Abstract
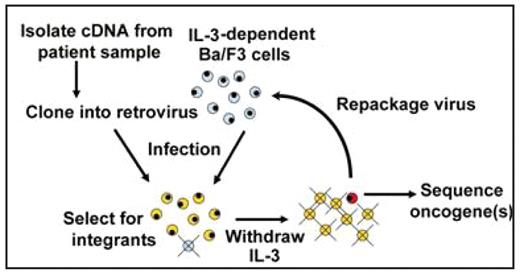
Approximately half of adult pre-B cell acute lymphocytic leukemias (pre-B ALL) harbor well-characterized chromosomal rearrangements known to affect prognosis and/ or treatment. In the 50% of cases with normal cytogenetics, pathogenesis is very poorly understood. Recent studies identified several somatic gene mutations in pre-B ALL samples. However, the functional significance of nearly all of these mutations remains unclear. We developed an approach to efficiently identify functionally-important, cryptic alterations directly from patient samples by establishing retroviral cDNA libraries. Streamlined methods for cDNA cloning and retroviral transduction have made it both feasible and cost-efficient to construct patient-specific libraries for oncogene identification. In our system (Figure), patient-specific retroviral cDNA libraries are used to infect the IL- 3-dependent B cell line, BaF3. Transformation by oncogenic kinases, transcription factors and other oncogenes results in IL-3 independent survival, allowing for the isolation of individual transformed clones and sequencing of integrated cDNA. cDNA is isolated from pre-B ALL samples and cloned into a retroviral backbone. Construction using clonase enzymes, rather than traditional restriction enzymes, maximizes efficiency and establishes representational cDNA libraries. BaF3 cells are infected with retroviral cDNA libraries then IL-3 is withdrawn. Viable, IL-3 independent clones are isolated and integrated cDNA are sequenced. If multiple cDNAs integrate into the same IL-3-independent clone, retrovirus is repackaged within that clone then applied at low titer to a new batch of IL-3- dependent BaF3 cells to identify the specific cDNA that confers IL-3 independence. With this approach, cryptic mutations, translocations and other rearrangements with functional significance can be identified. In addition, weaker oncogenes that require coordinated effects from other alterations can be elucidated by increasing the multiplicity of infection. To pilot the system, we assembled libraries using BCR-ABL-expressing K562 cells and peripheral blood lymphocytes from healthy donors. Restriction fragment analysis confirmed the presence of broadly heterogeneous cDNAs within the libraries. IL-3 independent cells were recovered after infection with the K562 libraries but not after infection with the healthy donor lymphocyte libraries. The IL-3 independent cells from the K562 library-infected population were confirmed to contain integrated BCR-ABL by PCR. We are currently characterizing transforming oncogenes recovered from pre-B ALL specimens with normal cytogenetics. Future efforts will characterize the biologic activity of these novel oncogenes and define the molecular epidemiology, as well as clinical features, associated with these alterations. In conclusion, large-scale sequencing efforts are underway at many centers to identify somatic mutations in tumor samples. Our functional approach offers an efficient alternative, focused on phenotypically meaningful, and therefore potentially drugabble, alterations. This approach could be easily applied to other tumor types, using either BaF3 or other cell lines that undergo oncogene-mediated transformation.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal