Abstract
Fanconi anemia (FA) is a complex genetic disorder characterized by congenital abnormalities, bone marrow failure, and myeloid malignancies. Identification of 13 FA genes has been instrumental to explore gene transfer technologies aimed at correction of autologous FA-deficient stem cells. To date, 3 human FA stem cell gene therapy trials with standard 4-day transduction protocols using gammaretroviral vectors failed to provide clinical benefit. In addition, 2- to 4 day ex vivo manipulation of bone marrow from mice containing a disruption of the homologue of human FANCC (Fancc) results in a time-dependent increase in apoptosis and a risk for malignant transformation of hematopoietic cells. Here, we show that a 14-hour transduction period allows a foamyviral vector construct expressing the human FANCC cDNA to efficiently transduce murine FA stem cells with 1 to 2 proviral integrations per genome. Functionally, the repopulating activity of Fancc−/− stem cells from reconstituted mice expressing the recombinant FANCC transgene was comparable with wild-type controls. Collectively, these data provide evidence that short-term transduction of c-kit+ cells with a foamyviral vector is sufficient for functional correction of a stem cell phenotype in a murine FA model. These data could have implications for future gene therapy trials for FA patients.
Introduction
Fanconi anemia (FA) is a complex recessive inherited disorder that is clinically characterized by variable congenital abnormalities, progressive bone marrow (BM) failure, and a high propensity to develop myeloid and epithelial malignancies.1-5 On a cellular level, FA is characterized by a profound hypersensitivity upon exposure to DNA cross-linking agents such as mitomycin-C (MMC) or diepoxybutane (DEB).6-9 Genetically, germ-line mutations in 13 genes (FANCA/B/C/D1/D2/E/F/G/I/J/L/M/N) result in the clinical phenotype of FA.2,8,10-14
Spontaneous genetic correction of a germ-line mutation leading to repopulation of the entire hematopoietic system with normal progeny has been identified in a few FA patients.15-19 These observations, in combination with the fact that the hematopoietic system can be functionally corrected in mice with targeted disruptions of FA genes by retroviral vectors expressing human analogues of the targeted mouse genes in stem cells,20-22 have led to 3 clinical stem cell gene therapy phase 1 studies in FA-A and FA-C patients. So far, neither long-term marking/correction of cells nor clinical benefits for the patients were observed.23,24 Due to the biologic characteristics of the gammaretroviral vectors used for transduction of stem cells,25,26 optimal gene transfer protocols for delivery of genes to mammalian stem cells require a prestimulation period of 1 to 2 days with cytokines that promote the proliferation and survival of stem/progenitor cells. This is followed by a 2- to 3-day exposure of the target cells to vector containing supernatant on the recombinant fibronectin fragment CH-296.27,28 This gene transfer protocol was successful in transducing hematopoietic stem cells in humans, primates/monkeys, and mice.29-31 However, in murine FA models, prolonged in vitro culture of Fancc−/− BM results in a length-of-culture–dependent reduction in myeloid progenitors and repopulating ability,32,33 and the surviving untransduced Fancc−/− repopulating cells have an increased risk of developing cytogenetic abnormalities and myeloid malignancies.22 Therefore, limiting the in vitro culture would be predicted to enhance both the efficacy and safety for genetic therapies of FA stem cells.
Wild-type foamy viruses are the only retroviruses that are not associated with any disease in their natural hosts or in accidentally infected human beings.34-36 It has been shown that vectors based on the prototype (formerly human) foamy virus (FV) can efficiently transduce hematopoietic stem cells from mice,37 dogs,38 and nonobese diabetic/severe combined immunodeficiency (NOD/SCID) repopulating human cells.39-41 Further, FV vectors are at least equally efficient at transduction of CD34+ umbilical cord blood cells engrafting in NOD/SCID mice as lentiviral vectors based on HIV-1.40 In the present study, we demonstrated for the first time the ability of FV vectors encoding the human FANCC transgene to completely correct Fancc−/− myeloid progenitors and repopulating hematopoietic stem cells in a 14-hour transduction protocol without prestimulation. This short gene transfer protocol resulted in 1 to 2 proviral integrations in the reconstituting stem cells and was not associated with the development of myelodysplastic syndromes (MDSs)/acute myeloid leukemia (AML) in Fancc−/− stem cells transduced with the reporter construct. These characteristics support the hypothesis that FV vectors are a viable strategy for stem cell gene transfer strategies in FA.
Methods
Mice
Fancc−/− and Fancc+/+ mice (C57Bl/6 × SV129) were backcrossed 10 generations into a C57Bl/6 stain (CD45.2+). Congenic C57Bl/6 strain (CD45.2+) and B6.SJL-Ptracpep3b/BoyJ (BoyJ) mice (CD45.1+) were originally purchased from The Jackson Laboratory (Bar Harbor, ME) and are maintained in our animal facility.22,32 All studies were approved by the institutional animal care and use committee of Indiana University.
Foamyviral vectors and virus production
The foamyviral constructs used in our studies were derivatives of the MD9 construct,42 a kind gift of Axel Rethwilm (Würzburg, Germany). In the MD9 vector, all foamyviral genes and also the enhancer elements in the 3′ U3 region (Figure 1) have been functionally inactivated by partial deletions. The remaining noncoding 5′ region of GAG and the 3′ region of POL are harboring the packaging signals (CAS I and II) and are therefore essential for the production of recombinant foamyviral particles.42 A linker was cloned into the NotI site 3′ of MD9 and then an expression cassette containing the encephalomyelocarditis virus (EMCV) internal ribosomal entry site (IRES) and the enhanced green fluorescence protein (EGFP) cDNAs introduced from S11IEG3 via BamHI and SpeI, thereby creating the MD9-EGFP construct. The human FANCC cDNA was cloned into the BamHI site resulting in the MD9-FANCC/EGFP vector.
FV-containing supernatant was generated in 293T cells with the FV helper plasmid pcgp1 and the FV envelope plasmid EM02 as previously described.40,42 The titers of the viral supernatant were 1 to 5 × 107 viral particles/mL for MD9-EGFP and 4 to 10 × 106 viral particles/mL for MD9-FANCC/EGFP construct after concentration.40
Foamy virus–mediated BM transduction and transplantation
BM cells were obtained from Fancc−/− or Fancc+/+ wild-type (WT) mice and purified for c-kit+/CD117+ cells, as described previously.33 CD117+Fancc−/− cells were transduced for 14 hours with the foamyviral vectors (MOI 20) on non–tissue culture–treated plates treated with the recombinant human fibronectin fragment CH296, RetroNectin (2 μg/cm2; TAKARA BIO, Otsu, Japan) as previously described27,28,40 in the presence of mIL-6 (200 U/mL) and mSCF (100 ng/mL; both from Peprotech, Rocky Hill, NJ). WT cells were transduced with MD9-EGFP only. Cells were harvested following transduction and washed twice with 10 to 15 volumes of phosphate-buffered saline (PBS), and 5 × 105 transduced CD117+ cells were injected into the tail vein of lethally irradiated 8- to 10-week-old C57Bl/6 WT recipients, as described previously.32,33 In parallel, an aliquot of the transduced cells was plated in semisolid medium that promotes clonogenic growth of myeloid progenitors to determine the transduction efficiency.
FANCD2 Western blot
Unaffected human or FANCC-deficient fibroblasts (GM3136) were cultured in Iscove modified Dulbecco medium (IMDM) containing 15% fetal calf serum and 1% penicillin G and streptomycin. They were transduced with foamyviral vectors encoding either the reporter gene only or MD9-FANCC/EGFP. Forty-eight hours following transduction, the cells were treated with irradiation (IR) (10 gray) and 3 hours later protein extracts were isolated. Protein extract (20 μg) was analyzed by Western blotting using a monoclonal antibody (1 μg/mL; Novus Biologicals, Littleton, CO) that recognizes FANCD2 and monoubiquitinated FANCD2. The protein was detected with an antimouse antibody conjugated to horseradish peroxidase and developed using the enhanced chemiluminescence (ECL)–Plus system (Amersham Biosciences, Piscataway, NJ).
Transplantation protocols for competitive repopulation assay
Transduced, washed CD117+ cells were mixed with a common pool of BoyJ mononuclear cell (MNC) competitors and transplanted into 8- to 10-week-old lethally irradiated C57Bl/6 mice as described.32,33 In cohort 1, 1.5 × 105 test cells were transplanted with 5 × 105 competitor cells. In cohort 2, twice as many test cells were administered to recipients that received a transplant of Fancc−/− cells transduced with the virus encoding EGFP only as in the other experimental groups in an attempt to equalize chimerism. Mean donor chimerism was analyzed to evaluate for significant differences between groups. CD45.1 and CD45.2 chimerism were analyzed monthly following transplantation as previously described.32,33 Repopulating units (RUs) were calculated as (competitor numbers × 105 × % donor chimerism)/(100 − % donor chimerism) as described previously.32,33 An unpaired Student t test was used to determine whether significant differences existed in chimerism between genotypes.
Hematopoietic progenitor assays and drug resistance
FV-transduced cells were plated in triplicate 35-mm plates (Becton Dickinson, Franklin Lakes, NJ) with increasing concentration of MMC or tumor necrosis factor alpha (TNF-α) as described.43,44 To determine the phenotypic correction or the gene transfer efficiency, the number of total colonies formed per plate was enumerated and the EGFP-expressing colonies were counted by the fluorescent microscopy.
Amplification of genomic and proviral DNA
Colonies of FV-transduced cells plated in progenitor assays were individually collected and suspended in PBS. The genomic DNA was isolated and polymerase chain reaction (PCR) for EGFP was performed: forward 5′-ATGGTGAGCAAGGGCGAGGAG-3′, reverse 5′-AAGTCGTGCTGCTTCATGTG-3′, with the following program: 95°C, 5 minutes; 95°C, 40 seconds; 55°C, 30 seconds; 72°C, 1 minute; cycled to step 2 for 31 cycles; 72°C, 10 minutes and then stored at 4°C and analyzed on a 1% agarose gel. The amplified product has a size of approximately 250 bp. In addition, PCR for the genotype of the progenitor cells was performed. Three primers were used: 5′-GAGCAACACAAATGGTAAGG-3′, 5′-CCTGCCATCTTCAGAATTGT-3′, and 5′-TTGAATGGAAGGATTGGAGC-3′, with the following program: 95°C, 5 minutes; 95°C, 30 seconds; 55°C, 2 minutes; 72°C, 1.5 minutes; cycled to step 2 for 31 cycles; 72°C, 10 minutes, and then stored at 4°C and analyzed on a 1% agarose gel. The amplified product of the WT copy of the Fancc gene is approximately 800 bp, whereas the knockout gene PCR product is approximately 600 bp.
Ligation-mediated polymerase chain reaction
For detection of FV integration sites, BM MNCs from 2 primary recipients of Fancc−/− mice transduced with MD9-FANCC/EGFP from the first cohort and 2 primary recipients from the second cohort were enriched for CD45.2+ cells by fluorescence-activated cell sorting (FACS) and then plated in standard progenitor assay. From each of the 4 mice, 20 progenitor colonies were picked and then subjected to ligation-mediated (LM)–PCR as described previously45 with minor variations. The restriction enzyme was HaeIII (New England Biolabs, Frankfurt, Germany). The biotinylated primer 5′biotin-GTACAATCTAGGTGACCACTTTC-3′ (407) was used in a one-step extension at 94°C, 15 minutes; 58°C, 2 minutes; 72°C, 10 minutes; 2 cycles. The 2 internal primers for the nested PCR were 5′-TCTCATCCCAGGTACGTCTATGA-3′ (404) and AP2 as previously described.45 The DNA from excised bands was cloned into pCR2.1 using the TOPO cloning kit (Invitrogen, Frederick, MD) and then sequenced on an ABI Gene Amp 3770 System (Applied Biosystems, Foster City, CA). As described previously,46 SeqMap (http://seqmap.compbio.iupui.edu, Indiana University School of Medicine) was used to map the sequences against the mouse genome. This was then confirmed by mapping the positions using the Ensembl website (http://www.ensembl.org) and mus musculus database release 42, December 2006,47 and the UCSC Genome Browser (http://genome.ucsc.edu).48
Southern blot for junctional fragment analysis
Genomic DNA from BM and spleen specimens was isolated using phenol-chloroform extraction and digested with XhoI (New England BioLabs, Ipswich, MA). Fragments were isolated via ethanol precipitation and run on a 1% agarose gel. The DNA was transferred to a nylon membrane using the TurboBlotter system (Schleicher & Schuell, Keene, NH). To generate the hybridization probe, the MD9 plasmid was digested with PstI (New England BioLabs, Ipswich, MA) and the 1655-bp fragment was isolated using QIAquick gel extraction kit (QIAGEN, Valencia, CA), labeled using the Prime-It II Random Primer labeling kit (Stratagene, La Jolla, CA) and purified using a microspin 30 column (Bio-Rad, Hercules, CA). The membrane was prehybridized for 2 hours at 42°C with the hybridization solution (6 × SSC, 50% formamide, 5 × Denhardt, 0.5% SDS in water) supplemented with 100 μg/mL denatured salmon sperm DNA (Stratagene). After prehybridization, the membrane was hybridized for 16 hours at 42°C with the hybridization solution supplemented with 100 μg/mL denatured salmon sperm DNA (Stratagene) and denatured labeled probe. The next day, the membrane was washed 4 times for 15 minutes at 42°C with the wash solution (2 × SSC, 0.1% SDS in water) and exposed to film (BioMax MS film; Kodak, Rochester, NY) at −80°C with a Cronex Lightning Plus intensifying screen (DuPont, Wilmington, DE).
Microscopy
Colony pictures (Figure 2A,B) were taken on a Zeiss Axiovert 25 (Zeiss, Ontario, NY) inverted microscope with an Epiplan 5×/0.13 numeric aperture (NA) objective using an X-cite 120 lamp (EXFO, Mississauga, ON). Images were captured using a SPOT RT Color camera, model 2.2.1 (Diagnostic Instruments, McHenry, IL) and edited using SPOT advanced software version 4.1.2 (Diagnostic Instruments).
The remaining pictures (Figure 5B) were taken on a Zeiss Axioscope (Zeiss) with the Plan Neofluar 40×/1.30 NA oil objective (Zeiss). Images were captured using a SPOT RT Color camera, model 2.2.1 (Diagnostic Instruments) and edited using SPOT Advanced software version 4.1.2 (Diagnostic Instruments). Slides were stained with hematoxylin and eosin (H&E) stain.
Results
Efficient transduction of hematopoietic progenitors from Fancc−/− mice by short-term exposure to FV vectors in the absence of prestimulation
The recombinant MD9 FV vector42 was used (Figure 1) to express the human FANCC and the EGFP cDNA linked via an EMCV IRES element. This expression cassette was under the transcriptional control of the spleen focus-forming virus (SFFV) promoter element, which had been sufficient to mediate expression of transgenes in NOD/SCID repopulating human CD34+ umbilical cord blood cells.40 An MD9 vector that expresses the EGFP transgene only was used as a control (Figure 1).
Structure of the WT foamyviral genome and the recombinant vectors. (A) Schematic representation of the prototype (formerly human) foamyviral genome with the viral genes gag, pol, env, Tas, and Bet. IP is the internal promoter for the expression of Tas. CAS I and II depict the discontinuous packaging signal residing in the cis-acting sequences I and II. (B) Structures of the recombinant FV self-inactivating vectors MD9-FANCC/EGFP and MD9-EGFP with deletions in gag, pol, env, and the U3 region of the 3′ LTR. SFFV U3 is the promoter of the spleen focus-forming virus U3 region driving the human FANCC and the EGFP cDNAs.
Structure of the WT foamyviral genome and the recombinant vectors. (A) Schematic representation of the prototype (formerly human) foamyviral genome with the viral genes gag, pol, env, Tas, and Bet. IP is the internal promoter for the expression of Tas. CAS I and II depict the discontinuous packaging signal residing in the cis-acting sequences I and II. (B) Structures of the recombinant FV self-inactivating vectors MD9-FANCC/EGFP and MD9-EGFP with deletions in gag, pol, env, and the U3 region of the 3′ LTR. SFFV U3 is the promoter of the spleen focus-forming virus U3 region driving the human FANCC and the EGFP cDNAs.
CD117+ cells from WT and Fancc−/− mice were transduced with foamyviral vectors in the presence of growth factors and viral supernatants on CH296-coated plates for 14 hours. Recovery of CD117+ cells after the overnight transduction protocol was comparable in all experimental groups and was consistently 70% to 80% of input cells (data not shown). The next day, cells were cultured in semisolid medium to determine the gene transfer efficiency into clonogenic myeloid progenitors. After 7 days, the proportion of EGFP-positive myeloid progenitors was scored (Figure 2). Collectively, short-term exposure of both WT and Fancc−/− CD117+ cells to FV supernatants on recombinant fibronectin resulted in a gene transfer efficiency of greater than 50% of clonogenic progeny based on EGFP expression in 4 independent experiments.
Transduction of WT and Fancc−/− CD117+ cells with FV vectors. (A,B) Light and dark field microscopy of a transduced EGFP-positive progenitor. (C) Transduction efficiency of progenitors cultured from CD117+Fancc−/− and WT cells following FV transduction with the indicated constructs. Data shown are mean transduction efficiency and error bars represent standard error of the mean of 3 replicate dishes. The number of colonies ranged from 90 to 120 per dish. Data are representative of 4 independent experiments with similar results.
Transduction of WT and Fancc−/− CD117+ cells with FV vectors. (A,B) Light and dark field microscopy of a transduced EGFP-positive progenitor. (C) Transduction efficiency of progenitors cultured from CD117+Fancc−/− and WT cells following FV transduction with the indicated constructs. Data shown are mean transduction efficiency and error bars represent standard error of the mean of 3 replicate dishes. The number of colonies ranged from 90 to 120 per dish. Data are representative of 4 independent experiments with similar results.
Foamy virus-mediated expression of FANCC corrects the DNA damage and inflammatory cytokine hypersensitivity of Fancc−/− myeloid progenitors
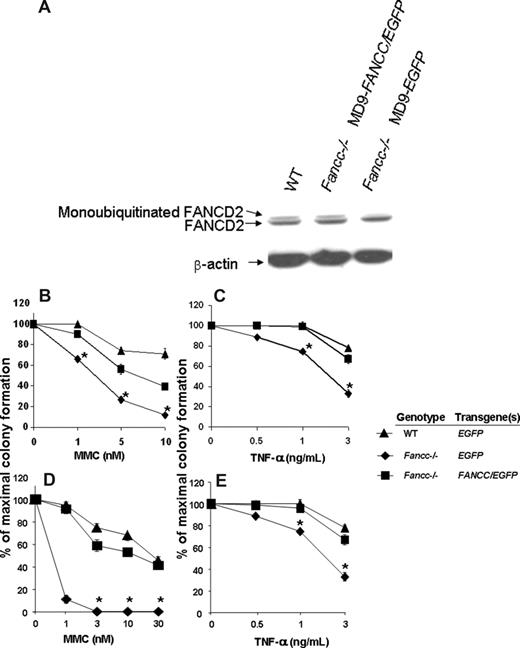
Since FANCC is a component of the FA core nuclear complex and is required for FANCD2 monoubiquitination in response to DNA damage and during S-phase,49,50 the detection of monoubiquitinated FANCD2 can be used as a measure of functional FANCC protein. Unaffected human cutaneous and FANCC-deficient fibroblast lines were transduced and 48 hours later the cells were treated with 10 Gy ionizing radiation. Three hours subsequently, cells were harvested and protein extracts were isolated. As expected, the FANCC-deficient fibroblasts transduced with the MD9-EGFP reporter construct do not express the monoubiquitinated form of FANCD2 (Figure 3A). In contrast, FANCC-deficient fibroblasts transduced with MD9-FANCC/EGFP can restore the assembly of the FA nuclear complex, leading to efficient monoubiquitination of FANCD2 (Figure 3A). These data provide biochemical evidence indicating that the FV vector encoding human FANCC expresses a functional recombinant protein that is sufficient to allow activation of the downstream effector FANCD2.
Biochemical and functional correction of FA-C cells. (A) FANCC-deficient or unaffected human fibroblasts were transduced with foamyviral vectors encoding either the reporter gene only or MD9-FANCC/EGFP. Forty-eight hours following transduction, the cells were treated with IR (10 gray) and 3 hours later protein extracts were collected. Protein extract (20 μg) was analyzed by Western blot for monoubiquitinated FANCD2. (B-E) Clonogenic progenitor cell growth in the presence of increasing concentrations of MMC (B,D) or TNF-α (C,E). (B,C) Triplicate cultures of transduced c-kit+ cells were cultured immediately after transduction. (D,E) Six months following transplantation, transduced bone marrow cells from each experimental group were analyzed in triplicate cultures (WT-MD9-EGFP, n = 3; Fancc−/−-MD9-EGFP, n = 3; Fancc−/−-MD9-FANCC/EGFP, n = 5). Data reflect the mean and standard error of the mean (SEM) of all recipients examined in each experimental group. *P < .05 comparing Fancc−/− EGFP to the other experimental groups.
Biochemical and functional correction of FA-C cells. (A) FANCC-deficient or unaffected human fibroblasts were transduced with foamyviral vectors encoding either the reporter gene only or MD9-FANCC/EGFP. Forty-eight hours following transduction, the cells were treated with IR (10 gray) and 3 hours later protein extracts were collected. Protein extract (20 μg) was analyzed by Western blot for monoubiquitinated FANCD2. (B-E) Clonogenic progenitor cell growth in the presence of increasing concentrations of MMC (B,D) or TNF-α (C,E). (B,C) Triplicate cultures of transduced c-kit+ cells were cultured immediately after transduction. (D,E) Six months following transplantation, transduced bone marrow cells from each experimental group were analyzed in triplicate cultures (WT-MD9-EGFP, n = 3; Fancc−/−-MD9-EGFP, n = 3; Fancc−/−-MD9-FANCC/EGFP, n = 5). Data reflect the mean and standard error of the mean (SEM) of all recipients examined in each experimental group. *P < .05 comparing Fancc−/− EGFP to the other experimental groups.
To determine whether the SFFV promoter–mediated expression of FANCC was sufficient to phenotypically correct Fancc−/− primitive clonogenic cells, CD117+ cells were transduced with MD9-FANCC/EGFP or the reporter construct, and clonogenic assays of myeloid progenitors were established in the presence of a range of concentrations of MMC, a bifunctional alkylating agent, or the inhibitory cytokine TNF-α. Consistent with established work,43,44 Fancc−/− progenitors transduced with the reporter construct were hypersensitive to both MMC and TNF-α. In contrast, progenitors transduced with the construct expressing FANCC were corrected to WT levels (Figure 3B,C). As an initial assessment of in vivo function of the expressed FANCC protein, transduced CD117+ cells were transplanted into lethally irradiated syngeneic recipients to allow long-term reconstitution of the hematopoietic system by genetically modified stem cells. Six months after transplantation, BM low-density MNCs from the reconstituted mice were isolated and clonogenic cells from the respective experimental groups were cultured in a range of concentrations of MMC or the inhibitory cytokine TNF-α. Consistent with previous studies,43,44 and with the clonogenic assays conducted prior to transplantation (Figure 3B,C), the Fancc−/− progenitors transduced with the reporter construct were hypersensitive to both MMC and TNF-α (Figure 3D,E). In contrast, progenitors isolated from mice reconstituted with Fancc−/− cells expressing the FANCC transgene had sensitivities to MMC and TNF-α that were comparable to progenitors from WT mice (Figure 3D,E). These findings support the hypothesis that FV expression of human FANCC introduced via this short-term transduction protocol is sufficient for transduction and functional com-plementation of Fancc−/− long-term repopulating cells to cytotoxic agents.
Foamyviral transfer of FANCC restores the repopulating ability of Fancc−/− stem cells to WT levels
Competitive repopulation is an established quantitative measure of stem cell repopulating activity that allows a direct comparison of the proliferative activity of reconstituted stem cells of different genotypes via their relative proliferation of myeloid and lymphoid lineages to a common pool of competitor cells.32,51-55 Since murine Fancc−/− BM cells have reduced repopulating ability compared with WT cells,32 and a prevalent phenotype in FA patients is BM failure,1,2,4 we used this methodology to assess the potential of the foamyviral vector MD9-FANCC/EGFP to correct the repopulating ability of Fancc−/− stem cells to that of syngeneic WT stem cells. CD117+ WT and Fancc−/− BM cells were isolated, transduced with either the MD9-FANCC/EGFP or the control vector, and then cotransplanted with a common pool of CD45.1+ competitors into lethally irradiated recipients using previously established methods.32,33,53 As shown in Figure 4A, the test cell chimerism of individual recipients from 1 of 2 independent experiments with 6 to 8 primary recipients per experimental group was followed on serial measurements over the course of one year. Consistent with previous results,32,33 recipients reconstituted with mock-transduced Fancc−/− cells have a reduced repopulating ability compared with recipients reconstituted with WT cells. In contrast, mice reconstituted with Fancc−/− cells expressing the FANCC cDNA after foamyviral gene transfer have a test cell chimerism that is comparable to recipients that received a transplant of WT cells at all time points measured (Figure 4A). At time of killing, the repopulating activities of the respective test cell populations were assessed by measuring the number of repopulating units (RUs) as originally described by Haneline et al22,32 and Harrison.53 The results demonstrate that the repopulating ability of Fancc−/− cells expressing the EGFP reporter transgene are only approximately 17% of WT marrow, whereas the RUs of Fancc−/− cells expressing the recombinant FANCC transgene were comparable to WT marrow (Table 1). Comparable results were obtained from a second independent experiment also containing 6 to 8 recipients in each experimental group (data not shown). The improvement of the repopulating ability in Fancc−/− cells transduced with MD9-FANCC/EGFP construct compared with the activity of WT test cells supports the hypothesis that foamyviral expression of the transgene is correcting the repopulating activity of Fancc−/− stem cells.
Donor chimerism of mice that received a transplant of transduced test cells in competitive repopulation assays. Peripheral blood donor chimerism was determined by flow cytometry after staining with antibodies against CD45.1 and CD45.2. (A) Donor chimerism of individual mice in the first cohort over a time period of 12 months. The mean donor chimerism of mice that received a transplant of mock-transduced Fancc−/− control cells was significantly lower than that of mock-transduced WT control or MD9-FANCC/EGFP–transduced Fancc−/− cells (*P < .05). (B) Peripheral blood cell chimerism of recipient in secondary transplants receiving transduced cells from the second cohort 6 months after the initial transplantation. (C,D) Analysis of genotype and transduction efficiency of myeloid progenitors. CD45.2+ BM cells were sorted and cultured in semisolid medium to grow myeloid progenitors. DNA from individual colonies was isolated and amplified to detect the genotype (C) and presence of the provirus (D). Representative gels of the amplified PCR products are shown. The test cell genotype and the provirus used are indicated.
Donor chimerism of mice that received a transplant of transduced test cells in competitive repopulation assays. Peripheral blood donor chimerism was determined by flow cytometry after staining with antibodies against CD45.1 and CD45.2. (A) Donor chimerism of individual mice in the first cohort over a time period of 12 months. The mean donor chimerism of mice that received a transplant of mock-transduced Fancc−/− control cells was significantly lower than that of mock-transduced WT control or MD9-FANCC/EGFP–transduced Fancc−/− cells (*P < .05). (B) Peripheral blood cell chimerism of recipient in secondary transplants receiving transduced cells from the second cohort 6 months after the initial transplantation. (C,D) Analysis of genotype and transduction efficiency of myeloid progenitors. CD45.2+ BM cells were sorted and cultured in semisolid medium to grow myeloid progenitors. DNA from individual colonies was isolated and amplified to detect the genotype (C) and presence of the provirus (D). Representative gels of the amplified PCR products are shown. The test cell genotype and the provirus used are indicated.
Repopulating units 12 months after transplantation
| Fancc genotype . | Vector . | No. of mice . | Repopulating units . |
|---|---|---|---|
| +/+ | MD9-EGFP | 7 | 6.11 ±0.96 |
| −/− | MD9-EGFP | 5 | 1.03 ±0.11* |
| −/− | MD9-FANCC/EGFP | 8 | 7.01 ±0.95 |
| Fancc genotype . | Vector . | No. of mice . | Repopulating units . |
|---|---|---|---|
| +/+ | MD9-EGFP | 7 | 6.11 ±0.96 |
| −/− | MD9-EGFP | 5 | 1.03 ±0.11* |
| −/− | MD9-FANCC/EGFP | 8 | 7.01 ±0.95 |
Quantities shown are mean plus or minus SEM.
P < .05 compared with either WT MD9-EGFP or Fancc−/− MD9-FANCC/EGFP.
To confirm that the correction in repopulating ability is associated with transduction of the transgene, it would have been ideal to evaluate expression of the EGFP transgene directly in CD45.2+ cells. Unfortunately, we found that EGFP expression was downmodulated in the bicistronic vector containing both EGFP and FANCC, consistent with previous work by us and others.22,46,56 Therefore, to confirm that the CD45.2+ test cells were of the predicted genotype and contained the transgene, CD45.2+ BM cells were sorted by FACS and plated in standard progenitor assays. DNA from individual progenitors was isolated and amplified to assess the genotype and the presence of the transgene. The PCR analysis of 70 colonies from 7 recipients containing Fancc−/− MD9-FANCC/EGFP test cells demonstrated that 67 (96%) of 70 colonies were positive for the Fancc−/− genotype. A representative analysis is shown in Figure 4C. A high proportion of those same Fancc−/− test progenitors (90%) also contained the provirus. A representative blot demonstrating this is shown in Figure 4D. The majority of progenitors (25/29) analyzed from CD45.2+ bone marrow cells of 3 representative recipients that received a transplant of Fancc−/− cells transduced with MD9-EGFP also were Fancc−/−. Given the stochastic nature of the hematopoietic system, the high proportion of Fancc−/− progenitors that contain the MD9-FANCC/EGFP provirus, and the 6- to 7-fold difference in repopulating activity observed in Fancc−/− test cells containing the provirus encoding FANCC/EGFP versus EGFP only, we conclude that the test cell chimerism in recipients of Fancc−/− cells transduced with the MD9-FANCC/EGFP provirus reflects the functional correction of the FA pathway.
To assess that the transgene was transduced into a stem cell with repopulating ability as well as the long-term proliferative ability assessed in primary recipients, low-density MNCs from selected primary recipients were transplanted into lethally irradiated recipients. The chimerism 6 months following transplantation into secondary recipients did not change, and the test cell chimerism of mice reconstituted with corrected Fancc−/− cells remained comparable to that of recipients of transplanted WT cells (Figure 4B), indicating that the transgene was functional in repopulating stem cells. In that same experiment, recipients reconstituted with Fancc−/− cells transduced with the FANCC transgene had comparable numbers of “test” colony-forming units/femur (2.8 ± 0.1 × 104) as WT recipients (2.4 ± 0.2 × 104), whereas recipients reconstituted with Fancc−/− test cells encoding EGFP only had significantly reduced test progenitors (1.5 ± 0.3 × 104).
Evaluation for evidence of myelodysplasia in mice reconstituted with Fancc−/− cells
We have previously found in 2 separate studies involving 5 cohorts of mice that in vitro culture of uncorrected Fancc−/− BM for 2 to 4 days prior to transplantation predisposes the recipients of those cells to MDS.22,33 A characteristic of the MDS phenotype observed in Fancc−/− mice is that myeloid progenitors are resistant to the apoptotic signals induced by TNF-α in progenitor assays.33 To determine whether this indicator of malignant transformation was present in recipients reconstituted with mock-transduced Fancc−/− cells, CD45.2+ cells from primary recipients 12 months following transplantation were sorted using FACS and myeloid progenitors were cultured. Analyses revealed that the Fancc−/− progenitors retained the characteristic hypersensitivity to TNF-α (Figure 5A). In addition, the BM and spleen of 7 primary recipients and 4 secondary recipients reconstituted with Fancc−/− cells expressing the EGFP transgene only were examined at postmortem and had normal architecture (Figure 5B). Similar histologic data were found in primary and secondary recipients receiving MD9-FANCC/EGFP–transduced Fancc−/− cells (data not shown). Collectively, all functional and histologic data fail to detect evidence of transformation of uncorrected Fancc−/− stem cells in primary or secondary recipients.
TNF-α hypersensitivity is maintained in primary recipients of mock-transduced Fancc−/− stem cells 6 months after transplantation. (A) Low-density mononuclear cells from Fancc−/− MD9-EGFP (n = 6) and WT-MD9-EGFP (n = 5) recipients were cultured in clonogenic assays in triplicate to determine their respective sensitivity to TNF-α at 12 months following transplantation. The number of colonies ranged from 80 to 120 per dish. The genotypes are indicated. Data represent the mean and standard error of the mean growth of all recipients in each respective group. *P < .05 and statistical significance between the 2 experimental groups. (B) Representative histologic analysis of BM and spleen of a recipient reconstituted with Fancc−/− or WT test cells previously transduced with MD9-EGFP.
TNF-α hypersensitivity is maintained in primary recipients of mock-transduced Fancc−/− stem cells 6 months after transplantation. (A) Low-density mononuclear cells from Fancc−/− MD9-EGFP (n = 6) and WT-MD9-EGFP (n = 5) recipients were cultured in clonogenic assays in triplicate to determine their respective sensitivity to TNF-α at 12 months following transplantation. The number of colonies ranged from 80 to 120 per dish. The genotypes are indicated. Data represent the mean and standard error of the mean growth of all recipients in each respective group. *P < .05 and statistical significance between the 2 experimental groups. (B) Representative histologic analysis of BM and spleen of a recipient reconstituted with Fancc−/− or WT test cells previously transduced with MD9-EGFP.
Evaluation of proviral integrants
We next wanted to assess the number of integrations that are present in progeny of the transduced Fancc−/− stem cells. BM low-density MNCs from 4 primary recipients were harvested at the time of secondary transplantations, sorted for CD45.2 expression, and then plated in methylcellulose assays. After one week, 20 colonies were picked for each mouse and subjected to standard LM-PCR analysis for identifying the location of the provirus in the genome. As described previously,46 the position of the transgene from each colony was evaluated using the SeqMap website (http://seqmap.compbio.iupui.edu) and confirmed using the http://genome.ucsc.edu and http://www.ensembl.org websites. From each mouse, one proviral integration was detected (Table 2). Each integration site detected was independent of the others and was contained either in an intron or outside of a gene, consistent with other studies of FV integration patterns.37,38,46,57-60 Because progenitors provide limited amounts of DNA for LM-PCR analysis and proviral integrants may not always be detected, integrations were also evaluated from whole BM and spleen samples by Southern analysis in a replicate set of experiments using similar transduction conditions. Results by Southern blot revealed one junctional fragment per recipient for 9 of 12 mice and 2 junctional fragments per recipient for 3 of 12 mice evaluated. These data are summarized in Table 3.
Location of observed proviral integration sites on the mouse genome
| Mouse . | Chromosome . | Location . | Gene ID . |
|---|---|---|---|
| 91 (cohort 1) | 16 B4 | Upstream of Zbtb20 and Ndufs5 | NM 019778 and BC002163 |
| 93 (cohort 1) | 5 E3 | No nearby gene | |
| 1 (cohort 2) | 10 B3 | Intron 2 of Pkib | NM 008863 |
| 2 (cohort 2) | 13 A3.1 | No nearby gene |
| Mouse . | Chromosome . | Location . | Gene ID . |
|---|---|---|---|
| 91 (cohort 1) | 16 B4 | Upstream of Zbtb20 and Ndufs5 | NM 019778 and BC002163 |
| 93 (cohort 1) | 5 E3 | No nearby gene | |
| 1 (cohort 2) | 10 B3 | Intron 2 of Pkib | NM 008863 |
| 2 (cohort 2) | 13 A3.1 | No nearby gene |
Proviral integration number in the mouse genome
| Genotype . | Transgene . | No. of mice . | No. of mice with 1 integrant . | No. of mice with 2 integrants . |
|---|---|---|---|---|
| Fancc−/− | FANCC/EGFP | 6 | 5 | 1 |
| Fancc−/− | EGFP | 3 | 1 | 2 |
| WT | EGFP | 3 | 3 | 0 |
| Genotype . | Transgene . | No. of mice . | No. of mice with 1 integrant . | No. of mice with 2 integrants . |
|---|---|---|---|---|
| Fancc−/− | FANCC/EGFP | 6 | 5 | 1 |
| Fancc−/− | EGFP | 3 | 1 | 2 |
| WT | EGFP | 3 | 3 | 0 |
Discussion
The only long-term cure for the BM failure in FA patients is transplantation of normal hematopoietic stem cells, ideally from an HLA-matched sibling.4,61-64 Allogeneic BM or cord blood transplantation is not without subsequent risk as the conditioning regimens cause genotoxic stress that predispose patients to an increased incidence of squamous cell carcinomas especially when compounded by the presence of chronic graft-versus-host disease.65,66 Thus, the transduction of autologous, genetically corrected stem cells in the absence of genotoxic myelopreparation could provide a therapy that does not expose the patient to these ongoing potential sequelae.67
Natural reversions of inherited germ-line mutations seen in a small population of FA patients strongly suggest that an oligoclonal population or perhaps a single hematopoietic stem cell is sufficient to correct the hematopoietic system.15-19 Unfortunately, long-term multipotential cell transduction leading to hematologic improvement in the patient has not been observed with gammaretroviruses in FA clinical trials to date.23,24 One possible reason for this failure of genetic therapy is the low number of stem cell targets available for gene transfer.68 In addition, even in the presence of saturating concentrations of growth factors, FA cells have an increased propensity to undergo apoptosis.33 Therefore, the prolonged culture required to induce stem cells into cycle and allow gammaviruses to integrate into the target cell genome predisposes untransduced FA stem cells to undergo apoptosis.33
In studies here, we found that foamy virus vectors expressing the human FANCC cDNA can functionally correct multiple defects in Fancc−/− stem cells capable of repopulating primary and secondary recipients. Due to their specific biology, the use of FV vectors can theoretically address several problems associated with somatic stem cell gene therapy in FA. First, long-term serial evaluation of a cohort of humans (median follow up of 22 years) who have been chronically infected by primate FVs provides epidemiologic evidence that FV is not associated with any disease in humans.69 Therefore, this gene delivery system appears to be especially attractive if gene integration into the target cell genome is needed for the therapeutic effect. Second, in contrast to gammaretroviruses that have a half-life of only 4 to 6 hours in vivo,70 nonintegrated FV virions retain the potential for stable integration in quiescent cells for up to 30 days following transduction.71 Thus, cells need to be exposed in vitro to virions for only a short time, allowing cycling of the target cells and proviral integration to occur in vivo. Since FA murine and human FA stem cells have increased in vitro time-dependent induction of apoptosis and at least Fancc−/− cells develop in vitro dependent clonal transformation,22,33 this abbreviated transduction protocol appears optimal for genetic correction of FA stem cells. Here, we demonstrate that in vitro manipulation of Fancc−/− CD117+ cells for 14 hours in the absence of prestimulation is sufficient for FV-mediated delivery of FANCC to stem cells and correction of stem cell repopulating activity in primary and secondary transplantations. In preliminary studies, we have now found that efficient transduction can be observed following in vitro culture in as few as 8 hours (data not shown).72
We observed 1 to 2 proviral integrants in the repopulating cells of reconstituted mice. There are at least 3 possibilities for this observation. It is possible that analogous to rare cases of apparent spontaneous correction of FA stem cells,15-19 correction of a subpopulation of Fancc−/− cells with a functional transgene leads to an in vivo selection of the corrected cells. Future studies mixing ratios of transduced and untransduced cells into irradiated recipients may allow formal testing of this hypothesis. Alternatively, previous studies have shown that hematopoiesis in reconstituted lethally irradiated mice tends to be oligoclonal in nature.73,74 This correlates with our data, which show a low number of integrants, regardless of the transgene or stem cell genotype. In contrast to our studies in mice, a recent report using foamyviral vectors to treat canine leukocyte adhesion deficiency (CLAD) found multiple integrants in this large-animal model.57 A third possibility that we considered was that of clonal selection leading to MDS. To test for this possibility, we examined the hematopoietic organs and evaluated clonogenic growth from the myeloid progenitors of primary and secondary recipients over a total of 18 months. From multiple recipients, no pathological abnormalities were observed. The lack of pathological sequelae is consistent with the study in CLAD dogs where foamyviral vector integrants were found within genes or near oncogenes at a much lower frequency as reported for retroviral vectors.57
A potential limitation to the use of FV vectors in the clinic to date is that the fusogenic capacities of FV envelope proteins75,76 and packaging of the gag-pol helper plasmid may have toxicity on the target cells,42 especially when cells are exposed to higher virus titers and for prolonged periods of time. Although toxicity was not problematic in the murine studies reported here, we are currently evaluating the envelope toxicity on human cells by testing different FV envelope proteins with point mutations of functional elements. In addition, we are separating the Gag and Pol genes onto different helper plasmids and mutating the Cas I and Cas II packaging sequences to prevent expression of the gag and pol proteins in the target cells.
Collectively, these studies demonstrate that the functional expression of a transgene using a FV construct can cure a disease phenotype in a murine model of a human disease. We hypothesize that with additional modifications this gene delivery system may be applicable for therapeutic use in humans.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Karen Pollok, Laura Haneline, and Ken Cornetta for numerous helpful discussions and Dirk Lindemann and Axel Rethwilm for sharing unpublished data and expert advice on foamy virus biology.
This work was supported by the National Institutes of Health (Bethesda, MD) PPG-P01-HL533586 (D.W.C., H.H., and S.M.) and the Deutsche Forschungsgemeinschaft (Bonn, Germany) SPP1230 (HA2322/2-1; H.H.). The work was also supported by the Riley Children's Foundation (Indianapolis, IN) and the Indiana University NCI-designated Cancer Center (Indianapolis, IN).
National Institutes of Health
Authorship
Contribution: Y.S., A.C.P., Y.L., S.C., C.L., J.Y., O.E., and S.F. performed experiments; Y.S., A.C.P., S.M., H.H., and D.W.C analyzed results; Y.S. and A.C.P. made the figures; Y.S., H.H., and D.W.C. designed the research; and Y.S., A.C.P., H.H., and D.W.C. wrote the paper.
Conflict-of-interest disclosure: H.H. may receive royalties based on a license agreement between Indiana University and Takara Shuzo, Ltd, resulting from the sale of the fibronectin fragment CH296 (RetroNectin). All other authors declare no competing financial interests.
Correspondence: D. Wade Clapp, Cancer Research Institute, 1044 W Walnut St, R4 402A, Indianapolis, IN 46202-5254; e-mail: dclapp@iupui.edu; or Helmut Hanenberg, Children's Hospital, Heinrich Heine University, Moorenstr. 5, 40225 Duesseldorf, Germany; e-mail: hanenberg@uni-duesseldorf.de.
References
Author notes
*Y.S. and A.C.P. contributed equally to this study.