Abstract
Hematopoietic stem cells (HSCs) show pronounced heterogeneity in self-renewal and differentiation behavior, which is reflected in their repopulation kinetics. Here, a single-cell–based mathematical model of HSC organization is used to examine the basis of HSC heterogeneity. Our modeling results, which are based on the analysis of limiting dilution competitive repopulation experiments in mice, demonstrate that small quantitative but clonally fixed differences of cellular properties are necessary and sufficient to account for the observed functional heterogeneity. The model predicts, and experimental data validate, that competitive pressures will amplify small clonal differences into large changes in the number of differentiated progeny. We further predict that the repertoire of HSC clones will evolve over time. Last, our results suggest that larger differences in cellular properties have to be assumed to account for genetically determined differences in HSC behavior as observed in different inbred mice strains. The model provides comprehensive systemic and quantitative insights into the clonal heterogeneity among HSCs with potential applications in predicting the behavior of malignant and/or genetically modified cells.
Introduction
Hematopoietic stem cells (HSCs) are characterized functionally by their ability to generate mature cells of all hematopoietic lineages. Differentiation capacity, together with self-renewal, is critical for the engraftment and long-term persistence of HSCs following transplantation (eg, in the treatment of hematologic diseases), as supportive therapy following marrow ablation, or as vehicle for gene therapies (for reviews see, for example, Devine and DeMeyer1 and Klein and Baum2 ). In all these settings, it is essential to control and possibly predict clonal repopulation dynamics. However, little is known about the regulatory mechanisms underlying the engraftment of HSC clones and their competitive growth potential
Many studies have demonstrated that HSCs are heterogeneous with respect to a number of properties, such as differentiation and engraftment potential, self-renewal capacity, and surface marker phenotype.3-10 Part of this heterogeneity derives from the response of HSCs to extrinsic stimuli such as changing microenvironmental and signaling conditions.4,11-13 However, there is also evidence that the HSC compartment consists of functionally distinct subsets of HSCs, each with their own qualitatively predictable repopulation behavior.7,14
The strongest support for a predetermined behavior of HSC subsets comes from serial, long-term transplantation experiments. Muller-Sieburg et al15-18 and later others10 showed that although HSCs differ considerably in their engraftment behavior, the repopulation and differentiation patterns within the progeny of individual HSCs are remarkably similar. This suggests that HSCs have distinct, cell-intrinsically determined self-renewal and differentiation capacities. Additional support for predetermined properties of HSCs comes from in vivo data and modeling studies showing that genetic factors determine many aspects of the behavior of HSCs.15,19-22
It is the objective of this paper to provide insight into the following questions: (1) can one explain the heterogeneity of engraftment kinetics without the assumption of clonally fixed differences in HSC properties; (2) if clone-specific heterogeneity in cellular properties is necessary, how much do these properties vary in the population of HSCs; and (3) is the concept of clonally fixed HSC characteristics consistent with the observation of phenotypic and functional flexibility and reversibility of HSCs?
To address these questions, we apply a mathematical model of HSC organization.23,24 This model has previously been validated in different murine systems22,25,26 and was valuable for the understanding of disease dynamics and treatment effects in human chronic myeloid leukemia.27,28 The model features a stochastic single-cell–based approach to analyze stem-cell dynamics on the population as well as on the single-cell level. This makes it ideally suited for investigating the dynamic behavior of individual HSC clones.
Methods
Experimental procedure
The experimental procedure for clonal repopulation has already been described elsewhere.15,18 Briefly, individual HSCs were obtained either after in vitro limiting dilution on the stromal cell line S17 or after injecting limiting numbers of freshly explanted bone marrow (BM) cells. All animals were of the C57BL/6 (B6) background and donor type cells were either congenic for the allelic forms of CD45 or they expressed green fluorescent protein (GFP) as a transgene. Hosts were either sublethally irradiated W41/W41 mice,29 lethally irradiated B6 mice, or B6 mice in which the CD45 gene had been ablated by homozygous recombination.30 All mice were bled every other month, and the percentage of donor type cells of the lymphoid and myeloid lineages were assessed by flow cytometry. Secondary transplantations were done at least 5 months after transplanting the primary HSCs. For serial transplantations, 5 × 106 unseparated BM cells from the primary host were injected into multiple (“paired”) secondary, lethally irradiated hosts. For competitive secondary repopulation experiments, 5 × 106 cells each from 2 different clonally repopulated primary hosts were injected either individually or in a 1:1 mixture (107 cells total). The 2 donor clones differed in GFP and CD45 expression. All experiments were approved by the Institutional Animal Care and Use Committee of the Sidney Kimmel Cancer Center, San Diego.
Model description
All simulation results are based on a single-cell–based model of HSC organization as previously described.23,27 To summarize the model assumptions: stem cells are able to reversibly switch between 2 signaling contexts (A, Ω) that induce different functional states. Whereas cells in A are quiescent (ie, in G0), cells in Ω are proliferating (with cell-cycle duration τc). The affinity of individual cells to reside in A is characterized by the variable a. Whereas cells in Ω gradually decrease a (controlled by model parameter d), cells in A regenerate a up to a maximum value amax (controlled by model parameter r). Cells with a < amin are assumed to have lost their affinity to change into context A and are considered as differentiated cells that are finally removed from the system after a fixed differentiation/maturation phase (λ). Transition of HSCs between A and Ω is characterized by sigmoid functions fα and fω, which describe the probability of transition, dependent on the number and on the state of HSCs. This basic model has been extended to accommodate the analysis of clonal heterogeneity. Earlier versions assumed equal properties for all cells of one genetic background regarding proliferative activity, differentiation, and regeneration potential with identical model parameters τc, d, and r. The same is true for the transition characteristics fα and fω. We now allow for heterogeneity of these parameters among individual HSC clones (ie, on initialization each cell is assigned a particular set of parameter values randomly chosen from a prespecified range). These values are inherited from mother to daughter cells, thus representing a clonally fixed characteristic. All model parameters have been chosen on the basis of the previously derived reference values for B6 mouse systems22 and are given in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Figure S1 provides a graphic representation of the model structure as well as of the transition functions fα and fω.
Simulation procedure
Primary recipients
Model assumptions.
Representing a limiting dilution setting, we assume that only one HSC is contained in any donor graft. Host systems (W41/W41 mice) are assumed to contain only few competitive HSCs. In particular, we used 4 model HSCs in all host systems. The results do not qualitatively depend on the exact number of host HSCs as long as it is small enough (< 8; data not shown).
Implementation.
Empty model systems are initiated with one donor HSC and 4 competing host HSCs. The initial affinity a of these cells is randomly chosen from a uniform distribution over the interval [0.1; 1.0]. To distinguish donor and host cells, they are labeled as cell type D and H, respectively. Engraftment levels are determined in silico by tracking the proportion of differentiated type D cells over a period of 7 months.
Secondary recipients
Model assumptions.
Because residual primary host W41/W41 HSCs do not engraft in secondary recipients,16 we assume that the chimerism in secondary hosts is initiated by clonally derived primary donor (D) HSCs and by residual B6 host (H) HSCs. For the competitive repopulation experiments, we assume the cotransplantation of 2 donor clones (from different primary hosts: D1, D2) competing with residual B6 host (H) HSCs. Primary donor grafts are assumed to consist of 50 × p type D HSCs, with p denoting the donor engraftment level in the primary hosts at time of transplantation. This is motivated by an estimated number of one HSC in 105 freshly explanted, unmanipulated bone marrow (BM) cells31,32 and the transplantation of 5 × 106 BM cells from a clonally repopulated primary host.
Implementation.
Empty model systems are “cotransplanted” with 50 × p type D HSCs and 4 type H HSCs. Affinity a of donor cells is sampled from the distribution of type D cells in the primary host at the time of transplantation. For type H, it is chosen according to a uniform distribution from the interval [0.1; 1.0]. The particular choice of the a values does not qualitatively change the results (data not shown). Competitive secondary repopulation experiments have been modeled by coinitiating empty systems with 50 × p1 D1 and 50 × p2 D2 cells (p1, p2 denoting the proportions of donor type cells in the 2 primary hosts) together with 4 H stem cells. The relative engraftment levels of the 2 donor types are determined in silico by tracking the relative proportion of types D1 and D2 with respect to the total number of differentiated cells (ie, D1 + D2 + H) over a period of 7 months.
Statistical analysis
To quantify the temporal trend of engraftment kinetics, we consider the change of the engraftment levels within predetermined intervals. For a qualitative characterization, the trend is denoted as positive or negative if the engraftment level at the end of the interval is 10 percentage points larger or smaller than at the beginning of this interval, respectively. Otherwise, the engraftment is considered unchanged and the trend is denoted by the symbol π. To statistically test differences in the heterogeneity of engraftment levels and trends, we used the Bartlett test of homogeneity of variances. All statistical calculations and graphic representations have been preformed using the statistical programming environment R.33
Parameter selection and fitting algorithm
To determine a parameter configuration that generates an optimal fit of the experimental and the simulated distribution of engraftment kinetics, we applied a 2-step strategy. First, we identified model parameters that sensitively affect the qualitative pattern of engraftment kinetics in the clonal repopulation setting using a coarse-grained parameter variation. We systematically varied each of 8 critical model parameters (d, r, τc, amax, fα(Ñ/2), fα(Ñ), fω(Ñ'/2), and fω(Ñ')). We considered about 300 parameter configurations to determine the qualitative effect of each parameter on the engraftment dynamics. It turned out that the parameters d and fα(Ñ) most sensitively affect the engraftment dynamics. In the second step, we simulated primary host engraftment kinetics for a fine-grained variation of d (additive step size of 0.0005 with d ∈ [1.03; 1.13]) and fα(Ñ) (multiplicative step size of 1/0.975 with fα(Ñ) ∈ [0.0001; 0.015]). Within this 2D parameter range (consisting of 55 074 grid points), we chose rectangular regions of different size and determined the frequency distribution of qualitatively different engraftment kinetics according to 9 different classes. To compare those to the experimentally observed frequency distribution, we calculated χ2 statistics
with ei denoting the experimentally observed proportion and si denoting the simulated proportion of engraftment kinetics in class i. Whereas an optimal concordance of experiment and simulation would result in χ2 = 0, high χ2 values indicate a worse fit.
Results
Explaining engraftment heterogeneity
As described previously,15,18 transplants of primary HSCs on the clonal level show extensive heterogeneity in their engraftment kinetics (Figure 1A). However, daughter HSCs derived from a single HSC showed remarkable similarities in their repopulation kinetics when retransplanted (Figure 1B). This suggests the existence of distinct, presumably epigenetically fixed, functional characteristics of HSC clones. However, it is unclear how much the properties of these clones must differ to account for the results. To quantify such differences, we perform extensive simulation studies based on this described mathematical model.
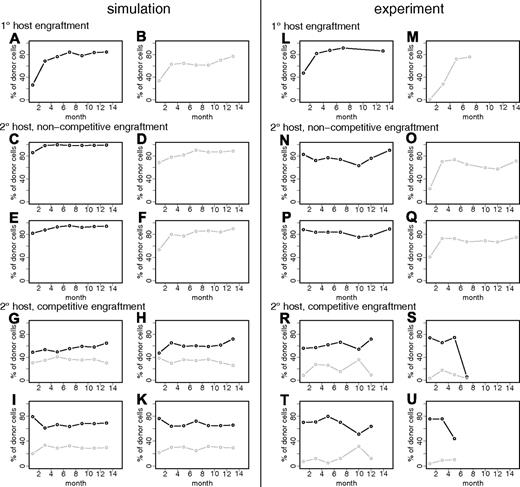
Experimentally observed and simulated engraftment kinetics in primary and secondary recipients. (A) The graphic illustrates experimentally observed engraftment kinetics, as determined in the peripheral blood (PB), generated by different, clonally derived transplants in primary hosts (different colors indicate different clones). At month 7, HSCs from these primary hosts were transplanted into secondary hosts. (B) Experimentally observed engraftment kinetics in the PB of secondary hosts. Corresponding colors were used to indicate the relationship of primary HSCs and its clonal progeny revealed in multiple secondary hosts. Data in panels A and B are taken from Muller-Sieburg et al.16 (C,D) Representative examples of simulated engraftment kinetics in primary (C) and secondary (D) host assuming no predetermined differences of individual clones. Identical color codes indicate identical clonal origin in primary and secondary recipients. Gray lines give more examples of primary kinetics to better illustrate the distribution. They are not shown in secondary hosts. (E,F) See panels C and D, but this time assuming small differences between individual HSCs with respect to parameter d. d has been randomly sampled from a normal distribution with mean 1.07 (reference value) and standard deviation 0.01. The particular d values for the given example realizations are 1.06052 (black), 1.06501 (red), 1.06624 (yellow), 1.06708 (green), and 1.07337 (blue). (G,H) See panels C and D but with large differences in parameter d. d has been randomly sampled from the 2 intervals [1.038, 1.049] and [1.091, 1.102], which relate to deviations of plus or minus 2% to 3% from the reference value 1.07. The particular d values for the given example realizations are 1.04705 (black), 1.04392 (red), 1.09377 (green), 1.02802 (blue), and 1.04761 (light blue). To facilitate the comparison with the experimental data, simulated engraftment levels are shown on a bimonthly scale, although they were actually simulated using a 1-hourly scale.
Experimentally observed and simulated engraftment kinetics in primary and secondary recipients. (A) The graphic illustrates experimentally observed engraftment kinetics, as determined in the peripheral blood (PB), generated by different, clonally derived transplants in primary hosts (different colors indicate different clones). At month 7, HSCs from these primary hosts were transplanted into secondary hosts. (B) Experimentally observed engraftment kinetics in the PB of secondary hosts. Corresponding colors were used to indicate the relationship of primary HSCs and its clonal progeny revealed in multiple secondary hosts. Data in panels A and B are taken from Muller-Sieburg et al.16 (C,D) Representative examples of simulated engraftment kinetics in primary (C) and secondary (D) host assuming no predetermined differences of individual clones. Identical color codes indicate identical clonal origin in primary and secondary recipients. Gray lines give more examples of primary kinetics to better illustrate the distribution. They are not shown in secondary hosts. (E,F) See panels C and D, but this time assuming small differences between individual HSCs with respect to parameter d. d has been randomly sampled from a normal distribution with mean 1.07 (reference value) and standard deviation 0.01. The particular d values for the given example realizations are 1.06052 (black), 1.06501 (red), 1.06624 (yellow), 1.06708 (green), and 1.07337 (blue). (G,H) See panels C and D but with large differences in parameter d. d has been randomly sampled from the 2 intervals [1.038, 1.049] and [1.091, 1.102], which relate to deviations of plus or minus 2% to 3% from the reference value 1.07. The particular d values for the given example realizations are 1.04705 (black), 1.04392 (red), 1.09377 (green), 1.02802 (blue), and 1.04761 (light blue). To facilitate the comparison with the experimental data, simulated engraftment levels are shown on a bimonthly scale, although they were actually simulated using a 1-hourly scale.
First, we simulated primary and secondary transplantations without assuming any clone-specific differences (ie, applying identical model parameters for all HSCs). The parameter values are based on previously derived reference values for B6 mice (Table S1). We found that the model is able to explain a considerable degree of clonal variability in the engraftment levels in primary hosts (Figure 1C). The heterogeneity is induced by stochastic fluctuations in the clonal contribution shortly after transplantation when only very few HSCs compete against each other. However, the experimentally observed engraftment pattern is only incompletely reproduced. As illustrated in Figure 2A, the variability of engraftment levels at 7 months of the simulated primary host kinetics is significantly smaller than that for the experimentally observed kinetics (P < .001). Similarly, the long-term engraftment trends (ie, the difference in engraftment levels between months 3 and 7 after transplantation) vary significantly less (P < .001) in the simulations of primary host repopulation compared with the experimental data (Figure 2B). Moreover, even though the observed similarity in the “paired” secondary engraftment kinetics could be reproduced without assuming clonally fixed differences (Figure 1D), the variability of long-term trends is again significantly reduced (P < .001) in the simulations (Figure 2C). Specifically, changes in the engraftment levels of more than 20 percentage points after month 3, as occasionally observed in the experiments (compare with Figure 1B), are not generated in the simulations assuming no clone specific differences (Figure 1D). It should be noted that the experimentally observed heterogeneity with respect to engraftment levels and trends can also be explained by random alterations of the model parameters (eg, generated during each cell division). However, this is not the case for the similarity of “paired” secondary engraftment kinetics (data not shown). Collectively, these results show that simulations without assuming clone specific differences recreates some but not all aspects of the experimental data.
Experimentally and simulated distributions of engraftment levels and trends. (A) Distributions of engraftment levels at 7 months after transplantation in primary hosts. (B,C) Distributions of engraftment trends (ie, differences in engraftment levels between month 3 and month 7 after transplantation) in primary (B) and secondary (C) hosts. According to the number of experimentally determined engraftment kinetics, we simulated the repopulation of n = 87 primary host and 8 pairs (n = 16) of secondary hosts. Boxplots show the median (black line), the interquartile range (box), and the total range (whiskers, or circles in case of “outliers,” ie, data points that deviate from the median more than 1.5 times the box range). P values are given for the Bartlett test of equality of variances.
Experimentally and simulated distributions of engraftment levels and trends. (A) Distributions of engraftment levels at 7 months after transplantation in primary hosts. (B,C) Distributions of engraftment trends (ie, differences in engraftment levels between month 3 and month 7 after transplantation) in primary (B) and secondary (C) hosts. According to the number of experimentally determined engraftment kinetics, we simulated the repopulation of n = 87 primary host and 8 pairs (n = 16) of secondary hosts. Boxplots show the median (black line), the interquartile range (box), and the total range (whiskers, or circles in case of “outliers,” ie, data points that deviate from the median more than 1.5 times the box range). P values are given for the Bartlett test of equality of variances.
Next, we introduced small differences in the model parameters of individual HSCs. These parameters are preserved during cell division so that daughter HSCs within a clone receive and maintain the same parameter set. Initially, only the differentiation coefficient d, was varied. Small, clonally preserved differences in d (less than 1% of d) permit the simulations to more closely match the experimentally observed heterogeneity (Figures 1E,F, 2). Larger differences (already 2% to 3% of d) will, in most cases, induce abrupt changes in the engraftment levels with quickly arising dominance or extinction of donor contribution (Figures 1G,H, 2). This, however, has only been observed infrequently in the experiments.
Estimating the degree of clonal heterogeneity
A total of 54 qualitatively and quantitatively different classes of engraftment kinetics of HSC have been considered previously to describe clonal heterogeneity.18 To accommodate the comparison of the experimental data with simulation results and to permit a statistical analysis, we chose a modified scheme which uses only 9 qualitatively distinct classes of engraftment kinetics. Specifically, we consider an early (months 1 to 3) and a late (months 3 to 7) regeneration phase. Within these 2 phases, the engraftment trend is characterized as increasing (+), decreasing (−), or unchanged (π). Figure 3A shows the distribution of the 87 previously published18 clonal repopulation kinetics according to the 9-way classification. Analogously to the results of the earlier report,18 this classification shows underrepresentation of some engraftment patterns. Particularly, no engraftment kinetics with an initial decrease followed by a long-term increase has been observed. Moreover, the majority of kinetics, namely 46 of 87, showed an initial increase.
Qualitative classification of individual engraftment kinetics. (A) Shown are 87 previously published18 repopulation kinetics of mice transplanted with clonally derived HSC grafts obtained by in vitro or in vivo limiting dilution. Shaded areas illustrate early (light) and late (dark) repopulation phases. Whereas +/− indicate an increase or decrease in the engraftment levels of more than 10% between the first and the last measurement in the given period, π refers to a constant (ie, change of less than 10%) time progression. (B) Shown are 87 representative simulated engraftment kinetics according to the given classification scheme. Underlying model parameters d and fα(Ñ) are randomly sampled from the best-fit region shown in Figure 4B.
Qualitative classification of individual engraftment kinetics. (A) Shown are 87 previously published18 repopulation kinetics of mice transplanted with clonally derived HSC grafts obtained by in vitro or in vivo limiting dilution. Shaded areas illustrate early (light) and late (dark) repopulation phases. Whereas +/− indicate an increase or decrease in the engraftment levels of more than 10% between the first and the last measurement in the given period, π refers to a constant (ie, change of less than 10%) time progression. (B) Shown are 87 representative simulated engraftment kinetics according to the given classification scheme. Underlying model parameters d and fα(Ñ) are randomly sampled from the best-fit region shown in Figure 4B.
To reconcile the biased engraftment patterns seen in vivo with our general model of stem cell organization, we systematically varied all model parameters within a coarse grid of different values to test which of them critically affects the variability and the trend of the engraftment kinetics (data not shown). The results showed that particularly the differentiation coefficient d and the transition characteristic fα affect the qualitative alteration of engraftment kinetics within the time scale considered. Whereas d characterizes the velocity of differentiation (ie, the loss of repopulating capability), fα determines the dynamic control of the transition probability of HSCs from the proliferation and differentiation promoting into the quiescence and regeneration promoting signaling context. Biologically, fα could be interpreted as the probability of an HSC to home to a stem cell–supporting niche environment.
Based on these results, we now focused on d and fα. We made the simplifying assumption that all other parameters are fixed and equal for all HSCs. Because the transition characteristic fα is determined by 5 parameters (Figure S1B), we additionally assumed that only one of these 5 parameters is changed. Specifically, we varied the value fα(Ñ), which induces alterations in the steepness of the sigmoid function fα. This leaves us with 2 degrees of freedom to explain clonal heterogeneity. Because the variation of each of these 2 parameters alone did not explain the observed repertoire of engraftment kinetics, large-scale simulations with a systematic variation of d and fα(Ñ), using more than 55 000 quantitatively different combinations, were performed. Classifying the simulated kinetics according to the 9 classes permits visualization of characteristic regions in this parameters space, each of these representing a preferential pattern of engraftment (Figure 4A). Engraftment was measured by a competitive in silico repopulation assay. Herein, the model systems were initiated with 2 small competing HSC clones, exactly as described in Simulation Procedure/Primary Recipients. For each of the d − fα(Ñ) combinations (with all other parameters fixed to B6 reference values; Table S1) an individual donor HSC was transplanted. The competing host clones always consist of 4 HSCs with identical B6 reference values for all parameters.
In silico engraftment kinetics depending on model parameters of donor HSCs. (A) Graphic representation of the qualitative differences of simulated engraftment kinetics depending on the choice of model parameters d and fα(Ñ). Each individual color dot illustrates the qualitative pattern of the engraftment kinetic observed in a simulation using the particular parameter combination. In total, 55 074 different parameter combinations are shown in the figure. Simulations were initiated with one donor cell with the particular d and fα(Ñ) values (according to the position in the diagram) and 4 competing host cells with reference parameters d = 1.07 and fa(Ñ) = 0.01. All other parameters had been fixed to B6 reference values, identically for donor and host cells. (B) Parameter region (depicted as white square in panel A; comprising 11 682 simulation runs) that yields the best fit of the simulated to the experimentally observed distribution of engraftment kinetics (compare with Figure S2). The 2 black lines indicate the reference values for d and fa(Ñ) used for the competing host cells. Color coding for engraftment kinetics (identical in panels A and B): maroon [+, +], red [+, π], pink [π, +], dark green [−, −], medium green [−, π], light green [π, −], purple [π,π], blue [+, −], yellow [−, +], and black (no donor engraftment).
In silico engraftment kinetics depending on model parameters of donor HSCs. (A) Graphic representation of the qualitative differences of simulated engraftment kinetics depending on the choice of model parameters d and fα(Ñ). Each individual color dot illustrates the qualitative pattern of the engraftment kinetic observed in a simulation using the particular parameter combination. In total, 55 074 different parameter combinations are shown in the figure. Simulations were initiated with one donor cell with the particular d and fα(Ñ) values (according to the position in the diagram) and 4 competing host cells with reference parameters d = 1.07 and fa(Ñ) = 0.01. All other parameters had been fixed to B6 reference values, identically for donor and host cells. (B) Parameter region (depicted as white square in panel A; comprising 11 682 simulation runs) that yields the best fit of the simulated to the experimentally observed distribution of engraftment kinetics (compare with Figure S2). The 2 black lines indicate the reference values for d and fa(Ñ) used for the competing host cells. Color coding for engraftment kinetics (identical in panels A and B): maroon [+, +], red [+, π], pink [π, +], dark green [−, −], medium green [−, π], light green [π, −], purple [π,π], blue [+, −], yellow [−, +], and black (no donor engraftment).
As illustrated in Figure 4A, the different classes of engraftment kinetics are restricted to bounded regions of the parameter space. Notably, whereas the engraftment kinetics [−, +], which has not been observed in the experiments, rarely occurs for fα(Ñ) values smaller than 0.01, the engraftment kinetic [+, −] almost exclusively appears for small fα(Ñ) values. Also regarding the d-dimension of the parameter space, marked differences can be observed. Whereas predominantly decreasing donor engraftments (ie, [−, π], [π, −], [−, −]) are restricted to high values of d, the opposite is true for predominantly increasing kinetics (ie, [+, π], [π, +], [+, +]). Thus, qualitatively different classes of engraftment kinetics can be consistently described by quantitatively different parameters values.
Subsequently, we determined the parameter region that is best compatible with the experimentally observed spectrum of engraftment kinetics in primary hosts (Figure 3A). To do this, we considered d and fα(Ñ) within variable intervals around their reference values. For each of these parameter regions, the compatibility of the observed and the simulated distributions of engraftment kinetics had been quantified by calculating a goodness-of-fit measure χ2 (for a graphic illustration, see Figure S2). Figure 4B gives a detailed picture of the parameter region that generated the best fit (χ2 = 8.4) to the experimental observations for the clonal repopulation of primary hosts.
As illustrated by Table 1 and substantiated graphically in Figure 3B and Figure S3A,C and D, the model predicts that the determined bounded parameter region (Figure 4B) fully accounts for the experimentally observed heterogeneity in the engraftment kinetics of primary hosts. Furthermore, the estimated parameter heterogeneity consistently accounts for the engraftment patterns, including trends and similarity of “paired” repopulations in the secondary host situation (Figure S3B,E).
Distribution of engraftment kinetics
| Early repopulation . | Late repopulation . | |||||
|---|---|---|---|---|---|---|
| Increasing (+) . | Constant (π) . | Decreasing (−) . | ||||
| Observed . | Simulated . | Observed . | Simulated . | Observed . | Simulated . | |
| Increasing (+) | 11.5 (5.7; 20.1) | 13.7 | 21.8 (13.7; 32.0) | 22.5 | 19.6 (11.8; 29.4) | 17.2 |
| Constant (π) | 4.6 (1.3; 11.4) | 2.9 | 24.1 (15.6; 34.5) | 17.7 | 6.9 (2.6; 14.4) | 7.3 |
| Decreasing (−) | 0.0 (0; 4.2) | 1.6 | 6.9 (2.6; 14.4) | 10.3 | 4.6 (1.3; 11.4) | 6.8 |
| Early repopulation . | Late repopulation . | |||||
|---|---|---|---|---|---|---|
| Increasing (+) . | Constant (π) . | Decreasing (−) . | ||||
| Observed . | Simulated . | Observed . | Simulated . | Observed . | Simulated . | |
| Increasing (+) | 11.5 (5.7; 20.1) | 13.7 | 21.8 (13.7; 32.0) | 22.5 | 19.6 (11.8; 29.4) | 17.2 |
| Constant (π) | 4.6 (1.3; 11.4) | 2.9 | 24.1 (15.6; 34.5) | 17.7 | 6.9 (2.6; 14.4) | 7.3 |
| Decreasing (−) | 0.0 (0; 4.2) | 1.6 | 6.9 (2.6; 14.4) | 10.3 | 4.6 (1.3; 11.4) | 6.8 |
Given are experimentally observed and simulated frequencies of engraftment kinetics within the 9 classes that are characterized by early (regenerating phase) and late (reconstituted phase) engraftment behavior: decreasing (−), increasing (+), and constant (π) donor contribution. Experimental results are given by class percentage out of 87 successfully engrafted animals (data taken from Sieburg et al18 ) complemented by the corresponding 95% confidence intervals. Simulation results represent class percentages estimated from 9820 individual runs (11 682 total simulations minus 1862 nonengrafting systems). Confidence intervals are negligibly small due to the high number of simulation replicates.
We tested whether this variability has to be completely attributed to clone-specific characteristics or whether it can also be explained by differences in the host systems. Therefore, we assumed only one parameter (either d or fα(Ñ)) to be clonally fixed, while the other parameter is chosen randomly from the estimated region for each host system. Our simulations demonstrate that a nonclonal but host-specific d is not compatible with the experimental observations because it significantly reduces the similarity of “paired” secondary repopulation kinetics (data not shown). Also, the assumption of a nonclonal fα(Ñ) leads to a reduction of this similarity (data not shown). This effect, however, is not as pronounced as for parameter d. To conclusively disentangle cellular and microenvironmental components in the regulatory control of HSCs, further studies are needed.
Predicting competitive differences for HSCs with qualitatively identical engraftment potential
The model leads to the testable prediction that HSC clones that exhibit similar engraftment kinetics in (noncompetitive) clonal repopulating settings can behave differently in situations where they directly compete against each other. Particularly, our model predicts that subtle, quantitative differences of clonal properties can be revealed in a competitive setting where 2 or more individual HSC clones contribute to hematopoiesis. We simulated 2 lethally irradiated primary hosts that underwent transplantation separately with single HSCs that are characterized by slightly different parameter values (d = 1.0655 vs 1.0665; fα(Ñ) = 0.0075 vs 0.008). Both HSC clones show qualitatively identical engraftment kinetics if transplanted into a lethally irradiated model host (Figure 5A,B; Figure S4A,B). In silico transplantation of cells from either of these primary hosts into secondary host systems leads to similar engraftment patterns (Figure 5C-F; Figure S4C,D). In contrast, when a mixture of both clones is transplanted into secondary hosts, the difference between the 2 clones is revealed, as one of them shows a systematic bias toward a higher engraftment level (Figure 5G-K; Figure S4E-Q).
Simulation results and experimental data on individual clone competition. (A,B) Simulated primary host engraftments of 2 clones with slightly differing parameters: (A) d = 1.0655, fα(Ñ) = 0.0075; (B) d = 1.0665, fα(Ñ) = 0.008. (C,E) 2 representative examples of secondary engraftments of the clone shown in panel A (ie, in silico transplantation of 50 ×p1 primary HSCs together with 4 residual secondary host HSCs). p is denoting the proportion of primary engraftment level, which is p1 = 0.85 in this particular example. (D,F) See panels C and E, but for the clone shown in panel B, p2 = 0.75. (G-K) A total of 4 representative examples of in silico cotransplantation of 50 × p1 cells of the clone shown in panel A plus 50 × p2 cells of the clone shown in panel B together with 4 residual secondary host cells. Further simulation examples of these settings can be found in Figure S4. (L,M) Experimentally observed engraftment kinetics (twice-monthly measurements) of 2 clonally repopulated mice. (N-Q) Engraftment kinetics for the transplantation of 5 × 106 BM cells of either one or the other clone (shown in panels L and M, respectively) in 2 lethally irradiated secondary B6 hosts. (R-U) Engraftment kinetics for cotransplantation of 2 × (5 × 106) BM cells from both primary hosts into 4 lethally irradiated secondary B6 hosts (Cho et al34 ). Color (black/gray) always illustrates relationship to primary host.
Simulation results and experimental data on individual clone competition. (A,B) Simulated primary host engraftments of 2 clones with slightly differing parameters: (A) d = 1.0655, fα(Ñ) = 0.0075; (B) d = 1.0665, fα(Ñ) = 0.008. (C,E) 2 representative examples of secondary engraftments of the clone shown in panel A (ie, in silico transplantation of 50 ×p1 primary HSCs together with 4 residual secondary host HSCs). p is denoting the proportion of primary engraftment level, which is p1 = 0.85 in this particular example. (D,F) See panels C and E, but for the clone shown in panel B, p2 = 0.75. (G-K) A total of 4 representative examples of in silico cotransplantation of 50 × p1 cells of the clone shown in panel A plus 50 × p2 cells of the clone shown in panel B together with 4 residual secondary host cells. Further simulation examples of these settings can be found in Figure S4. (L,M) Experimentally observed engraftment kinetics (twice-monthly measurements) of 2 clonally repopulated mice. (N-Q) Engraftment kinetics for the transplantation of 5 × 106 BM cells of either one or the other clone (shown in panels L and M, respectively) in 2 lethally irradiated secondary B6 hosts. (R-U) Engraftment kinetics for cotransplantation of 2 × (5 × 106) BM cells from both primary hosts into 4 lethally irradiated secondary B6 hosts (Cho et al34 ). Color (black/gray) always illustrates relationship to primary host.
We tested this prediction experimentally by cotransplanting BM from 2 clonally repopulated primary hosts into lethally irradiated secondary recipients.34 If transplanted alone, the selected HSC clones show similar repopulation kinetics, both in the primary and in the secondary hosts (Figure 5L-Q). Yet, a distinct and reproducible bias toward one clone could be detected when these 2 HSC clones directly competed against each other (Figure 5R-U). Although the number of biological replicates was small, this suggests that quantitative differences in the clonal properties of HSCs determine their relative competitive potential.
Predicting differences between epigenetically and genetically determined HSC properties
In contrast to these functionally relevant but small within-strain interclone differences, genetically determined between-strain differences are expected to be considerably larger. To analyze the effect of different genetic backgrounds, we simulated HSCs from the D2 background as described previously.22 We assume that D2 HSCs exhibit the same degree of (epigenetically determined) intraindividual clonal heterogeneity as do B6 mice. Technically, this is achieved by considering the same range of variability with respect to clonal parameters (Figure 4B) for both strains. However, this range is “centered” around the corresponding reference parameter values of the B6 and D2 strains, respectively (Table S1). Whereas the reference value of the differentiation coefficient d is assumed to be identical for D2 and B6 HSCs, the transition functions fα differ considerably between the strains (Figure S5).
Based on these assumptions, our model analysis predicts that the pattern of repopulation kinetics will change only slightly when D2 cells are transplanted into syngeneic hosts. In contrast, a significant change in a competitive D2 versus B6 repopulation is expected. Particularly, we predict an increase of fast-repopulating donor cells ([+, π] kinetics) and a decrease of continuously rising [+, +] kinetics, as well as an increase of initially rising but long-term declining donor contributions ([+, −] kinetics). These shifts are mainly explained by the growth advantage in the early repopulation phase (ie, few HSCs) and the growth disadvantage in fully repopulated systems (ie, many HSCs) of D2 versus B6 cells induced by the different fα characteristics. An additional prediction is the almost complete loss of initially decreasing (ie, [−, −], [−, π], [−, +]) kinetics in the competitive scenario, which is consistent with experiments using cotransplantations of (nonclonal) D2 and B6 HSCs.21,22,35 However, the validity of the predicted distribution of engraftments still has to be tested experimentally with clonal transplants. Figure S6 provides an overview of these model predictions.
Predicting the evolution of clonal composition
Due to its stochastic nature, our model predicts an evolution of the clonal composition of the HSC population over time. Although the number of HSCs in a homeostatic system is kept at a stable (mean) level, the number of clones (ie, the number of cells with different ancestor cells) will decrease over time. This is true even if all clones have identical properties. Based on a simulation study, we previously estimated the magnitude of this effect in a homeostatic reference system, predicting a reduction to one-tenth of the initial clone numbers within the lifespan of a B6 mouse.26
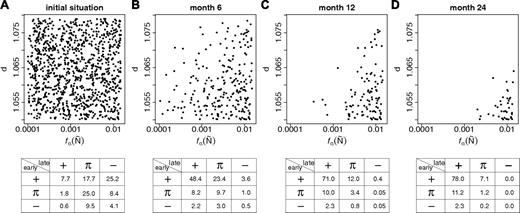
Although the process of clonal selection is predicted to be enhanced by differences in the growth characteristics of individual cells,26 the estimated within-strain heterogeneity of clonal properties does not cause a significant acceleration of this process (data not shown). Nevertheless, our model predicts the exhaustion of clones with competitive disadvantages. Consequently, the total number of different clones and therefore also the heterogeneity of engraftment kinetics is predicted to reduce over time. To quantify the conversion effect, we analyzed the clonal composition in an “aging” B6 model system. We estimated the average frequency of clones with a particular parameter configuration (again regarding d and fα(Ñ)) over time (Figure 6). It can be seen that the decreasing number of contributing clones is complemented by the loss of clones with particular parameters. Hence, we predict that the clonal repertoire of aged mice is restricted. It should be emphasized that the predicted enrichment of clones with a highly competitive engraftment potential does not account for the possibility that molecular effects associated with aging and/or senescence36-39 might impede the repopulation potential of HSCs.
Simulation results on clonal conversion during aging. Given is the clone distribution (characterized by clone specific d / fα(Ñ) values) within a model system starting from the initial situation (A) at different time points: 6 months (B), 12 months (C), and 24 months (C). The system had been initiated with 1000 clonally distinguishable HSCs. Each dot in the graphs (A-D) represents one particular clone. The corresponding frequency tables show the distributions of engraftment patterns obtained from 10 000 simulations, each initiated by the transplantation of 1 randomly chosen cell from the given stem cell pool in competition with 4 host cells.
Simulation results on clonal conversion during aging. Given is the clone distribution (characterized by clone specific d / fα(Ñ) values) within a model system starting from the initial situation (A) at different time points: 6 months (B), 12 months (C), and 24 months (C). The system had been initiated with 1000 clonally distinguishable HSCs. Each dot in the graphs (A-D) represents one particular clone. The corresponding frequency tables show the distributions of engraftment patterns obtained from 10 000 simulations, each initiated by the transplantation of 1 randomly chosen cell from the given stem cell pool in competition with 4 host cells.
Discussion
Our results demonstrate that clonally fixed differences in the repopulating potential are necessary to consistently explain the experimentally observed engraftment behavior of HSCs. Beyond this, we show that the concept of a self-organizing HSC population with flexibly and reversibly changing stem cell states22-24,26,27 is compatible with clonally determined cellular characteristics. Previous versions of the model assumed identical repopulation potentials (ie, identical model parameters) for all HSCs. We have now updated the model to allow for heterogeneity in HSC potentials by introducing clone-specific parameters. These clonally fixed potentials still allow for reversible changes of the functional status of individual cells and are therefore compatible with observations on phenotypic and functional flexibility.4,11-13,40
We show that varying only 2 model parameters is sufficient to account for the experimentally observed heterogeneity of clonal repopulation data. However, this result does not exclude the possibility of a higher-dimensional heterogeneity. There are other cellular characteristics, such as lineage potential, that are clonally determined.10,17,34 However, to avoid another level of model complexity, we intentionally decided to neglect these phenomena in the current study. We refrained from predicting a detailed picture of particular cellular properties that could contribute to the functional heterogeneity. Rather, our minimalistic approach serves as proof-of-principle that clonally fixed differences of HSCs are essential improvements of our theory of HSC organization. It should be added that our results on random variation of the transition characteristic fα in principle allows for attributing the predicted parameter variability partially to differences between the host systems. Therefore, our current model analysis cannot rule out the possibility that the predicted parameter heterogeneity partially comprises a nonclonal component.
Our simulations show that small quantitative differences in the clonal makeup of the repopulating potential of HSC clones can cause considerable changes in their competitive behavior. Most of these differences, however, can only be detected in situations of direct competition of individual HSC clones. In the (theoretical) situation of a noncompetitive repopulation (ie, no residual host stem cells at all) or when transplanting a large number of clonally derived cells into lethally irradiated systems, all HSCs with properties from within the estimated clonal repertoire are predicted to exhibit similar repopulation kinetics (Figure S7).
Beyond the clonally determined properties of HSCs, our model suggests that there is another independent variance component underlying the observed heterogeneity of engraftment levels. We predict that random fluctuations in the contribution of HSC clones cause a considerable degree of variability in the established engraftment levels (even without the assumption of clonal differences) if the number of HSCs within competing clones is small. These effects might be important even when large numbers of HSCs are transplanted, such as in clinical transplantation settings. If the transplant represents a polyclonal cell population with a number of small clones, stochastic fluctuations might cause the extinction of clones, even if these clones actually have a relative growth advantage. To experimentally test the predicted clone-size–dependent component in the engraftment variability, the secondary repopulation experiments would have to be performed at clonal cell densities. Contrasting the results shown in Figure 1B, the model predicts an increased variability in the secondary engraftment levels within the same clonal background.
To account for genetically determined between-strain differences in the repopulating potential of HSCs, we modeled interstrain differences by assuming larger quantitative differences of the same model parameters than the ones used to model clonal differences within mice of one strain. Although this assumption gives plausible results, it is an open question whether or not clonal differences within the same genetic background and differences between mouse strains are determined by qualitatively distinct regulatory mechanisms.
The predicted quantitatively small but clonally fixed differences in the properties of HSCs have important implications for experimental strategies aiming to identify the molecular determinants of HSC function and for the prospective selection of highly potent clones. Our results imply that the repopulating potential of HSCs, as imprinted in the genetic and the epigenetic (clonal) fixation of cellular properties, is in general predictable. However, it should be pointed out that the “quality” of a HSC clone, in terms of its actual repopulation behavior, can only be appraised relative to its competitor clones in the system. Although a particular clone might have a relative competitive advantage in one particular situation, it might just as well be inferior if it is faced with “better” competitor clones. Because exact knowledge of the competitive properties of all clones is unlikely, predicting the actual repopulating behavior of a particular clone is only possible in a probabilistic sense. This also has implications for the situation of acquired clonal differences (eg, due to mutations). Whether or not a mutated clone will become dominant might considerably depend on the context of competing cells, particularly if the potentially induced growth advantage is small.
Our results imply that strategies for identifying molecular (eg, epigenetic) determinants of HSC potential have to rely on the comparison of clonally derived cells. Pooling of HSCs will average and thereby mask the results. However, the differences are inherited from mother to daughter cells. Thus, a longitudinal pooling of clonally derived HSCs could be feasible. It might even be advantageous, because it can highlight the clonally fixed properties while leveling out other effects, such as regulatory processes acting on short time scales.
The quantification of clonal heterogeneity and the prediction of individual clone dynamics are relevant for several clinical applications. For example, drug-resistant clones of leukemic cells41-43 or stem cell clones with properties altered by insertional mutagenesis after gene vector–mediated manipulations show varying and sometimes unexpected growth kinetics.44-46 As also shown by others,47-54 mathematic modeling approaches can contribute to a deeper understanding of the mechanisms driving individual clone dynamics as well as to a better prediction of the in vivo behavior of different types of stem cell and/or cancer clones. The single-cell–based model that we developed is particularly suited to describe effects that are induced by the competition of clones with distinct characteristics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work has been supported by the German Research Foundation (DFG) within the Research Priority Program SPP1230, grant RO3500/1-1 to I.R., and by grants DK048015 and AG023197 from the National Institutes of Health (Bethesda, MD) to C.M.S.
National Institutes of Health
Authorship
Contribution: I.R. and C.M.S. designed the research; I.R., C.M.S., H.B.S., and M.L. wrote the paper; C.M.S. and R.C. performed experiments; I.R. and K.H. performed simulations; and K.H. analyzed the results and made the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ingo Roeder, Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Haertelstrasse 16-18, D-04107 Leipzig, Germany; e-mail: ingo.roeder@imise.uni-leipzig.de.


![Figure 1. Experimentally observed and simulated engraftment kinetics in primary and secondary recipients. (A) The graphic illustrates experimentally observed engraftment kinetics, as determined in the peripheral blood (PB), generated by different, clonally derived transplants in primary hosts (different colors indicate different clones). At month 7, HSCs from these primary hosts were transplanted into secondary hosts. (B) Experimentally observed engraftment kinetics in the PB of secondary hosts. Corresponding colors were used to indicate the relationship of primary HSCs and its clonal progeny revealed in multiple secondary hosts. Data in panels A and B are taken from Muller-Sieburg et al.16 (C,D) Representative examples of simulated engraftment kinetics in primary (C) and secondary (D) host assuming no predetermined differences of individual clones. Identical color codes indicate identical clonal origin in primary and secondary recipients. Gray lines give more examples of primary kinetics to better illustrate the distribution. They are not shown in secondary hosts. (E,F) See panels C and D, but this time assuming small differences between individual HSCs with respect to parameter d. d has been randomly sampled from a normal distribution with mean 1.07 (reference value) and standard deviation 0.01. The particular d values for the given example realizations are 1.06052 (black), 1.06501 (red), 1.06624 (yellow), 1.06708 (green), and 1.07337 (blue). (G,H) See panels C and D but with large differences in parameter d. d has been randomly sampled from the 2 intervals [1.038, 1.049] and [1.091, 1.102], which relate to deviations of plus or minus 2% to 3% from the reference value 1.07. The particular d values for the given example realizations are 1.04705 (black), 1.04392 (red), 1.09377 (green), 1.02802 (blue), and 1.04761 (light blue). To facilitate the comparison with the experimental data, simulated engraftment levels are shown on a bimonthly scale, although they were actually simulated using a 1-hourly scale.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/13/10.1182_blood-2008-05-155374/4/m_zh80240827740001.jpeg?Expires=1769152798&Signature=RpJoXAXmLMrjmp9dxIQ49o9tLHUUGf~KBQ9PQGnduabphcVKMPw1Ojeu2G6-bqioJrzwVuZoI3UKWFa6KBTjViEapwosUwoVFUn0LWyvpr-A6nWjCilAnT61Ov-dKTRSqnrg6raxQtayB2Ii4lJVWX235rlXdzBSBN2AOUrfx0Vmqhy70PI3uS2KRlaR6rDSurHqIdC1yfL0yr75og2wOa045PnWZW3n07tPmUMBlwtzXONa6Dy5fK6uVzqxkeBlZ5W~8ecLQtQg9IdQZCq3NlmVyZq1PuQ8~G37bB9yLAGTPTO~fMI3jjFA~~CB3l~HkH9LRL3SHH-7PwEG-zqJxw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 4. In silico engraftment kinetics depending on model parameters of donor HSCs. (A) Graphic representation of the qualitative differences of simulated engraftment kinetics depending on the choice of model parameters d and fα(Ñ). Each individual color dot illustrates the qualitative pattern of the engraftment kinetic observed in a simulation using the particular parameter combination. In total, 55 074 different parameter combinations are shown in the figure. Simulations were initiated with one donor cell with the particular d and fα(Ñ) values (according to the position in the diagram) and 4 competing host cells with reference parameters d = 1.07 and fa(Ñ) = 0.01. All other parameters had been fixed to B6 reference values, identically for donor and host cells. (B) Parameter region (depicted as white square in panel A; comprising 11 682 simulation runs) that yields the best fit of the simulated to the experimentally observed distribution of engraftment kinetics (compare with Figure S2). The 2 black lines indicate the reference values for d and fa(Ñ) used for the competing host cells. Color coding for engraftment kinetics (identical in panels A and B): maroon [+, +], red [+, π], pink [π, +], dark green [−, −], medium green [−, π], light green [π, −], purple [π,π], blue [+, −], yellow [−, +], and black (no donor engraftment).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/13/10.1182_blood-2008-05-155374/4/m_zh80240827740004.jpeg?Expires=1769152798&Signature=jWFLIbrvPjpoJsBRcJWCDnPjKoXsc5g6Kacf1Y5ibbIYoqwn9C6x4PgW2P5pD3~dEtoMUV~vSNABvdr9GXXScv4sYYeS~3oPINqTNq8pEVnzBqR8ohRsulbmUba2-B5Lkm1g3EtsSXcrdQTZdvRdYNBFQsUcJYgMyE567JAe~xubKsi4WTdn0sl3m3Oiq86nGJO~0jkWJ~efaVUgNPHwvRupCVlmX937963UCHBnoY~49h03VmRSh2nDDF6dTmBa5~Rl5WY9NbdDdxUeyyF4FHtN~A75pniUcu-lT5F3718oWKJd98pa9rC6sUT3-j9R5n295qFYLiVjtbCXwrOAAQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal