Abstract
Introduction. Chronic myeloid leukaemia (CML) is a malignant clonal myeloproliferative disorder characterised by the presence of the Philadelphia chromosome (Ph) and expression of the BCR-ABL fusion protein with constitutive tyrosine kinase activity and multi-target dysregulation. Standard therapy employs the use of tyrosine kinase inhibitors, such as Imatinib mesylate, with a high level of success. However, in a proportion of patients this approach is sub-optimal necessitating the use other treatment modalities. Allogeneic stem cell transplantation (allo-SCT) is established and potentially curative via the immunologically mediated graft-versus-leukaemia (GvL) effect, but it is associated with significant morbidity, long term effects and high mortality rate (30%). An alternative approach is to employ peptide vaccines directed against leukaemia associated antigens (LAA) in order to evoke a curative anti-leukaemia immune response without the risk of graft versus host disease. Prevalent among these LAAs are WT1 (Wilms Tumour 1) and PRAME (preferentially expressed antigen in melanoma). WT1 encodes a transcription factor of the zinc-finger family, and is known to be over-expressed in many types of leukaemia. PRAME (PRA) is a cancer testis antigens over-expressed in haematological malignancies including CML, making it an attractive target for immunotherapy. HLA-A2 and A-24 restricted epitopes derived from PRA have been shown to be immunogenic by a number of groups. Here, we studied the value of PRA and WT1 measurement in assessing minimal residual disease (MRD) in CML compared to BCR-ABL.
Materials & Methods. cDNA from PB samples from 76 diagnostic samples and 127 follow-up samples from 114 patients were tested. The follow-up samples were subdivided into 3 groups according to BCR-ABL expression: MRD1 >10%, MRD2: 1%-10%, MRD3 <1%. All samples were analysed for WT1 and PRA, except for the diagnostic samples, where only 6 were available for PRA analysis due to a lack of material. All expression data was obtained by quantitative real-time PCR (Q-PCR) on the ABI 7500 platform. The BCR-ABL was expressed as a percentage ratio against ABL, using absolute quantification. WT1 and PRA expression was derived using the relative expression dCt method, normalised against a house keeping gene (GUS), after ensuring equivalent Q-PCR efficiencies.
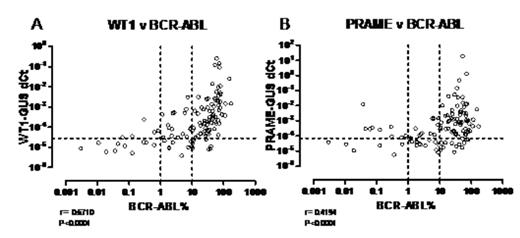
Results. WT1 and PRA expression was detectable in all samples analysed. A non-parametric correlation, performed on all data points, found a positive correlation between expression of BCR-ABL and expression of WT1 (r= 0.6710, P<0.0001) and PRA (r= 0.4194, P<0.0001) (Figure 1). The median WT1 and PRA expression of the remission samples was calculated and used as a cut-off to define over-expression (2.89E-05 and 7.04E-05, respectively). This was then used to assess the proportion of samples from the diagnostic and follow-up groups that over-expressed WT1 (Diag= 96.1%, MRD1= 90.0%, MRD2= 66.7%, MRD3= 33.3%) and PRA (Diag= 100%, MRD1= 82.5%, MRD2= 41.7%, MRD3= 60.0%). The differences in over-expression between the groups were found to be highly significant by 2×4 contingency Chi square test (P<0.0001 for both WT1 and PRA).
Discussion. This analysis shows that both WT1 and PRA expression positively correlate with BCR-ABL level in CML patients, and can be reliably quantitated using Q-PCR. This technique can therefore provide useful information for treatment monitoring of immunotherapy directed against such antigens. The median expression of the remission samples can be used as a useful cut-off for assessing over-expression for WT1, but maybe of less value in respect to PRA, due to a lack of discrimination at low BCR-ABL levels shown by the discordantly high value of positivity in the MRD3 group.
Expression of A: WT1 and B: PRAME in CML, subdivided by BCR-AEL expression (vertical lines) and over-expression thresold (horizontal line).
Expression of A: WT1 and B: PRAME in CML, subdivided by BCR-AEL expression (vertical lines) and over-expression thresold (horizontal line).
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal