Abstract
Endoglin is an accessory receptor for TGF-β signaling and is required for normal hemangioblast, early hematopoietic, and vascular development. We have previously shown that an upstream enhancer, Eng −8, together with the promoter region, mediates robust endothelial expression yet is inactive in blood. To identify hematopoietic regulatory elements, we used array-based methods to determine chromatin accessibility across the entire locus. Subsequent transgenic analysis of candidate elements showed that an endothelial enhancer at Eng +9 when combined with an element at Eng +7 functions as a strong hemato-endothelial enhancer. Chromatin immunoprecipitation (ChIP)–chip analysis demonstrated specific binding of Ets factors to the promoter as well as to the −8, +7+9 enhancers in both blood and endothelial cells. By contrast Pu.1, an Ets factor specific to the blood lineage, and Gata2 binding was only detected in blood. Gata2 was bound only at +7 and GATA motifs were required for hematopoietic activity. This modular assembly of regulators gives blood and endothelial cells the regulatory freedom to independently fine-tune gene expression and emphasizes the role of regulatory divergence in driving functional divergence.
Introduction
Transcriptional regulation is a key mechanism controlling the formation and subsequent behavior of hematopoietic stem cells (HSCs).1 We have previously shown that clustering of Ets and Gata sites can be exploited in genome-wide computational screens to identify new hematopoietic stem/progenitor elements.2-5 These computational screens were performed using the Ets/Ets/Gata (E/E/G) signature (2 conserved Ets and 1 conserved Gata site within a 50-base-pair sequence window with defined spacing and orientation constraints) using the Scl +19 HSC enhancer as a template.3 Given these constraints, the predicted E/E/G elements have all been clustered within a single hematopoietic enhancer. The possibility that the E/E/G HSC signature could be assembled across multiple independent enhancers had not been tested.
During development, endothelium and blood arise from common or closely related progenitors, and there are several signaling molecules and transcription factors (TFs) that are key regulators of both.6 Several lines of evidence support a role for endoglin in blood and blood vessel development: endoglin (ENG or CD105) is an accessory receptor for members of the transforming growth factor-beta (TGF-β) superfamily, including TGFβ1/3, activin, and BMP2/7, and is expressed on the surface of proliferating endothelial cells and adult bone marrow HSCs7,8 ; Eng−/− mice have severely anemic yolk sacs by embryonic day (E) 9.5 and die at E10.5 to 11.5 with cardiac and vascular abnormalities9-11 ; endoglin has also been shown to mark blast-colony forming cells (BL-CFCs), the in vitro equivalent of the hemangioblast, the embryonic progenitor of hematopoietic and endothelial lineages; Eng−/− BL-CFCs are reduced in number and have limited hematopoietic potential.12 Differential expression of endoglin in various hematopoietic progenitor compartments has recently been used to define a functional hierarchy for myelo-erythroid progenitors.13
Despite its functional relevance, the only comprehensive survey of tissue-specific control of endoglin expression has been limited to the endothelium. We have previously shown that Ets TFs act on the Eng promoter and a −8kb enhancer to regulate endoglin expression in the endothelium.14 However, the TFs and cis-regulatory elements that regulate endoglin expression during blood development are not known. Taking advantage of new array-based methodologies to identify chromatin accessibility and TF binding over large genomic regions, we investigated the transcriptional regulation of endoglin during mouse embryonic hematopoiesis. We also sought to address the following general issues. First, can array-based methods be used to identify functionally valid hematopoietic enhancers? Second, can the E/E/G HSC signature be assembled across multiple distinct hematopoietic enhancers rather than being confined to a single enhancer? Third, do components of the Fli1/Gata2/Scl triad, which operates during early blood development and regulates master hematopoietic transcription factors such as Runx1, also regulate expression of endoglin?
Methods
In situ hybridization
In situ hybridization (ISH) was performed on paraffin or cryo sections using digoxigenin-labeled riboprobes and detected with an alkaline phosphatase-conjugated antidigoxigenin antibody as previously described.15 Photo micrographs of tissue sections were prepared by capturing images using an Axio Imager, Axio Cam MrC5 camera, and Axio Vision 4 LE software from Zeiss (Jena, Germany) and Adobe Photoshop (Adobe Systems, San Jose, CA).
Cell culture
HPC-7 cells were maintained as described.16 Bry-GFP embryonic stem (ES) cells were maintained and differentiated as described.17 Day 3 embryoid bodies (EBs) were harvested, trypsinized, stained for GFP (Bry) and Flk1 expression, and sorted for gene expression analyses. MS1 and BW5147 cells were maintained in DMEM and RPMI, respectively, each supplemented with 10% fetal calf serum.
In vitro colony assays
Sorted E11.5 fetal liver (FL) cells were counted and cultured in cytokine-supplemented Methocult GF-3434 for erythroid and myeloid colony formation. Six plates each were seeded with 2 × 103 FDG+ or FDG− cells from transgenic FLs. Six plates of 2 × 103 FDG-treated WT FL cells were also set up as a control. The plates were cultured at 37°C, and colonies were scored at 10 days.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed on HPC-7, BW5147, and MS1 cells. Enrichment was measured by real-time polymerase chain reaction (PCR) using Sybr Green (Stratagene-Agilent, Santa Clara, CA) or labeled for array hybridization as described. The levels of enrichment were normalized to that obtained with a control rabbit antibody and were calculated as a fold increase over that measured at a control region, Scl + 21. Methods and Primers are listed in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
DNaseI hypersensitivity assay
DNA templates were prepared as detailed elsewhere.18 Briefly, nuclei were prepared from 5 million cells and incubated with 120 units of DNaseI (Ambion-Applied Biosystems, Foster City, CA) on ice for 1 hour. DNA was then extracted, RNase A treated and repaired with T4 polymerase (NE Biolabs, Ipswich, MA), to provide double-stranded blunt ends. The DNA was then precipitated and resuspended for ligation to a double-stranded asymmetric biotinylated linker. After ligation, the DNA was reprecipitated, resuspended, and primer-extended. Primer-extended template was then purified from linker and digested genomic DNA, using streptavidin beads (Dynal-Invitrogen, Carlsbad, CA) and labeled for array hybridization as described.
Microarray fabrication
The array design has been deposited in ArrayExpress under the accession A-MEXP-1020 and A-MEXP-1021. See Document S1 for details.
Hybridization
Immunoprecipitated DNA was labeled with the Cy3 dye, mixed with Cy5-labeled control DNA, and then precipitated with cot-1 DNA. After prehybridization of the array slides with herring sperm DNA/cot-1 DNA, the samples were resuspended in hybridization buffer and hybridized to the slides for 45 hours using an automated TECAN 400 (Männedorf, Switzerland) hybridization station. After hybridization, slides were scanned using an Agilent DNA microarray scanner. Mean spot intensities from images were quantified using GenePix 6.0 with background subtraction. The array results have been deposited in ArrayExpress under E-TABM-455. See Document S1 for details
Statistical analysis
Statistical analysis information is detailed in Document S1.
Transgenic analysis
Candidate enhancer sequences were PCR-amplified from human genomic DNA. Mutations were generated by PCR using oligonucleotides with mismatches and verified by DNA sequencing. F0 transgenic mouse embryos and the −8/P/lacZ/ +7/ +9 line were generated by pronuclear injection of lacZ reporter fragments.19 For histology, the embryos were embedded in paraffin, sectioned, and counter-stained with brazilin. Photo micrographs of whole mount embryos were prepared by capturing images using an SMZ stereomicroscope and digital camera from Nikon (Tokyo, Japan) and Adobe Photoshop (Adobe Systems). Primers are listed in Document S1.
Flow cytometry
Single-cell suspensions of E11.5 and E12.5 FLs (L1091xWT) were stained with anti-CD105/PE (R&D Systems, Minneapolis, MN), anti-CD 150/APC (BioLegend, San Diego, CA), anti-CD48/PE, anti-CD41/PE, anti-cKit/APC, 7-AAD (all from BD Biosciences, Franklin Lakes, NJ) and the fluorescent substrate FDG (fluorescein di-β-D galactopyranoside; F-2756; Sigma-Aldrich, St Louis, MO), analyzed on a FACSCalibur (BD Biosciences) and reported using FlowJo software (TreeStar, Ashland, OR).
Expression analysis
Gene expression relative to βactin was quantified using SYBR-Green (Stratagene) or TaqMan (Roche, Indianapolis, IN) real-time PCR. Primers are listed in Document S1.
Results
Endoglin is expressed in hematopoietic tissues in the developing embryo
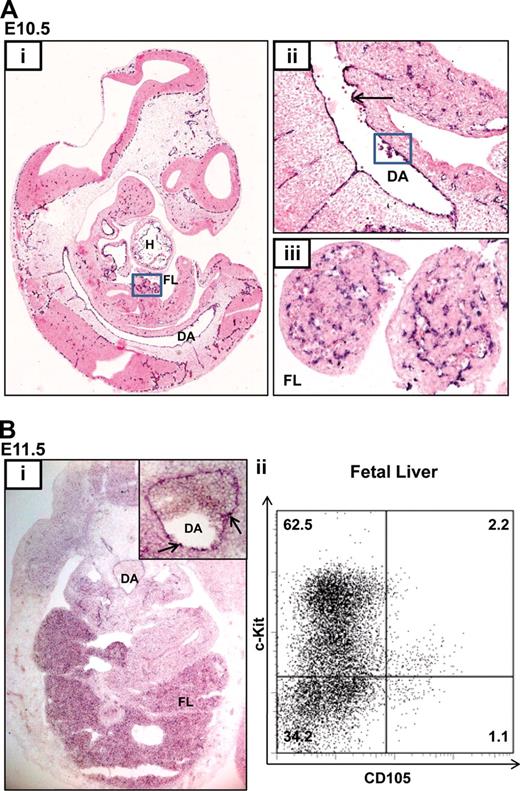
Although endoglin is expressed in adult HSCs and can be used to enrich adult bone marrow HSCs, its expression profile during fetal hematopoiesis is not known.13,20 During mouse development, the first long-term reconstituting HSCs are thought to be generated from the floor of the dorsal aorta (DA) in the aorta-gonad-mesonephros (AGM) region at E10.5.6 These putative HSCs are recognized as blood clusters and are thought to originate from the aortic endothelium or the underlying mesenchyme. HSC numbers are subsequently amplified in the FL and other sites. Endoglin expression is first evident at E6.5 in the amniotic fold and developing allantois and at E7.5 and E8.5 in the yolk sac, dorsal aorta, and primitive heart tube (Figure S1 and Jonker21 ). At E10.5, endoglin transcripts are present in the endocardium (H), endothelia of small and large blood vessels including the DA (Figure 1Ai), in hematopoietic intra-aortic clusters located in the floor of the DA (Figure 1Ai), and in the FL (Figure 1Aiii). At E11.5, endoglin expression persists in the hemogenic DA and in the FL (Figure 1Bi) where approximately 3% of FL hematopoietic cells are endoglin (CD105) positive, a majority of which also express the progenitor marker, c-kit (Figure 1Bii).
Endoglin is expressed in blood and endothelium in the developing embryo. (A) In situ hybridization (ISH) for Eng RNA in an E10.5 embryo. (i) Sagittal section of a paraffin embedded whole-mount embryo (×4); endoglin (black-purple stain) is expressed in the lining of the cardiac chambers (H), DA, FL (boxed), and vasculature of multiple tissues. (ii) Magnified view (×20) of the DA showing concentration of Eng RNA in the hemogenic endothelium (↑) and in blood clusters (boxed). (iii) Magnified view (×40) of the FL. (B) Endoglin expression at E11.5. (i) ISH for Eng RNA. Transverse cryosection (×20) of an embryo at the level of the AGM shows expression in FL and DA. The inset shows a magnified view (×40) of the DA with Eng RNA concentrated in blood clusters (↑). (ii) Flow cytometry plots showing Eng (CD105) expression in approximately 3% of FL cells. DA, dorsal aorta; FL, fetal liver; H, heart.
Endoglin is expressed in blood and endothelium in the developing embryo. (A) In situ hybridization (ISH) for Eng RNA in an E10.5 embryo. (i) Sagittal section of a paraffin embedded whole-mount embryo (×4); endoglin (black-purple stain) is expressed in the lining of the cardiac chambers (H), DA, FL (boxed), and vasculature of multiple tissues. (ii) Magnified view (×20) of the DA showing concentration of Eng RNA in the hemogenic endothelium (↑) and in blood clusters (boxed). (iii) Magnified view (×40) of the FL. (B) Endoglin expression at E11.5. (i) ISH for Eng RNA. Transverse cryosection (×20) of an embryo at the level of the AGM shows expression in FL and DA. The inset shows a magnified view (×40) of the DA with Eng RNA concentrated in blood clusters (↑). (ii) Flow cytometry plots showing Eng (CD105) expression in approximately 3% of FL cells. DA, dorsal aorta; FL, fetal liver; H, heart.
Array-based mapping of chromatin accessibility across the endoglin locus in hematopoietic and endothelial cells
We have previously identified, using multispecies alignments of the Eng locus, 9 regions (E1-E9; Figure 2A) with high-sequence conservation across a wide range of mammals.14 Transgenic analysis demonstrated that E1 as a −8kb enhancer (Eng −8) together with the Eng promoter (E3) targeted endoglin expression to the endothelium in transgenic mice.14 However, Eng −8 did not target blood progenitors in developing embryos (Figure S2). To identify gene regulatory elements that direct endoglin expression to blood progenitors, we mapped histone H3 acetylation status and DNaseI hypersensitivity sites in hematopoietic and endothelial cell lines (Figure 2).
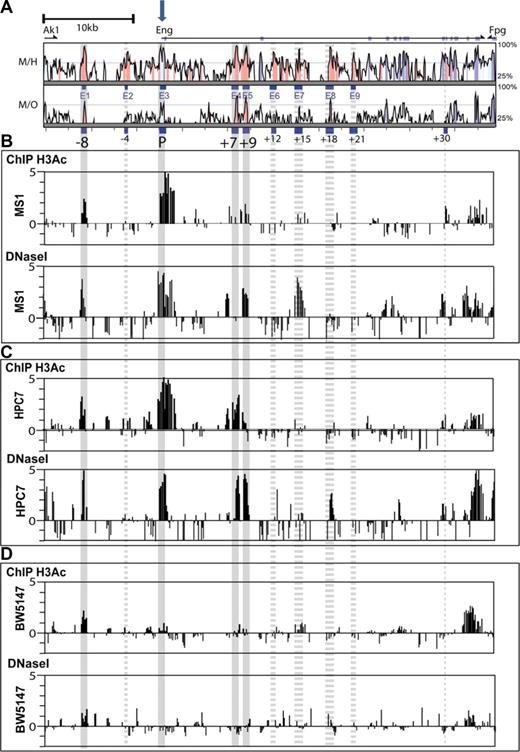
Chromatin accessibility profiles across the Eng locus of endothelial and blood progenitor cell lines. (A) Vista plots of sequence alignments of mammalian Eng loci. M, Mus musculus; H, Homo sapiens, and O, Monodelphis domestica. Conserved regions are displayed relative to their positions in the mouse genome (horizontal axis). Segments that show more than 66% sequence identity (indicated on the vertical axis) at the nucleotide level over a 100-bp window are highlighted in pink (noncoding regions) or cyan (coding exons). Exons are displayed above the comparison plots in cyan. Eng exon1 is marked with a block arrow. (B-D) Array-based histone acetylation state (H3Ac ChIP-chip) and DNaseI hypersensitive site profiles across the Eng loci of MS1 endothelial cells, HPC-7 hematopoietic progenitor cells, and BW5147 T-cells. Samples were hybridized in triplicate and fold enrichment over nonenriched input (normalized to the median of values across the Eng locus) is plotted (log2) against genomic position in kilobases. The width of each bar represents the width of each spotted oligonucleotide on the array. The gray longitudinal bars highlight regions of chromatin accessibility that overlap with genomic regions that are highly conserved in mammals. Accessibility at these conserved regions was either consistent (solid bars) or not (dashed bars) between ChIP-chip and DNaseI hypersensitivity. (B) In endoglin-expressing MS1 endothelial cells, significant enrichments (ie, chromatin accessibility) was noted on both profiles at the Eng promoter (P), the −8kb endothelial enhancer (−8), and also at a 500-bp region 9 kb downstream of the promoter (+9). (C) In endoglin-expressing HPC-7 cells chromatin accessibility was noted on both profiles at the Eng promoter (P) and also at the −8, +7 (∼ 500-bp region 7 kb downstream of the promoter) and +9 regions. (D) In endoglin-nonexpressing BW 5147 cells the Eng promoter (P) was not accessible.
Chromatin accessibility profiles across the Eng locus of endothelial and blood progenitor cell lines. (A) Vista plots of sequence alignments of mammalian Eng loci. M, Mus musculus; H, Homo sapiens, and O, Monodelphis domestica. Conserved regions are displayed relative to their positions in the mouse genome (horizontal axis). Segments that show more than 66% sequence identity (indicated on the vertical axis) at the nucleotide level over a 100-bp window are highlighted in pink (noncoding regions) or cyan (coding exons). Exons are displayed above the comparison plots in cyan. Eng exon1 is marked with a block arrow. (B-D) Array-based histone acetylation state (H3Ac ChIP-chip) and DNaseI hypersensitive site profiles across the Eng loci of MS1 endothelial cells, HPC-7 hematopoietic progenitor cells, and BW5147 T-cells. Samples were hybridized in triplicate and fold enrichment over nonenriched input (normalized to the median of values across the Eng locus) is plotted (log2) against genomic position in kilobases. The width of each bar represents the width of each spotted oligonucleotide on the array. The gray longitudinal bars highlight regions of chromatin accessibility that overlap with genomic regions that are highly conserved in mammals. Accessibility at these conserved regions was either consistent (solid bars) or not (dashed bars) between ChIP-chip and DNaseI hypersensitivity. (B) In endoglin-expressing MS1 endothelial cells, significant enrichments (ie, chromatin accessibility) was noted on both profiles at the Eng promoter (P), the −8kb endothelial enhancer (−8), and also at a 500-bp region 9 kb downstream of the promoter (+9). (C) In endoglin-expressing HPC-7 cells chromatin accessibility was noted on both profiles at the Eng promoter (P) and also at the −8, +7 (∼ 500-bp region 7 kb downstream of the promoter) and +9 regions. (D) In endoglin-nonexpressing BW 5147 cells the Eng promoter (P) was not accessible.
Modifications to histones that package the genome are thought to contribute to the orchestration of gene expression and cellular state, by determining higher order chromatin structure.22 Histone acetylation can increase TF access to chromatin by counteracting higher order chromatin folding that may mask cis-elements.23 Potential gene regulatory regions can therefore be identified by ChIP using an antibody to acetylated histone H3. Alternatively, cis-regulatory elements within actively transcribed gene loci can be mapped by their hypersensitivity to DNaseI digestion. By incorporating array technology, both ChIP and DNaseI hypersensitivity mapping have been successfully adapted to studying large genomic regions.24,25 We hypothesized that those regulatory elements that direct endoglin expression to blood or endothelial cells would be accessible to TF binding and as such show active chromatin marks by one or both assays in representative cells.
In endoglin-expressing MS1 endothelial cells (Figure S3), active chromatin marks were seen at P (Eng promoter) and at −8 by both H3Ac ChIP-chip and array-based DNaseI hypersensitivity mapping (ADHM; Figure 2B). A conserved region at +9 was also accessible by both assays. By contrast, 2 ADHM sites, one at a poorly conserved region 5′ to the highly conserved +7 region and another at the + 15 conserved region, did not show corresponding accessibility by H3Ac ChIP-chip. The HPC7 cell line expresses endoglin (Figure S3), requires stem cell factor for growth and has multi-lineage differentiation capacity in transplantation assays.16 As with MS1 endothelial cells, prominent active chromatin marks were seen at the Eng promoter (P) and Eng-8 enhancer (−8) in HPC-7 hematopoietic cells (Figure 2C) by both H3Ac ChIP-chip and ADHM. In addition, enrichments were also seen at genomic coordinates corresponding to +7 and +9 conserved regions in both assays. The DNaseI hypersensitive mark at + 18 in HPC-7 cells did not correlate with enrichment of acetylated histones at this site. By contrast, BW5147 T-cells, which do not express endoglin (Figure S3), lack active chromatin marks at the Eng promoter (P; Figure 2D). Interestingly somewhat diminished ‘active’ marks remain at −8 but are absent from +7 and +9 regions. Taken together, these results show first, that promoter accessibility correlates with endoglin expression irrespective of cell type; and second, that −8 and +9 noncoding conserved regions are accessible in both blood and endothelial cells whereas the +7 region is accessible in blood progenitors but not in endothelial cells. These results are summarized in Figure S4.
The +7 and +9 conserved noncoding regions target FL hematopoietic cells in vivo
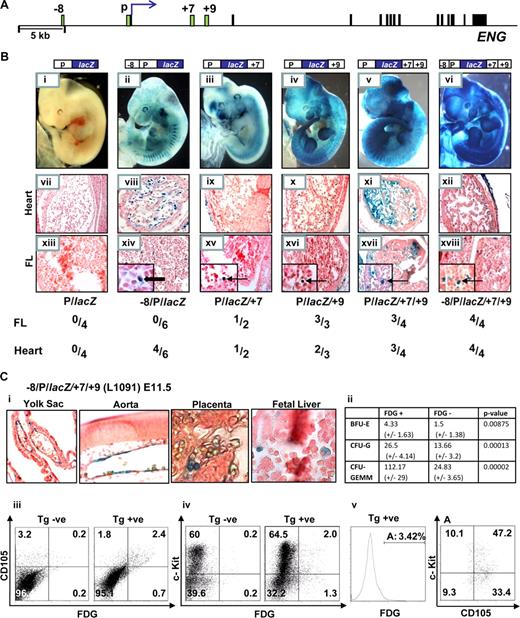
To test whether the genomic regions identified by profiling chromatin accessibility represent bona fide hematopoietic regulatory elements, we performed in vivo transgenic assays. The +7 and +9 regions are located in the first intron approximately +7 kb and +9 kb from the translation start site of human ENG (Figure 3A). We have previously established that the Eng promoter (P) targets lacZ reporter expression to subsets of endothelial cells, albeit weakly, and that the Eng −8 region significantly enhances promoter activity in the endothelium (Pimanda et al14 and Figure 3B, compare wholemount embryos and sections in ii, viii, and xiv with i, vii, and xiii). To evaluate the roles of the +7 and +9 regions as potential enhancers, we inserted 483 bp (+7431/+7913) and 500 bp (+8974/+9473) fragments corresponding to these regions, alone or in combination, downstream of either the Eng promoter (P/lacZ) or the Eng promoter/enhancer (−8/P/lacZ) reporter constructs (Figure 3B). Reporter gene expression was evident in FL hematopoietic cells in P/lacZ/+7 and P/lacZ/+9 transgenic embryos and both the frequency and intensity of expression was increased by combining +7 and +9 (Figure 3B, compare whole-mount embryos in v with iii and iv and FL sections in xvii with xv and also with xvi). By adding the −8 enhancer to P/lacZ/+7/+9 and thus incorporating the complete chromatin accessibility profile seen in HPC-7 hematopoietic cells into a single reporter construct, the specificity of hemato-endothelial expression was enhanced (Figure 3Bvi,xii,xviii).
The Eng +7 and Eng +9 regions target blood progenitors in developing embryos. (A) Schematic diagram of the human ENG locus. The exons and enhancer fragments are drawn to scale and are represented as black and green rectangles, respectively. (B) F0 transgenic embryos generated using various ENG fragments subcloned into ENG (P) promoter lacZ constructs. (i-vi) representative X-Gal stained whole-mount E11.5 embryos. (vii-xii) Sections (magnified ×20) through the hearts of corresponding embryos; (vii) no endocardial staining; (viii-xii) variable degrees of endocardial staining. (xiii-xviii) Sections (magnified ×20; insets ×100) through the livers of corresponding embryos; (xiii) no staining in FL cells; (xiv) staining of flat endothelial cells ( ); (xv-xviii) staining of round hematopoietic cells (
); (xv-xviii) staining of round hematopoietic cells ( ). A table summarizing the number of X-Gal stained F0 transgenic embryos that showed staining in the heart and/or liver out of the number of transgenic embryos generated with each construct is also shown. (C) Analyses of E11.5 embryos from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. (i) Tissue sections from X-Gal stained embryos showing reporter activity in yolk sac and dorsal aorta endothelium and blood cells in the placenta and fetal liver (magnified ×20, ×40, ×100, respectively). (ii) A table summarizing results from in vitro colony-forming assays using sorted cell fractions from FDG treated E11.5 FLs. (iii-v) Flow cytometry of FDG treated nontransgenic and transgenic E11.5 FLs from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. The transgene targets 3% to 4% of FL cells; the majority of which are (iii) endoglin positive, (iv) c-Kit positive, (v) endoglin and c-Kit dual positive. BFU-E, burst-forming unit-erythroid; CFU-G, colony-forming unit-granulocyte; CFU-GEMM, colony-forming unit granulocyte/erythroid/macrophage/megakaryocyte.
). A table summarizing the number of X-Gal stained F0 transgenic embryos that showed staining in the heart and/or liver out of the number of transgenic embryos generated with each construct is also shown. (C) Analyses of E11.5 embryos from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. (i) Tissue sections from X-Gal stained embryos showing reporter activity in yolk sac and dorsal aorta endothelium and blood cells in the placenta and fetal liver (magnified ×20, ×40, ×100, respectively). (ii) A table summarizing results from in vitro colony-forming assays using sorted cell fractions from FDG treated E11.5 FLs. (iii-v) Flow cytometry of FDG treated nontransgenic and transgenic E11.5 FLs from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. The transgene targets 3% to 4% of FL cells; the majority of which are (iii) endoglin positive, (iv) c-Kit positive, (v) endoglin and c-Kit dual positive. BFU-E, burst-forming unit-erythroid; CFU-G, colony-forming unit-granulocyte; CFU-GEMM, colony-forming unit granulocyte/erythroid/macrophage/megakaryocyte.
The Eng +7 and Eng +9 regions target blood progenitors in developing embryos. (A) Schematic diagram of the human ENG locus. The exons and enhancer fragments are drawn to scale and are represented as black and green rectangles, respectively. (B) F0 transgenic embryos generated using various ENG fragments subcloned into ENG (P) promoter lacZ constructs. (i-vi) representative X-Gal stained whole-mount E11.5 embryos. (vii-xii) Sections (magnified ×20) through the hearts of corresponding embryos; (vii) no endocardial staining; (viii-xii) variable degrees of endocardial staining. (xiii-xviii) Sections (magnified ×20; insets ×100) through the livers of corresponding embryos; (xiii) no staining in FL cells; (xiv) staining of flat endothelial cells ( ); (xv-xviii) staining of round hematopoietic cells (
); (xv-xviii) staining of round hematopoietic cells ( ). A table summarizing the number of X-Gal stained F0 transgenic embryos that showed staining in the heart and/or liver out of the number of transgenic embryos generated with each construct is also shown. (C) Analyses of E11.5 embryos from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. (i) Tissue sections from X-Gal stained embryos showing reporter activity in yolk sac and dorsal aorta endothelium and blood cells in the placenta and fetal liver (magnified ×20, ×40, ×100, respectively). (ii) A table summarizing results from in vitro colony-forming assays using sorted cell fractions from FDG treated E11.5 FLs. (iii-v) Flow cytometry of FDG treated nontransgenic and transgenic E11.5 FLs from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. The transgene targets 3% to 4% of FL cells; the majority of which are (iii) endoglin positive, (iv) c-Kit positive, (v) endoglin and c-Kit dual positive. BFU-E, burst-forming unit-erythroid; CFU-G, colony-forming unit-granulocyte; CFU-GEMM, colony-forming unit granulocyte/erythroid/macrophage/megakaryocyte.
). A table summarizing the number of X-Gal stained F0 transgenic embryos that showed staining in the heart and/or liver out of the number of transgenic embryos generated with each construct is also shown. (C) Analyses of E11.5 embryos from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. (i) Tissue sections from X-Gal stained embryos showing reporter activity in yolk sac and dorsal aorta endothelium and blood cells in the placenta and fetal liver (magnified ×20, ×40, ×100, respectively). (ii) A table summarizing results from in vitro colony-forming assays using sorted cell fractions from FDG treated E11.5 FLs. (iii-v) Flow cytometry of FDG treated nontransgenic and transgenic E11.5 FLs from a litter of −8/P/lacZ/+7/+9 (L1091) x WT crosses. The transgene targets 3% to 4% of FL cells; the majority of which are (iii) endoglin positive, (iv) c-Kit positive, (v) endoglin and c-Kit dual positive. BFU-E, burst-forming unit-erythroid; CFU-G, colony-forming unit-granulocyte; CFU-GEMM, colony-forming unit granulocyte/erythroid/macrophage/megakaryocyte.
Transgenic embryos generated with Eng promoter constructs that included the +9 fragment showed strong endothelial/endocardial and hematopoietic reporter gene expression (Figure 3Biv-vi). This is consistent with the ADHM profiles in Figure 2 that show chromatin accessibility at the +9 region in both MS1 endothelial and HPC-7 hematopoietic cells. Also consistent with the chromatin accessibility profiles in Figure 2, transgenic embryos generated with the P/lacZ/ +15 construct showed strong endothelial expression but lacked hematopoietic expression (Figure S5) and those generated with the P/lacZ/ +7 construct showed blood but relatively weak endothelial staining. Finally, despite the prominent chromatin mark at Eng +18 in the ADHM profile in HPC-7 cells, P/lacZ/+18 transgenics did not show reporter gene expression in FL blood cells (Figure S6). Therefore, by filtering chromatin accessibility profiles using in vivo transgenics, we were able to identify bona fide hematopoietic enhancers in the Eng locus. This emphasizes the need to correlate chromatin accessibility profiles in individual cell types with appropriate in vivo models so that potential enhancers are tested not in isolation but with due regard to tissue development in the whole embryo.
To explore the surface phenotype of FL cells targeted by the Eng +7 and Eng +9 enhancers, we generated a stable mouse line expressing the −8/P/lacZ/+7/+9 transgene (L1091) that incorporated all of the regions with active chromatin marks identified by both H3Ac ChIP-chip and ADHM in HPC-7 cells (Figure 3Bvi). The transgene is expressed as early as E7.5, where it targets cells in the blood band region in the proximal yolk sac (Figure S7). At E11.5, embryos show reporter expression in the yolk sac endothelium, hemogenic dorsal aorta and blood cells in the placenta and fetal liver (Figure 3Ci). Fetal liver hematopoietic cells targeted by the −8/P/lacZ/+7/+9 transgene show greater multilineage growth potential assayed by in vitro colony-forming assays (compare FDG positive vs negative fractions in Figure 3Cii). Flow cytometry of FL cells from −8/P/lacZ transgenic embryos (L1082) showed no reporter gene expression when stained with the fluorescentβ-galactosidase substrate, FDG (Figure 7S2). However, when the Eng +7 and Eng +9 enhancers are added to the construct (-8/P/lacZ/+7/+9) approximately 3% of E11.5 FL cells do (Figure 3Ciii). Furthermore, the majority of FL cells targeted by the transgene were also endoglin (CD105) positive. Although most of the c-Kit positive cells in the FL do not express either CD105 or the −8/P/lacZ/+7/+9 transgene (Figures 1Bii and 3Civ), approximately 60% of cells targeted by the transgene are both CD105 and c-Kit positive (Figure 3Cv). As shown in Figure S8, cells that express the −8/P/lacZ/+7/+9 transgene are also enriched for CD150+ blood stem/progenitor cells and show a relative paucity of lineage committed CD150−/CD48−/CD41− cells.26 However, this activity does not persist in adult bone marrow (Figure S9) and could indicate that these elements may not be used to express endoglin in adult bone marrow or more likely that the transgene is silenced with time. Taken together these data show that the −8/P/lacZ/+7/+9 transgene targets FL blood progenitors and that in vivo activity of these noncoding conserved regions is consistent with endogenous endoglin expression.
Ets and Gata binding sites within the Eng +7 and Eng +9 enhancers are required for their in vivo activity
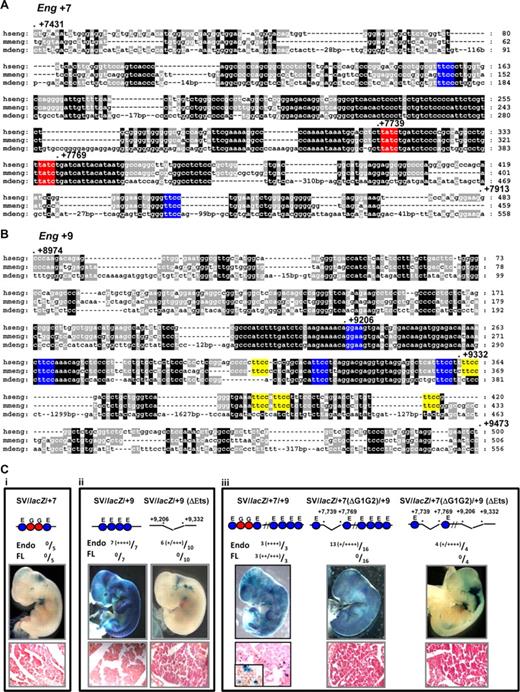
We have shown previously that clusters of highly conserved Ets and Gata sites are critical for the activity of early hematopoietic regulatory elements of the Scl, Lyl1, Fli1, Hhex and Gata2 genes.2-5,27 The Eng +7 enhancer has 2 conserved Gata binding sites within a large block of highly conserved sequence, flanked by 2 conserved Ets binding sites (Figure 4A). The Eng +9 enhancer has no Gata binding sites but has 4 Ets binding sites conserved down to opossum and an additional 5 conserved between human and mouse (Figure 4B). To establish the relative contribution of the various conserved Ets and Gata motifs, mutations were introduced and the activity of the enhancers tested in F0 transgenics (Figure 4C). Although the Eng +7 and Eng +9 enhancers in combination with the endogenous Eng promoter show hematopoietic activity, when combined with the SV promoter neither enhancer is sufficient on its own to target blood (compare Figure 4Ci and ii with Figure 3Biii and iv). However when combined in a single construct, Eng +7 and Eng +9 enhancers show robust activity in blood (compare Figure 4Ciii with Figure 3Bv). Eng +9 enhancer shows robust endothelial activity and deleting a core region with 4 highly conserved Ets binding sites reduces but does not eliminate this activity in all embryos (Figure 4Cii). This is probably due in part to the partially conserved Ets sites that are retained in Eng +9 (ΔEts; yellow blocks in Figure 4Bii). Importantly, the Gata binding sites in Eng +7 are critical for the hematopoietic activity of the SV/lacZ/+7/+9 construct although some endothelial activity remains in SV/lacZ/+7(ΔG1G2)/+9 and SV/lacZ/+7(ΔG1G2)/+9 (ΔEts) transgenic embryos (Figure 4Ciii).
Activity of the Eng +7 and Eng +9 enhancers require conserved Ets and/or Gata TF binding sites. (A) Nucleotide sequence alignment of the Eng +7 enhancer with conserved Ets binding sites marked in blue and Gata binding sites in red. The nucleotide numbering is relative to the first ATG. (B) Nucleotide sequence alignment of the Eng +9 enhancer with Ets binding sites conserved in human/mouse/opossum marked in blue and human/mouse but not opossum in yellow. (C) Representative X-Gal stained wholemount E11.5 F0 transgenic embryos generated with various wild-type or mutant Eng +7 and/or Eng +9 fragments subcloned into a SV40 promoter/lacZ reporter (SV/lacZ). Fully conserved Ets (E) and Gata (G) binding sites are represented as circles. The number of transgenic embryos with endothelial and/or fetal liver staining relative to number generated is also shown. The degree of X-gal staining is indicated as weak (+) to strong (+‖+‖+‖+). (i) SV/lacZ/ + 7 embryos show minimal blood/endothelial staining. (ii) SV/lacZ/+9 embryos show strong endothelial but little blood staining. SV/lacZ/+9 (ΔEts; missing region with fully conserved Ets binding sites in Eng +9) embryos show variable endothelial staining (some embryos show none) but no fetal liver staining. (iii) SV/lacZ/+7/+9 embryos show strong blood and endothelial staining. SV/lacZ7 (ΔG1G2)/+9 (missing region with conserved Gata binding sites in Eng +7) embryos show variable endothelial and no blood staining. SV/lacZ/+7(ΔG1G2)/+9 (ΔEts; missing region with conserved Gata sites in Eng +7 and fully conserved Ets sites in Eng +9) embryos show variable endothelial and no blood staining. Tissue sections magnified ×40; inset ×100.
Activity of the Eng +7 and Eng +9 enhancers require conserved Ets and/or Gata TF binding sites. (A) Nucleotide sequence alignment of the Eng +7 enhancer with conserved Ets binding sites marked in blue and Gata binding sites in red. The nucleotide numbering is relative to the first ATG. (B) Nucleotide sequence alignment of the Eng +9 enhancer with Ets binding sites conserved in human/mouse/opossum marked in blue and human/mouse but not opossum in yellow. (C) Representative X-Gal stained wholemount E11.5 F0 transgenic embryos generated with various wild-type or mutant Eng +7 and/or Eng +9 fragments subcloned into a SV40 promoter/lacZ reporter (SV/lacZ). Fully conserved Ets (E) and Gata (G) binding sites are represented as circles. The number of transgenic embryos with endothelial and/or fetal liver staining relative to number generated is also shown. The degree of X-gal staining is indicated as weak (+) to strong (+‖+‖+‖+). (i) SV/lacZ/ + 7 embryos show minimal blood/endothelial staining. (ii) SV/lacZ/+9 embryos show strong endothelial but little blood staining. SV/lacZ/+9 (ΔEts; missing region with fully conserved Ets binding sites in Eng +9) embryos show variable endothelial staining (some embryos show none) but no fetal liver staining. (iii) SV/lacZ/+7/+9 embryos show strong blood and endothelial staining. SV/lacZ7 (ΔG1G2)/+9 (missing region with conserved Gata binding sites in Eng +7) embryos show variable endothelial and no blood staining. SV/lacZ/+7(ΔG1G2)/+9 (ΔEts; missing region with conserved Gata sites in Eng +7 and fully conserved Ets sites in Eng +9) embryos show variable endothelial and no blood staining. Tissue sections magnified ×40; inset ×100.
Fli1, Pu.1, and Gata2 bind the Eng +7/Eng +9 hematopoietic enhancers in vivo
Blood and endothelium develop from closely related mesodermal progenitors (reviewed in Dzierzak6 ). Lineage tracing studies in zebrafish have confirmed earlier in vitro data that a proportion of blood and endothelial cells share a common progenitor, the hemangioblast.28 Nevertheless, after differentiation, ES cells in culture form colonies known as EBs that contain mesodermal (Brachyury+) progenitors that display both blood and endothelial potential (ie, BL-CFCs).29 This transient population (days 2.5-4 of differentiation) represents the in vitro equivalent of the yolk-sac hemangioblast. The ES/EB model system has been applied to an ES cell line with GFP targeted to the Brachyury locus (GFP-Bry ES).17 By sorting day 3 to 3.5 EBs based on GFP and Flk1 expression it is possible to identify 3 distinct cell populations, GFP−Flk1− (DN), GFP+Flk1− (SP) and GFP+Flk1+ (DP) that represent a developmental progression ranging from premesoderm (DN) to prehemangioblast mesoderm (SP) to the hemangioblast mesoderm (DP).
Endoglin is required for development of hemangioblast equivalents and early hematopoiesis12 and as shown in Figure 5Ai, expression levels increase approximately 3.5-fold as cells transit from prehemangioblast mesoderm (SP) to the hemangioblast mesoderm (DP). We have previously shown that Fli1 and Gata2, together with Scl, form a recursively wired transcriptional subcircuit that operates during early endothelial and HSC development in the embryo.5 Pu.1 is an Ets TF that is expressed in blood but not endothelial cells and is indispensable for the maintenance of HSCs and for normal myeloid and B-lymphoid development.30 To determine the role of Fli1, Gata2, Scl, and Pu.1 in regulating endoglin expression during blood specification, we measured variations in the levels of these TFs in cell fractions from sorted day 3.5 and in unsorted day 4.5 EBs. The levels of Fli1, Gata2, and Scl increased dramatically in DP cells (Figure 5Aii and Pimanda et al5 ). A day later, with further hematopoietic differentiation, the levels of these TFs continued to increase. Pu.1 levels lagged behind but increased at day 4.5. All 4 transcription factors are also present in FL cells at E11.5 (Figure 5B and Pimanda et al5 ).
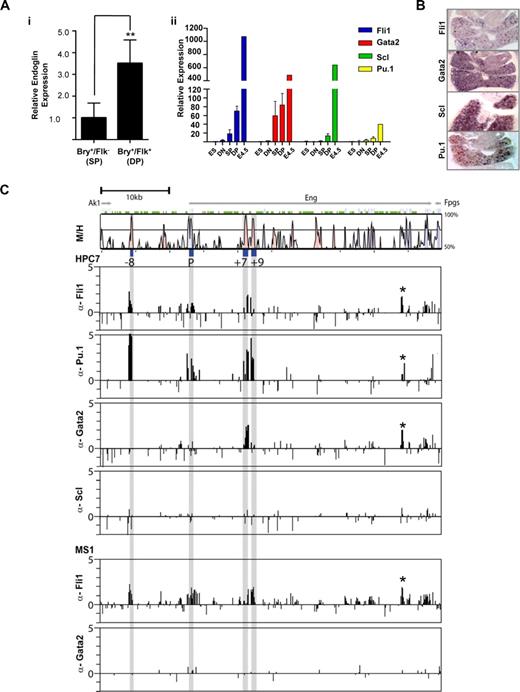
Fli1, Pu.1, and Gata2 bind the endoglin hematopoietic enhancers in vivo. (A) Variation in gene expression during in vitro differentiation of GFP-Bry ES cells to embryoid bodies. (i) Levels of endoglin expression in hemangioblast containing DP (Bry+/Flk+) cells relative to prehemangioblast SP (Bry+/Flk−) cells. (ii) TF levels in ES cells, cell fractions sorted from day 3.5 (DN, SP, and DP) and unsorted day 4.5 EBs. (B) In situ hybridization for Fli1, Gata2, Scl, and Pu.1 transcripts in fetal livers of E11.5 mouse embryos (magnified ×10). (C) ChIP-chip profiles of TF binding across the Eng locus of HPC-7 hematopoietic progenitor cells. Vista plots of sequence alignments of mouse and human Eng loci are shown in the top panel with ChIP-chip profiles shown below. The gray bars indicate the genomic positions of the conserved −8, P, +7 and +9 segments. ChIP assays were performed in HPC-7 hematopoietic progenitor cells with anti-Fli1, -Pu.1, -Gata2, and -Scl antibodies and MS1 endothelial cells with anti-Fli1 and Gata2 antibodies. Samples were hybridized in triplicate and fold enrichment over nonenriched input (normalized to the median of values across the Eng locus) is plotted (log2) against genomic position in kilobases. The width of each bar represents the width of each spotted oligonucleotide on the array. The HPC-7 plots show enrichment of Fli1 and Pu.1 at the Eng promoter and the Eng −8, Eng +7 and Eng +9 enhancers, enrichment of Gata2 at the Eng +7 enhancer but no enrichment of Scl at the Eng locus. The enrichment profile of Fli1 in MS1 cells is similar to that of HPC-7 cells but Gata2 is not enriched at the Eng locus in endothelial cells. EB, Embryoid Body; DN, Double Negative; SP, Single Positive; DP, Double Positive.
Fli1, Pu.1, and Gata2 bind the endoglin hematopoietic enhancers in vivo. (A) Variation in gene expression during in vitro differentiation of GFP-Bry ES cells to embryoid bodies. (i) Levels of endoglin expression in hemangioblast containing DP (Bry+/Flk+) cells relative to prehemangioblast SP (Bry+/Flk−) cells. (ii) TF levels in ES cells, cell fractions sorted from day 3.5 (DN, SP, and DP) and unsorted day 4.5 EBs. (B) In situ hybridization for Fli1, Gata2, Scl, and Pu.1 transcripts in fetal livers of E11.5 mouse embryos (magnified ×10). (C) ChIP-chip profiles of TF binding across the Eng locus of HPC-7 hematopoietic progenitor cells. Vista plots of sequence alignments of mouse and human Eng loci are shown in the top panel with ChIP-chip profiles shown below. The gray bars indicate the genomic positions of the conserved −8, P, +7 and +9 segments. ChIP assays were performed in HPC-7 hematopoietic progenitor cells with anti-Fli1, -Pu.1, -Gata2, and -Scl antibodies and MS1 endothelial cells with anti-Fli1 and Gata2 antibodies. Samples were hybridized in triplicate and fold enrichment over nonenriched input (normalized to the median of values across the Eng locus) is plotted (log2) against genomic position in kilobases. The width of each bar represents the width of each spotted oligonucleotide on the array. The HPC-7 plots show enrichment of Fli1 and Pu.1 at the Eng promoter and the Eng −8, Eng +7 and Eng +9 enhancers, enrichment of Gata2 at the Eng +7 enhancer but no enrichment of Scl at the Eng locus. The enrichment profile of Fli1 in MS1 cells is similar to that of HPC-7 cells but Gata2 is not enriched at the Eng locus in endothelial cells. EB, Embryoid Body; DN, Double Negative; SP, Single Positive; DP, Double Positive.
To correlate transgenic in vivo enhancer activity with in vivo TF occupancy, we performed ChIP-chip experiments with well-characterized antibodies in ES cell derived HPC-7 cells that fulfill many functional requirements defining HSCs.16 All 4 transcription factors are expressed at high levels in these cells (Pinto do et al16 and Figure S10). As shown in Figure 5C, Fli1 and Pu.1 are enriched at the Eng promoter (P), Eng −8, Eng +7, and Eng +9 enhancers. The enrichments of these Ets factors are consistent with both chromatin accessibility (Figure 2C) and the presence of conserved Ets binding sites (Pimanda et al14 and Figure 4A,B). Gata2 enrichment was most prominent at Eng +7, which has 2 highly conserved Gata binding sites (Figure 4A). Scl is expressed in HPC-7 cells (Figure S10 and Pinto do et al16 ) but is not enriched at the Eng locus despite the presence of a E-box element at Eng −8 that is highly conserved in mammals (mm9: Chr2: 32 493,775-800). Similar enrichment profiles were also seen in 416B blood progenitors (data not shown). By contrast, in MS1 endothelial cells, although the Fli1 enrichment profile mirrors that in HPC-7 cells, Gata2 is not enriched at the Eng locus (Figure 5C). Peaks of enrichment for Fli1, Pu.1, and Gata2 are also seen at Eng +30 (marked with an asterisk). The corresponding tile has a palindromic 8 bp region with 2 overlapping GATA and 2 Ets binding sites (ggatatcc). The corresponding region shows a degree of chromatin accessibility in HPC-7 and MS1 cells and is worthy of further study.
Discussion
In this report we show: First, that array-based chromatin accessibility surveys are useful in identifying tissue-specific enhancers but that in vivo validation is required to filter false positives. Second, that the Ets/Gata transcriptional program that regulates hematopoietic gene expression is not necessarily confined to a single hematopoietic enhancer and can be assembled across multiple tissue-specific enhancers. Third, that by distributing the Ets/Gata code across separate enhancers, hemangioblasts, blood stem/progenitors, and endothelial cells can regulate expression of genes such as endoglin that are functionally important to each, in a cell type–specific manner despite the overlap in their respective transcriptional programs. And fourth, that Pu.1 and Gata2 drive endoglin expression in blood stem/progenitors but not endothelial cells; a mechanism that could be a paradigm for expressing other genes shared between both tissues.
Transcriptional control of development
Characterization of transcriptional networks has become a major focus of developmental biology as these networks govern the spatial variation and temporal sequence of gene expression. Within these networks combinatorial interactions of transcription factors control the expression of key effector genes including those involved in signal transduction such as endoglin. Individual cis-regulatory elements are key components of transcriptional networks because they function as information processing units by integrating the inputs of their respective upstream regulators into tightly controlled spatiotemporal expression patterns. However, functional characterization of transcriptional regulatory elements has traditionally been a laborious process thus limiting our ability to reconstruct transcriptional regulatory networks.
Recent array-based methods have greatly facilitated chromatin accessibility surveys as a means to identify candidate gene regulatory regions spread across large genomic intervals. However, it remains unknown to what extent even comprehensive chromatin profiling across multiple tissues will be able to accurately identify tissue-specific regulatory elements. This comprehensive survey of the endoglin locus shows that although there is broad agreement between techniques, the chromatin accessibility sites identified by surveying histone modifications and DNaseI accessibility can differ. Some of these differences, and indeed the validity of the predicted elements as bona fide tissue-specific enhancers, can be resolved using in vivo transgenics. Indeed, the best predictor of tissue specificity of an element in transgenic assays was consistency between histone acetylation and DNaseI accessibility profiles. Our current data therefore suggest that although informative, recently reported genome-wide chromatin accessibility profiles31,32 should be interpreted with caution when drawing conclusions about true in vivo activities of any regions flagged as candidate regulatory elements.
