared with class 1 HD alleles; and (3) their activity is dependent on the integrity of the S2 cleavage site and the function of the γ-secretase complex. In the case of class 2 HD NOTCH1 mutants, aberrant S2 cleavage is induced by the insertion of extra amino acids immediately proximal to the classic S2 cleavage site, which is displaced closer to the membrane and out of reach from the structure of the LNR-HD complex (Figure 5). In contrast, insertions in the JME mutants are distal to the S2 cleavage site and displace the LNR-HD complex and the S2 site away from the membrane without altering the primary structure of any of these elements (Figure 5). These results suggest that moving the LNR-HD complex away from the cell surface facilitates aberrant metalloprotease S2 cleavage of the NOTCH1 receptor. Importantly, analysis of the amino acid sequence of the extracellular juxtamembrane region of different NOTCH receptors shows little sequence homology, but a more strict conservation on the length of the segment that separates the LNR-HD complex from the membrane. Thus, the S2 cleavage site is located 11 amino acids distant from the membrane in NOTCH2 and NOTCH3, while in NOTCH1 and NOTCH4 the extracellular juxtamembrane region is 12 amino acids long.
The specific structural basis for the strict requirement to maintain the LNR-HD complex in close proximity to the membrane to avoid spontaneous metalloprotease cleavage of the NOTCH1 receptor remains to be elucidated. It is possible that the LNR-HD complex interacts with membrane proteins that contribute to hold this structure together and that this interaction is disrupted upon displacement of the LNR-HD complex away from the membrane. Alternatively, it is possible that the proximity to the membrane restrains the ability of proteases to access the S2 cleavage site located within the HD-LNR complex and that displacement from the cell surface makes this sequence more accessible for protease cleavage.
Overall, these results identify a novel class of mutations involved in the aberrant activation of NOTCH1 signaling in T-ALL and provide further insight into the mechanisms that control the activation of the NOTCH1 receptor.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Jon Aster, Steve Blacklow, and Raphael Kopan for helpful comments and reagents.
This work was supported by the WOLF Foundation (Warren, NJ), the Swim Across America Foundation (Boston, MA), the Cancer Research Institute (New York, NY), the Leukemia & Lymphoma Society (White Plains, NY; grants 1287-08 and 6237-08), and National Institutes of Health (NIH, Bethesda, MD) grant CA120196 (A.A.F.). A.A.F. is a Leukemia & Lymphoma Society Scholar.
National Institutes of Health
Authorship
Contribution: M.L.S. performed research, analyzed data, and wrote the paper; O.W., T.P., V.T., S.P., P.J.R., and K.B. performed research; L.Z. and J.P.M. provided clinical samples and performed research; A.A.F. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adolfo A. Ferrando, Institute for Cancer Genetics-Columbia University, 1130 St Nicholas Avenue, Rm 505A, New York, NY 10032; e-mail: af2196@columbia.edu.