Control of early blood and endothelial development
Blood and endothelial cells share a close developmental link and, based on experiments in chick, the existence of a shared progenitor called the hemangioblast was proposed.33,34 Bipotential precursors have also been demonstrated in mouse and human ES cell cultures29,35 and from cells derived from the gastrulating mouse embryo.36 However, estimates of the in vivo frequency of these bipotential progenitors vary. On the one hand, single cell resolution fate mapping in zebrafish embryos showed that approximately 12.5% of labeled cells give rise to both blood and endothelial progeny and to no other cell type.28 On the other hand, fate-mapping analyses both by transplantation using early mouse embryos37 as well as in chimeras generated using differentially labeled mouse ES cells38 suggest that the majority of yolk-sac blood and endothelial cells develop independently from mesodermal precursors without necessarily originating from a shared progenitor. However, later during embryogenesis, blood stem cells are thought to be specified from hemogenic endothelium of the dorsal aorta,39 again suggesting a close relationship between endothelium and blood stem/progenitor cells.
Despite the above controversies likely to be caused by differences between what individual cells can do when placed in specific culture conditions as opposed to what they will do in an intact embryo, a wide range of genetic evidence supports a close biologic relationship between endothelial and early blood cells. Both gain- and loss-of-function studies for several genes result in phenotypes affecting both lineages40-42 and endoglin is of course a member of this growing list of genes.9 Extensive functional overlap between endothelial and blood stem/progenitor cells has also been identified at the level of transcriptional regulation. In particular, regulatory elements containing clusters of Ets and Gata binding sites are active in both lineages,2,27 and indeed an E/E/G signature has been used successfully for the computational identification of additional enhancers with this dual specificity.3,4 However, several regulatory elements with little or no hematopoietic activity have also been identified, and these are generally characterized by the presence of multiple Ets sites but no Gata sites.14,43
Unlike other genes studied to date that are coexpressed in endothelium and blood stem/progenitor cells, the endoglin locus does not contain a typical Ets/Gata responsive enhancer. Instead, the Ets/Gata regulatory code is assembled across distinct elements with the intriguing consequence that Gata2 control of endoglin expression becomes restricted to hematopoietic cells. Disentanglement of regulatory programs has recently emerged as a potential major driver of evolution in lower eukaryotes.44 Similarly, the modular assembly (and disassembly) of the Ets/Gata code across multiple tissue-specific regulatory elements appears to allow a degree of regulatory freedom so that when blood and endothelial cells coexpress a gene they can disentangle these enhancers and recruit only those elements they need to fine-tune gene expression to suit individual needs. This dynamic interaction and potential for alternative TF/cis-regulatory element interactions may be particularly important when expressing a functionally important gene against the changing transcriptional landscape of a progenitor transiting toward a definitive cellular phenotype.
Diversification of transcriptional programs during progenitor differentiation
Within the endoglin locus, Gata2 binding to the +7 enhancer is specific to the blood lineage and is required for hematopoietic activity of +9, an otherwise endothelial enhancer. It is striking that by co-opting the +7 region, the +9 endothelial enhancer is converted into a robust hemato-endothelial enhancer. Although the precise temporal and spatial emergence of blood and endothelial cells in the extra-embryonic yolk sac is debated, the endothelium predates the emergence of HSCs from the dorsal aorta. This raises the possibility that the regulatory network that specifies blood diverged from a preexisting endothelial regulatory network.
Gata2 is expressed in endothelial cells yet cannot access the +7 enhancer due at least in part to compacted chromatin. Accessibility at the +7 enhancer in the blood lineage is of fundamental importance and could be established in mesodermal precursors by pioneer transcription factors that are expressed in these progenitors and in blood but not in endothelium. FoxA, for example, opens compacted chromatin and by interacting with the albumin enhancer in endoderm precursors maintains enhancer accessibility in liver cells but not in tissues where FoxA is not expressed.45 Pu.1 has been shown to act as a pioneer transcription factor at the c-fms locus,46 is expressed in blood but not endothelium and binds the +7 enhancer with Gata2. Low-level Pu.1 expression is present in Brachyury+/Flk1+ mesodermal precursors and increases substantially with hematopoietic differentiation and could play a role in +7 enhancer accessibility in the blood lineage and Gata2 binding at this site. Potential synergism of Pu.1 and Gata2 in driving the early hematopoietic program is in stark contrast to the well established antagonism between Pu.1 and Gata1, widely seen as one of the key mechanisms driving lineage commitment of differentiating common myeloid progenitor cells.47 However, cooperative interactions between Pu.1 and Gata2 are not without precedent. For example, the differentiation of mast cells, which like stem cells depend on stem cell factor signaling, can be induced through cooperative action of Pu.1 and Gata2.48
The importance of endoglin levels in health and disease
Like endoglin, both Pu.1 and Gata2 play important roles in blood stem/progenitor cells, and it is tempting to speculate that at least some of the phenotypic consequences of loss of Pu.1 or Gata2 could be explained by reduced levels of endoglin. Moreover, it is interesting that Pu.1 when disrupted is leukaemogenic,49 as endoglin itself was first identified from a pre-B acute lymphoblastic leukemia cell line. Endoglin is also a marker of tumor vascularity, and soluble endoglin, a product of the native protein that is cleaved from the cell surface, is aetiologically linked with preeclampsia.50 Underlying both is the overexpression of endoglin in the vasculature. Unraveling the transcriptional programs that drive endoglin expression therefore not only sheds light on normal development but also opens up possible avenues to control the abnormal expression implicated in a range of pathologies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Michelle Hammett, Sandie Piltz, and Paula Braker for generating transgenic mice, Catherine Porcher (Oxford University) for the anti-Scl antibody used in ChIP, and Richard Auburn from FlyChip for printing the arrays.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council, the Leukemia Research Fund, Cancer Research UK, a C.J. Martin/R.G. Menzies fellowship from the National Health and Medical Research Council (Australia) to J.E.P., and fellowships from the Kay Kendall Leukemia Fund to K.O. and the Canadian Institutes for Health Research to J.R.L.
Authorship
Contribution: J.P. designed and performed research, analyzed data, and wrote paper; W.Y.I.C. performed research; N.K.W. performed research and analyzed data; A.M.S., S.K., K.K., and M.E.J. performed research; J.R.L. analyzed data; A.K.K. performed research; J.F. contributed essential reagents; D.T. analyzed data; K.O., G.A.F., G.L., and V.K. performed research and analyzed data; and B.G. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr John Pimanda, UNSW Cancer Research Centre Department of Haematology, Wallace Wurth Building, University of New South Wales, Sydney NSW 2052, Australia; e-mail: jpimanda@unsw.edu.au; or Dr Bertie Göttgens, Cambridge Institute for Medical Research, University of Cambridge, Cambridge CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk.
References
Author notes
*J.E.P. and W.Y.I.C. contributed equally to this paper.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal